Introduction
Gastric cancer is one of the most common types of
cancer and the second most common cause of cancer-related mortality
worldwide (1,2). Furthermore, gastric cancer is also one
of the most common malignant tumors and the second leading cause of
cancer-related mortality in China. Among malignant tumors of the
stomach, gastric adenocarcinoma (GA) is the most common
histological subtype, accounting for 95% of total morbidity.
Early-stage GA rarely causes symptoms or only causes mild symptoms.
However, once clinical symptoms eventually manifest, the chances of
successful treatment are reduced (3). Therefore, it is crucial to identify
landmark genes or small molecules that are associated with the
proliferation and metastasis of GA and may be used as diagnostic
biomarkers and drug targets for GA treatment (4,5).
Long non-coding (lnc) RNAs are transcripts >200
nucleotides in length that do not encode proteins. lncRNAs were
initially considered to be byproducts of RNA polymerase II
transcription (a type of genomic ‘noise’) and to have no
physiological function (6,7). However, recent studies revealed that
lncRNAs can in fact regulate gene expression in different
processes, including transcriptional and post-transcriptional
regulation, translation and epigenetic regulation (8,9). This
identification complements the traditional genetic law. Notably,
only ~1.5% of the genome is gene-coding, and the majority of these
genes are transcribed into non-coding sequences, which account for
9–11% of total RNA. According to their location relative to
protein-coding genes, lncRNAs may be divided into five types,
namely antisense, sense-overlapping, intronic, bidirectional and
intergenic (10). The positioning
of lncRNAs may help identify their function. Notably, various
lncRNAs are associated with human cancer. Previous studies have
indicated that the aberrant expression of lncRNAs may be associated
with the occurrence and development of tumors. For example, lncRNA
HOX transcript antisense RNA upregulation in GA increases gastric
cancer cell progression through recruitment of microRNA
(miR)-331-3p and regulation of human epidermal growth factor
receptor 2 gene expression by means of competing endogenous (ce)RNA
(11). In addition, inhibition of
lncRNA H19 can suppress the growth of myeloma cells through the
nuclear factor-κB signaling pathway (12). c-Myc gene-induced H19 can also
promote cell proliferation (13).
H19 has been shown to be able to promote gastric cancer cell
proliferation via miR-675 through the tumor suppressor runt domain
transcription factor 1 (14).
According to previous research (15), lncRNAs are stable in blood specimens
and can be examined using fluorescence quantitative polymerase
chain reaction (qPCR) technology. Although various
cancer-associated lncRNAs have been identified, their function
remains to be elucidated. Currently, it remains unclear how several
lncRNAs are involved in important biological processes, including
tumor growth, proliferation and differentiation.
GA is one of the most frequently occurring cancer
types in the Fujian area in China. Therefore, our previous study
aimed to investigate the lncRNA expression profile in tissue
samples from patients with GA using a high-throughput chip assay
technique (16). A number of
lncRNAs were found to be differentially expressed in GA tissues
compared with paired non-cancerous tissues. The results of the chip
studied by our group revealed that lncRNA RP1-163G9.1 was expressed
at low levels in GA tissue compared with its expression in adjacent
control tissues; however, its specific biological functions and
possible mechanisms of action remain unclear. To the best of our
knowledge, no previous research has indicated the role of lncRNA
RP1-163G9.1 in gastric cancer or other types of cancer. Therefore,
the aim of the present study was to assess the role of lncRNA
RP1-163G9.1 in GA.
lncRNA RP1-163G9.1 is located in chromosome
1:2976180-2978596, which contains three exons (1,697 bp in size).
Based on our previous data from microarray chip assays, lncRNA
RP1-163G9.1 was found to be downregulated in GA tissues. To the
best of our knowledge, the association of this lncRNA with GA has
not yet been reported. Therefore, it may be of value to study the
function and possible underlying mechanism of lncRNA RP1-163G9.1 in
GA. In the present study, the possible association of this lncRNA
with GA was investigated in vitro and in vivo and its
potential usefulness as a molecular diagnostic marker was also
evaluated.
Materials and methods
Specimen collection and
preservation
A total of 112 fresh clinically diagnosed GA
specimens and their paired non-cancerous tissues (collected at a
distance of ≥5 cm from the tumor) were collected between April 2014
and August 2016, and immediately placed into RNase-free
cryopreserved tubes containing RNAlater solution (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The specimens were numbered
with a unique ID code and labeled with specific information,
including storage time and tissue type. The specimens were stored
in a cryogenic refrigerator at −80°C for later use.
Data collection on clinicopathological
parameters
Patient information, including age, sex, tumor
location and size, depth of invasion, differentiation, lymph node
metastasis, histological type and immunohistochemical markers, was
collected following gastric cancer excision using the hospital's
Laboratory Information System, pathological diagnosis system and
database.
RNA extraction and reverse
transcription-qPCR (RT-qPCR)
Total RNA was extracted from fresh GA specimens and
GES-1, BGC-823, SGC7901, MGC803 and AGS cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 200 ng RNA
was used to synthesize cDNA with EasyScript One-Step gDNA Removal
and cDNA Synthesis SuperMix (cat. no. AE311; TransGen Biotech, Co.,
Ltd., Beijing, China). According to the known gene sequence,
specific primers of lncRNA RP1-163G9.1 and GAPDH were designed
using the National Center for Biotechnology Information online
primer design program Primer 3.0. All primers were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China). The sequences of
primers were as follows: GAPDH, forward 5′-ACCCACTCCTCCACCTTTGAC-3′
and reverse, 5′-TGTTGCTGTAGCCAAATTCGTT-3′; and lncRNA RP1-163G9.1,
forward 5′-CGCCTCTCACTGGTAAGTCC-3′ and reverse,
5′-AACTGAGTCCCCAAAGACCC-3′. The relative expression of lncRNA
RP1-163G9.1 was determined using RT-qPCR and SYBR-Green reagent
from a TransStartR qPCR Super Mix kit (cat. no. AQ131; TransGen
Biotech, Co., Ltd.). The qPCR conditions were as follows: 95°C for
3 min, 94°C for 5 sec and 58°C for 30 sec for 40 cycles, followed
by a dissociation stage at 95°C for 15 sec, 60°C for 15 sec and
95°C for 15 sec. The 2−ΔΔCq method was used to detect
the expression fold changes of lncRNA RP1-163G9.1. GADPH was used
as the internal control. The standard curve was constructed to
determine the amplification efficiency.
Fluorescence in situ hybridization
(FISH)
A total of 30 paired paraffin-embedded sections were
selected from the 112 pairs of GA specimens to determine the
expression localization of lncRNA RP1-163G9.1 using FISH. The
organization paraffin block was provided by the Department of
Pathology, Fuzhou General Hospital (Fuzhou, China). The probe
sequence of lncRNA RP1-163G9.1 was designed and synthesized by
GenePharma Co., Ltd. (Shanghai, China) and marked with 5′CY3. The
sequence was as follows: 5′-CTGCCGCCACCGTTCTACC-3′. Paraffin
sections were prepared by placing the paraffin block on ice for 1
h, cutting the samples into 3-µm slices and then naturally
drying the slices. Hybridization was conducted on a ThermoBrite
hybrid instrument (Qi Wei Industrial Co., Ltd., Shanghai, China)
overnight after sealing. The probe concentration was 5 µM.
The hybridization conditions were as follows: 75°C for 5 min
followed by incubation at 42°C for 16 h.
Ethics statement
The present study was approved by the Fuzhou General
Hospital Ethics Committee. All patients provided permission for the
use of their tissue samples for research purposes.
Scoring criteria of FISH
The cytoplasm was stained red and identified as
lncRNA RP1-163G9.1-positive expression. According to Bai et
al (17), staining intensity
was scored as follows: No staining [immunoreactive score (IRS): 0],
weakly positive (IRS: 1–2), moderately positive (IRS: 3–6),
strongly positive (IRS: 8–12). All results were determined by two
different members of staff at the same time.
Cell lines and culture conditions
Normal gastric epithelial cells (GES-1) were
obtained from Beijing Institute of Cancer Prevention (Beijing,
China), and the GA cell lines (BGC-823 and MGC-803) were obtained
from Cell Bank, Chinese Academy of Sciences (Shanghai, China). The
AGS and SGC-7901 GA cell lines were purchased from the American
Type Culture Collection (Manassas, VA, USA). GES-1 cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific,
Inc.). AGS cells were cultured in F12 medium with 10% FBS. BGC-823,
MGC-803 and SGC07901 cells were cultured in RPMI-1640 supplemented
with 10% FBS. The cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2 in culture flasks. The
medium was changed every 1–2 days.
Screening clones that stably express
pCDNA3.1-PR1 and pCDNA3.1
The vectors of pCDNA3.1-lncRNA RP1-163G9.1
(pCDNA3.1-PR1), pCDNA3.1-GFP and pCDNA3.1 were purchased from
GenePharma Co., Ltd. pCDNA3.1-PR1 vector, which overexpressed
lncRNA RP1-163G9.1, was constructed by sequence ligation of lncRNA
RP1-163G9.1 (1,697 bp) with pcDNA3.1 empty vector via BamHI
and EcoRI sites. In order to determine the most suitable
concentration, G418 (Nuoyang Biological Co., Ltd., Hangzhou, China)
was diluted from 300 to 1,100 µg/ml in nine consecutive
concentrations for drug screening with BGC-823 and SGC7901 cells.
BGC-823 and SGC7901 were digested with EDTA-trypsin, followed by
subculturing in 6-well plates at 1×105 cells/well. After
the cells had grown to 60–70% confluence, they were transfected
with pCDNA3.1-PR1, pCDNA3.1-GFP and pCDNA3.1 using Lipofectamine
2000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). pcDNA3.1-GFP was used to monitor transfection efficiency and
G418 was used at an appropriate concentration to determine stable
clone screening following a 24-h transfection. Culture solution and
G418 were changed every 3–4 days until cell clones could be
observed by the naked eye. Single clones were selected and culture
was continued with G418.
Cell proliferation experiment
Cells stably expressing pCDNA3.1-PR1 and empty
vector pCDNA3.1 were digested in a single-cell suspension solution,
seeded into 96-well plates at a density of 2,000 cells/well and
cultured at 37°C in an atmosphere containing 5% CO2. A
total of six replicate wells were used. According to the
manufacturer's instructions, 10 µl/well of Cell Counting
Kit-8 (CCK-8) reagent (Nuoyang Biological Co., Ltd., Hangzhou,
China) was added to the cells, followed by incubation at 37°C for 1
h. Absorbance was measured at 450 and 630 nm using a microplate
reader (Spectra Max 190; Molecular Devices, LLC, Sunnyvale, CA,
USA). The assays were performed on days 0 (following culture for 6
h), 1, 2, 3 and 4, and a cell proliferation curve was drawn.
Colony formation experiments
Single-cell suspension solutions of cells stably
expressing pcDNA3.1-PR1 and empty vector pcDNA3.1 were prepared and
subcultured in 6-well plates at a density of 1,000 cells/well.
Culturing was performed at 37°C in an atmosphere containing 5%
CO2 for 14 days. When a single colony contained ≥50
cells, the cells were fixed with methanol for 15 min and stained
with 0.1% crystal violet solution for 15 min. A total of 10 fields
of vision were randomly selecting per well and the number of
colonies was counted under a microscope. Three independent
experiments were conducted.
Detection of apoptosis and analysis of
the cell cycle
BGC823 cells, which stably expressed lncRNA
RP1-163G9.1 and pcDNA3.1, were digested by trypsin. A single-cell
suspension containing 5×105 cells was prepared.
According to the instructions of the manufacturer of Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis
detection kits (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China),
following staining for 5 min at room temperature away from light
with 5 µl PI and 5 µl Annexin V-FITC, apoptosis was
detected using a flow cytometry instrument (BD FACSCalibur; Becton,
Dickinson and Company, San Jose, CA, USA). A total of
5×105 cells in the logarithmic growth phase were
collected, resuspended and stained with 5 µl PI for 10 min
at room temperature away from light. Subsequently, the cell cycle
was analyzed using a flow cytometry instrument.
Transwell experiments
The Transwell chamber (EMD Millipore; Billerica, MA,
USA) was prepared with a BD Matrigel. The mixing ratio of DMEM and
BD Matrigel was 1:4. The chamber preparation experiment was
completed on ice and the temperature did not exceed 10°C. A total
of 100 µl cell suspension containing 5×104 cells
was added to the upper Transwell chambers. A total of 600 µl
complete medium containing serum supplemented with 10% FBS was
added to the lower chamber and the cells were cultured at 37°C in
an atmosphere containing 5% CO2. The experiments were
terminated following 48 h of incubation. The chamber was washed
three times with phosphate-buffered saline (PBS), fixed with
methanol for 20 min and then stained with 0.1% crystal violet
solution for 20 min. Excess dye was removed with PBS and cells in
the interior of the chamber were removed with a cotton swab. Five
fields of view were selected, and the number of cells was counted
using an inverted microscope (Olympus IX51; Olympus Corporation,
Tokyo, Japan).
In vivo experiments
A total of 20 BALB/c male nude mice (aged 4–6 weeks
and weighing 16–20 g) were purchased from Slac Laboratory Animal
Co., Ltd. (Shanghai, China). The mice were equally divided into two
groups. Single-cell suspensions using PBS and SGC7901 cells stably
overexpressing pCDNA3.1 and pCDNA3.1-PR1 were prepared and
subcutaneously inoculated in the right flank of nude mice
(8×106 cells/0.1 ml/mouse). Nude mice were observed
every day and the tumor volume was measured every week. Tumor
volume was calculated as follows: V (mm3) =
ab2/2, where ‘a’ is the long diameter and ‘b’ the short
diameter of the measured tumors. After 5 weeks, the nude mice were
euthanized by cervical dislocation, the tumors were removed and the
volumes and weights were determined, and hematoxylin and eosin
staining was performed on representative specimens.
Statistical analysis
The relative expression level of lncRNA RP1-163G9.1
was obtained by conducting statistical analysis with a paired
t-test using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla,
CA, USA). All clinicopathological data were assessed using SPSS
19.0 software (IBM Corp., Armonk, NY, USA). The correlation of
lncRNA RP1-163G9.1 expression with clinical pathological parameters
and immunohistochemical markers was assessed using analysis of
variance. Cell function experiments were conducted by a
double-sided t-test and P<0.05 was considered to indicate a
statistically significant difference. Data are presented as the
mean ± standard deviation calculated from three replicate
representative experimental results. In addition, the association
of lncRNA RP1-163G9.1 expression with the tumor-node-metastasis
(TNM) stage of the tumor, lymph node metastasis and tumor size was
performed using multi-factor logistic regression. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression levels of lncRNA
RP1-163G9.1 in GA and adjacent non-GA tissues
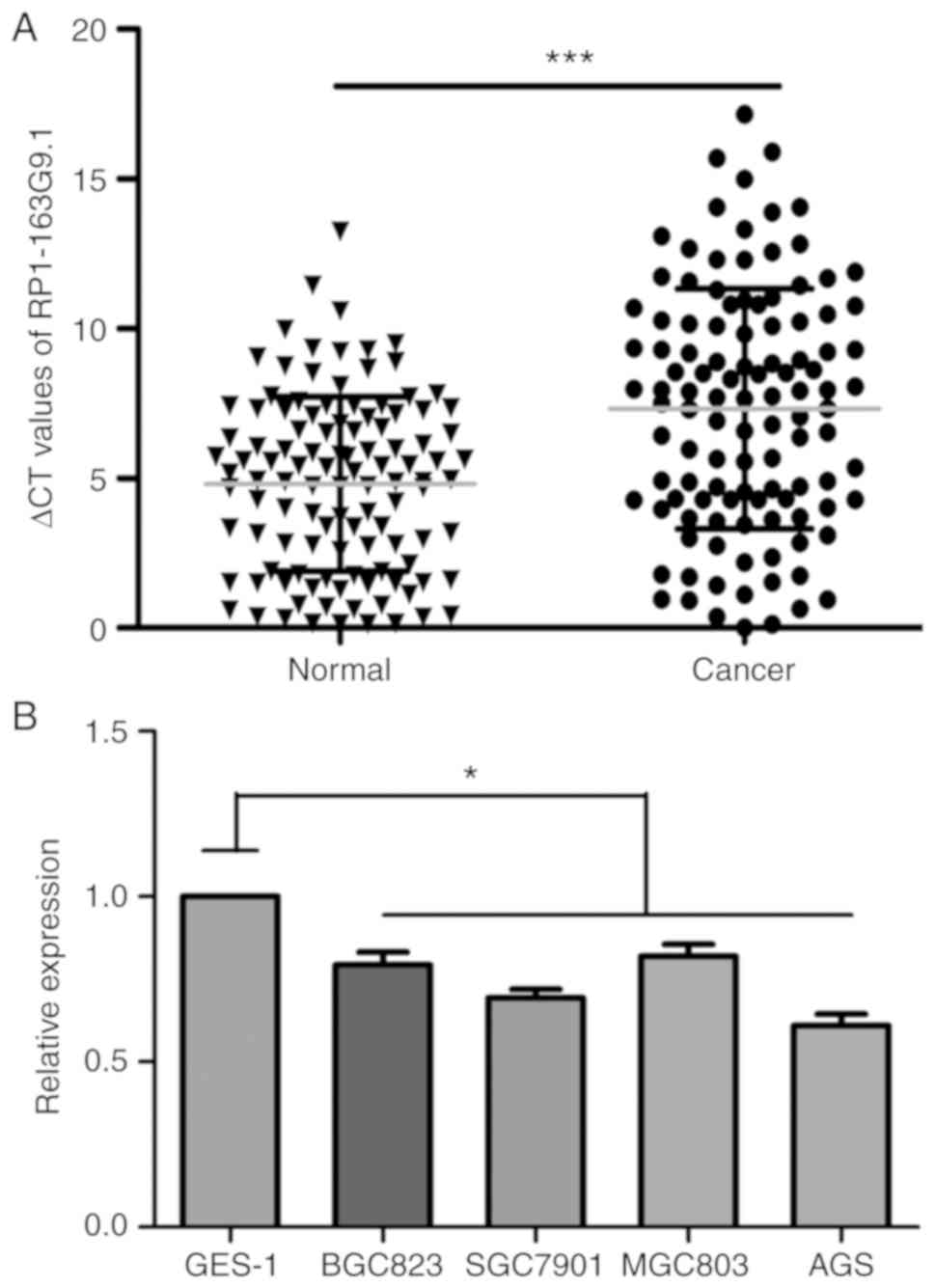
lncRNA RP1-163G9.1 was found to be downregulated in
GA tissues according to the results of the RT-qPCR analysis.
Notably, the Cq difference between the target lncRNA RP1-163G9.1
and the internal reference gene in cancer tissues was as follows:
ΔCq = 7.317±0.379. However, the Cq difference between paracancerous
tissues and the internal reference gene was as follows: ΔCq =
4.807±0.276. Statistical analysis indicated that the expression of
lncRNA RP1-163G9.1 in GA was significantly decreased compared with
that in adjacent non-GA tissues (P<0.001; Fig. 1A). The results of tissue validation
demonstrated that lncRNA RP1-163G9.1 expression was consistent with
the data of the chip analysis. The RT-qPCR results revealed that
lncRNA RP1-163G9.1 expression was downregulated in 85/112 cases of
GA (75.89%) by 23.94-fold. Notably, the expression of lncRNA
RP1-163G9.1 was also detected in gastric cancer cell lines (BGC823,
SGC7901, MGC803 and AGS) and the control gastric epithelial cells
(GES-1). lncRNA RP1-163G9.1 expression was also significantly
downregulated in the four gastric cancer cells compared with GES-1
cells (P<0.05; Fig. 1B).
Correlation analysis of lncRNA
RP1-163G9.1 expression with clinicopathological parameters and
immunohistochemical markers
Patients' detailed clinical data were collected and
correlation analysis of lncRNA RP1-163G9.1 expression with
clinicopathological parameters was performed. The results suggested
that decreased expression of lncRNA RP1-163G9.1 was significantly
associated with the depth of tumor invasion (P=0.001), lymph node
metastasis (P=0.009) and tumor size (P=0.037) (Table I). These findings suggested that low
lncRNA RP1-163G9.1 expression may promote tumor cell proliferation
and invasion. The correlation of the decreased expression of lncRNA
RP1-163G9.1 with immunohistochemical markers of GA was also
assessed. The results demonstrated that decreased lncRNA
RP1-163G9.1 expression was significantly associated with Ki-67
(P=0.010; Table II). Ki-67 is a
known cell proliferation marker. Notably, a higher Ki-67-positive
rate is indicative of faster tumor proliferation and a higher
degree of malignancy. Therefore, our results suggested that lncRNA
RP1-163G9.1 may be associated with tumor cell proliferation.
Multivariate logistic regression analysis also indicated that
lncRNA RP1-163G9.1 acted as a tumor-protective factor (P<0.05).
The adjusted variables were age, sex, tumor differentiation and
histological type (Table
III).
 | Table I.Analysis of the correlation between
lncRNA RP1-163G9.1 expression level and clinicopathological
parameters. |
Table I.
Analysis of the correlation between
lncRNA RP1-163G9.1 expression level and clinicopathological
parameters.
| Clinicopathological
parameters | Cases, n | ΔΔCqa | P-value |
|---|
| Age (years) |
|
| 0.643 |
|
<60 | 56 | 2.73±0.44 |
|
|
≥60 | 56 | 2.46±0.39 |
|
| Sex |
|
| 0.952 |
|
Male | 80 | 2.59±0.35 |
|
|
Female | 32 | 2.62±0.52 |
|
|
Differentiation |
|
| 0.074 |
|
Poor | 63 | 3.10±0.38 |
|
|
Moderate | 46 | 1.85±0.46 |
|
|
High | 3 | 4.36±1.52 |
|
| Depth of
invasion |
|
| 0.001 |
| T1 +
T2 | 23 | 0.39±0.56 |
|
| T3 +
T4 | 89 | 3.17±0.31 |
|
| Lymph node
metastasis |
|
| 0.009 |
| No | 35 | 1.49±0.58 |
|
|
Yes | 77 | 3.11±0.32 |
|
| Tumor size, cm |
|
| 0.037 |
|
<4 | 50 | 1.93±0.44 |
|
| ≥4 | 62 | 3.15±0.37 |
|
| Neural
invasion |
|
| 0.946 |
| No | 62 | 2.62±0.42 |
|
|
Yes | 50 | 2.58±0.39 |
|
| Vessel
invasion |
|
| 0.291 |
| No | 58 | 2.90±0.43 |
|
|
Yes | 54 | 2.28±0.38 |
|
 | Table II.Correlation analysis between lncRNA
RP1-163G9.1 expression level and immunohistochemical markers. |
Table II.
Correlation analysis between lncRNA
RP1-163G9.1 expression level and immunohistochemical markers.
| Immunohistochemical
markers | Cases, n | ΔΔCqa | P-value |
|---|
| α-fetoprotein |
|
| 0.607 |
|
Low | 102 | 2.55±0.31 |
|
|
High | 10 | 3.08±0.65 |
|
| Carcinoembryonic
antigen |
|
| 0.113 |
|
Low | 93 | 2.39±0.32 |
|
|
High | 19 | 3.62±0.72 |
|
| Carbohydrate
antigen 19-9 |
|
| 0.287 |
|
Low | 94 | 2.47±0.32 |
|
|
High | 18 | 3.31±0.30 |
|
| Vascular
endothelial growth factor |
|
| 0.715 |
|
Low | 34 | 2.76±0.49 |
|
|
High | 78 | 2.53±0.36 |
|
| C-erbB-2 |
|
| 0.932 |
|
Low | 98 | 2.61±0.31 |
|
|
High | 14 | 2.54±0.91 |
|
| Thymidine
synthase |
|
| 0.076 |
|
Low | 53 | 2.06±0.42 |
|
|
High | 59 | 3.09±0.40 |
|
| Breast cancer type
1 |
|
| 0.859 |
|
Low | 32 | 2.52±0.48 |
|
|
High | 80 | 2.63±0.36 |
|
| Excision repair
cross-complementation group 1 |
|
| 0.199 |
|
Low | 31 | 3.21±0.60 |
|
|
High | 81 | 2.37±0.33 |
|
| Protamine 1 |
|
| 0.566 |
|
Low | 77 | 2.49±0.35 |
|
|
High | 35 | 2.85±0.52 |
|
| Ki-67 |
|
| 0.010 |
|
Low | 37 | 1.54±0.48 |
|
|
High | 75 | 3.20±0.35 |
|
 | Table III.Logistic regression analysis of
lncRNA RP1-163G9.1 and tumor depth of invasion, LMN and tumor
size. |
Table III.
Logistic regression analysis of
lncRNA RP1-163G9.1 and tumor depth of invasion, LMN and tumor
size.
|
|
| Unadjusted |
Adjusteda |
|---|
|
|
|
|
|
|---|
| lncRNA RP1-163G9.1
expression | LNM (n) No/yes | OR | CI | P-value | OR | CI | P-value |
|---|
| Low | 20/65 |
| Reference |
|
| Reference |
|
| High | 15/12 | 0.25 | 0.10–0.61 | 0.001 | 0.21 | 0.08–0.57 | 0.002 |
|
|
|
|
Unadjusted |
Adjusteda |
|
|
|
|
|
| lncRNA
RP1-163G9.1 expression | Invasion depth
T1+T2/T3+T4 (n) | OR | CI | P-value | OR | CI | P-value |
|
| Low | 10/75 |
| Reference |
|
| Reference |
|
| High | 13/14 | 0.14 | 0.05–0.39 | 0.001 | 0.14 | 0.04–0.36 | 0.001 |
|
|
|
|
Unadjusted |
Adjusteda |
|
|
|
|
|
| lncRNA
RP1-163G9.1 expression | Tumor size, cm
<4/≥4 (n) | OR | CI | P-value | OR | CI | P-value |
|
| Low | 33/52 |
| Reference |
|
| Reference |
|
| High | 17/10 | 0.37 | 0.15–0.91 | 0.028 | 0.38 | 0.15–0.93 | 0.035 |
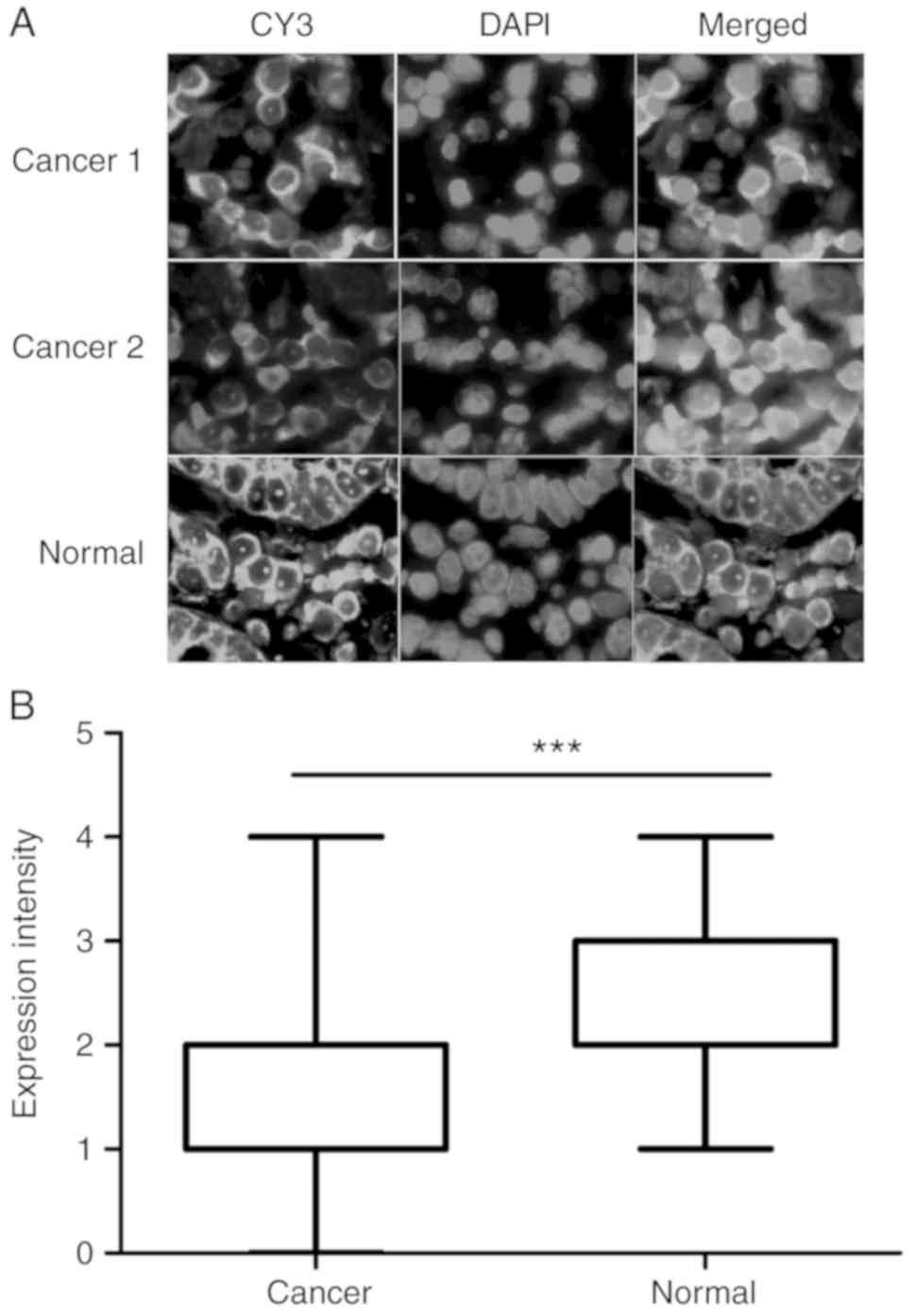
In situ expression and localization of
lncRNA RP1-163G9.1
A total of 30 pairs of GA paraffin-embedded tissues
were selected to conduct FISH. The results demonstrated that lncRNA
RP1-163G9.1 was primarily expressed in the cytoplasm, whereas no
expression was detected in the nucleus (Fig. 2A). Compared with paired control
tissues, the expression of lncRNA RP1-163G9.1 was significantly
lower compared with that of their paired control tissues when
analyzed with GraphPad Prism 5 software (P<0.001; Fig. 2B).
Screening of clones stably expressing
lncRNA RP1-163G9.1 in BGC-823 and SGC-7901 cells
G418 drug screenings of BGC823 and SGC7901 cells
were completed and the final concentration of G418 was 800 and 700
µg/ml, respectively. SGC7901 and BGC823 clones exhibiting
stable overexpression of lncRNA RP1-163G9.1 were successfully
constructed. The relative expression of lncRNA RP1-163G9.1 was
detected with RT-qPCR. Notably, the expression of lncRNA
RP1-163G9.1 in the pCDNA3.1-PR1 group was significantly increased
compared with the pCDNA3.1 empty vector stable expression groups in
SGC7901 and BGC823 cells (P<0.01).
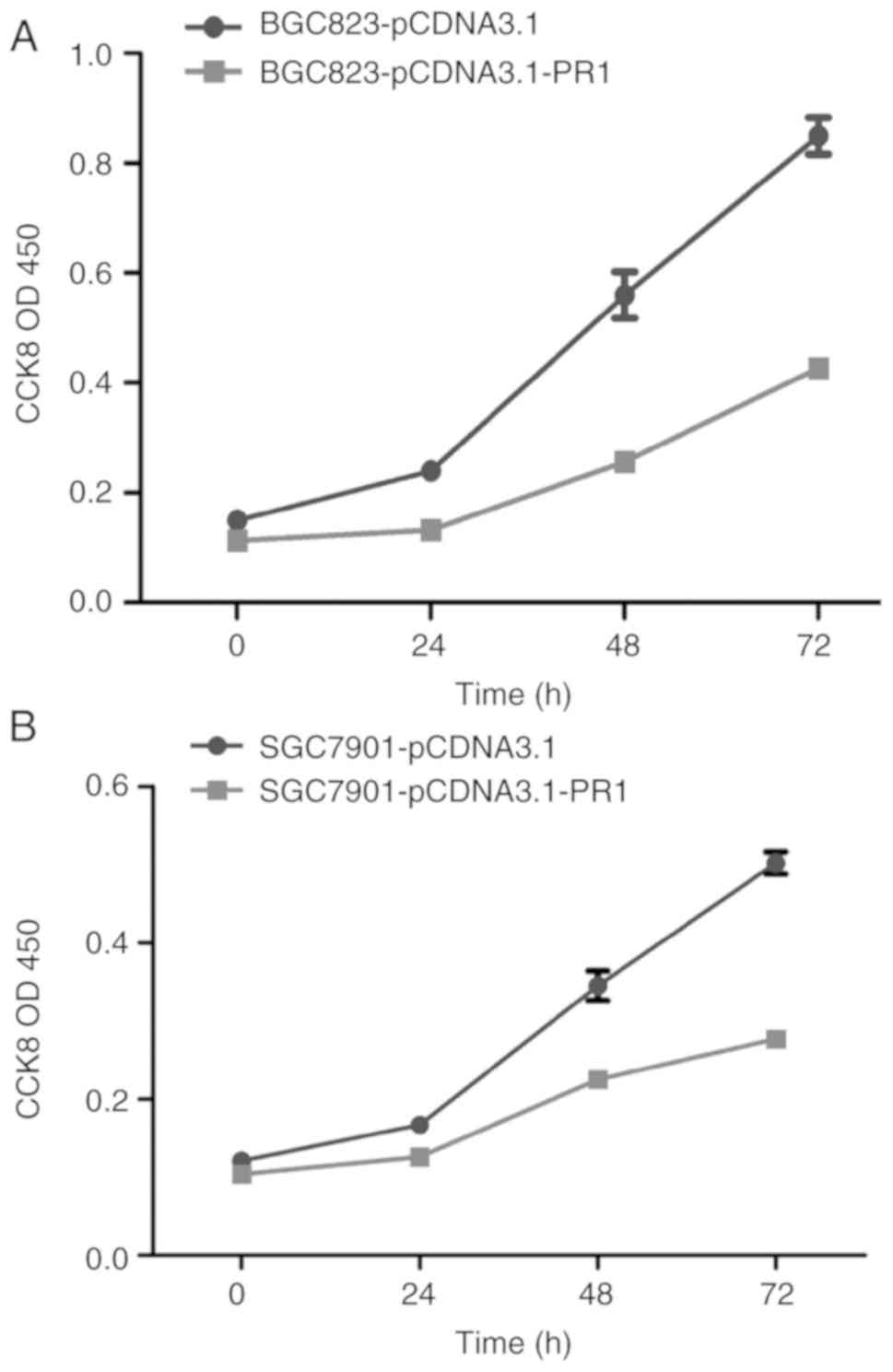
Overexpression of lncRNA RP1-163G9.1
inhibits tumor cell proliferation
CCK-8 proliferation experiments were performed in
BGC823 and SGC7901 cells overexpressing lncRNA RP1-163G9.1. The
results indicated that the proliferation ability of the
pCDNA3.1-PR1 clones was significantly lower compared with that of
the pCDNA3.1 empty vector clones, indicating that lncRNA
RP1-163G9.1 can inhibit the proliferation of GA cells at the
cellular level (P<0.01; Fig.
3).
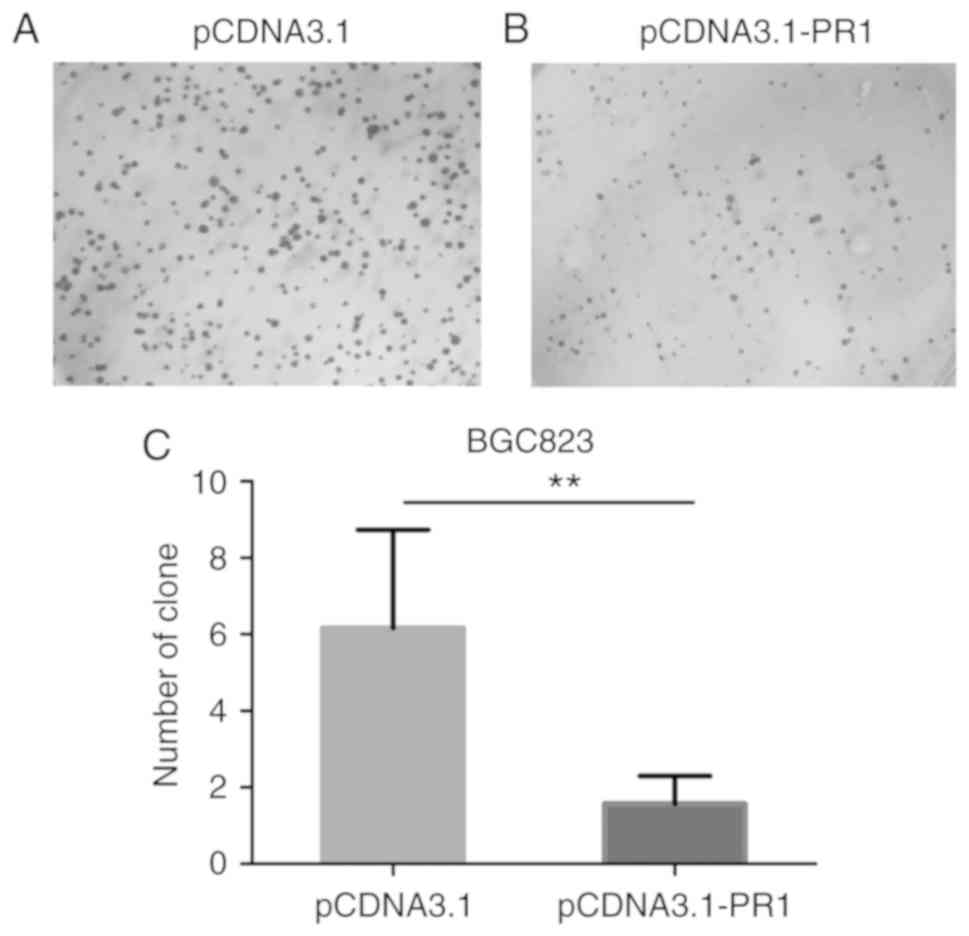
Overexpression of lncRNA RP1-163G9.1
inhibits the colony-forming ability of GA cells
Colony formation experiments were performed to
detect the ability of cells to form colonies. The colony-forming
ability of BGC823 cells was found to be decreased in pCDNA3.1-PR1
clones compared with the empty vector control (Fig. 4A and B). Statistical analysis
indicated that overexpression of lncRNA RP1-163G9.1 significantly
inhibited the proliferation of tumor cells (P<0.01; Fig. 4C).
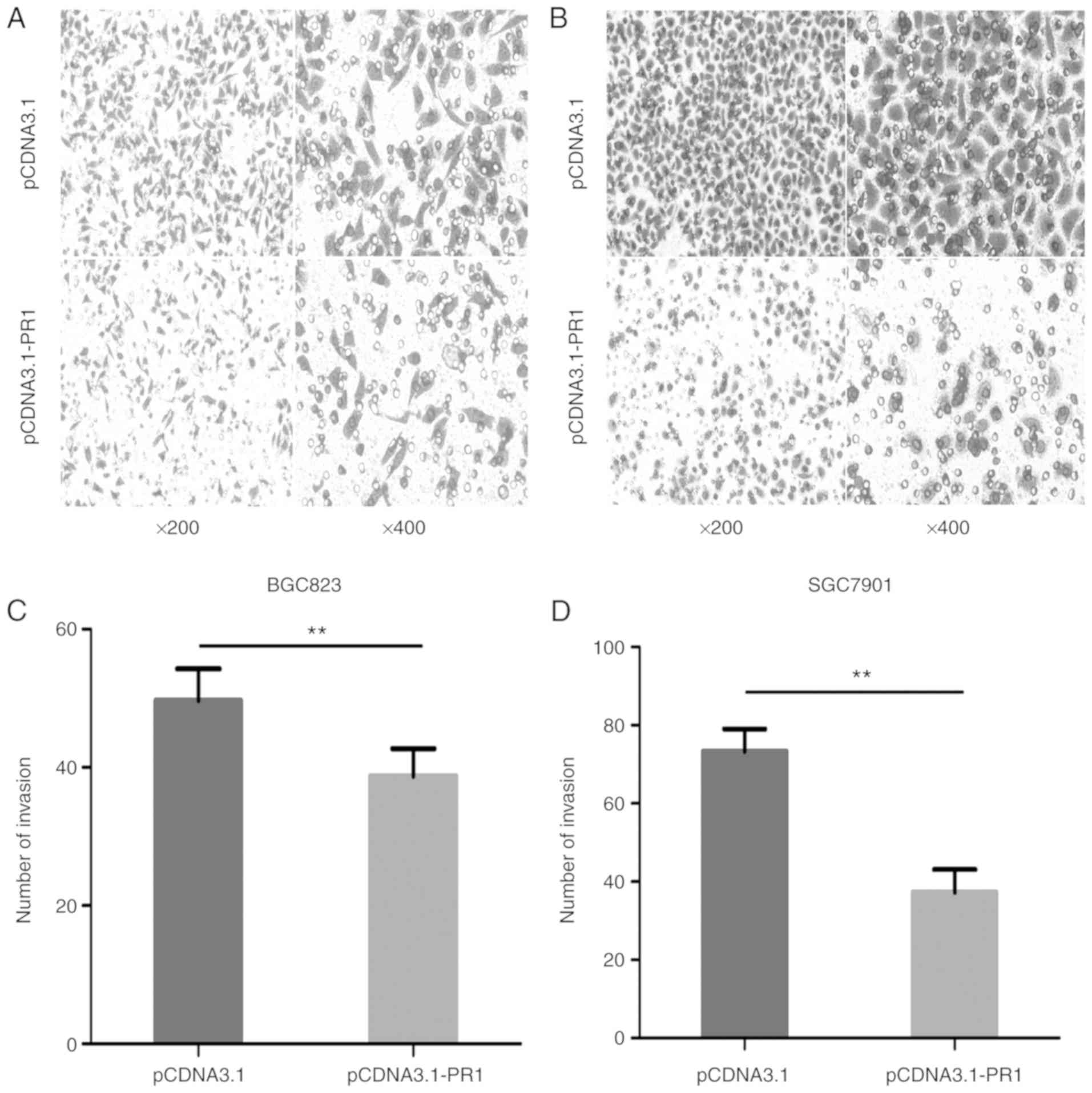
lncRNA RP1-163G9.1 overexpression
inhibits tumor cell invasion
In order to determine whether lncRNA RP1-163G9.1
affects the invasive ability of tumor cells, lncRNA RP1-163G9.1
stably overexpressing cell lines were used in Transwell experiments
(Fig. 5A and B). The invasive
ability of pCDNA3.1-PR1 was significantly decreased compared with
the pCDNA3.1 empty vector control group in BGC-823 and SGC-7901
cells (P<0.01; Fig. 5C and D).
These findings indicated that lncRNA RP1-163G9.1 may play a role in
tumor cell invasion and that it may exert a tumor-protective
effect.
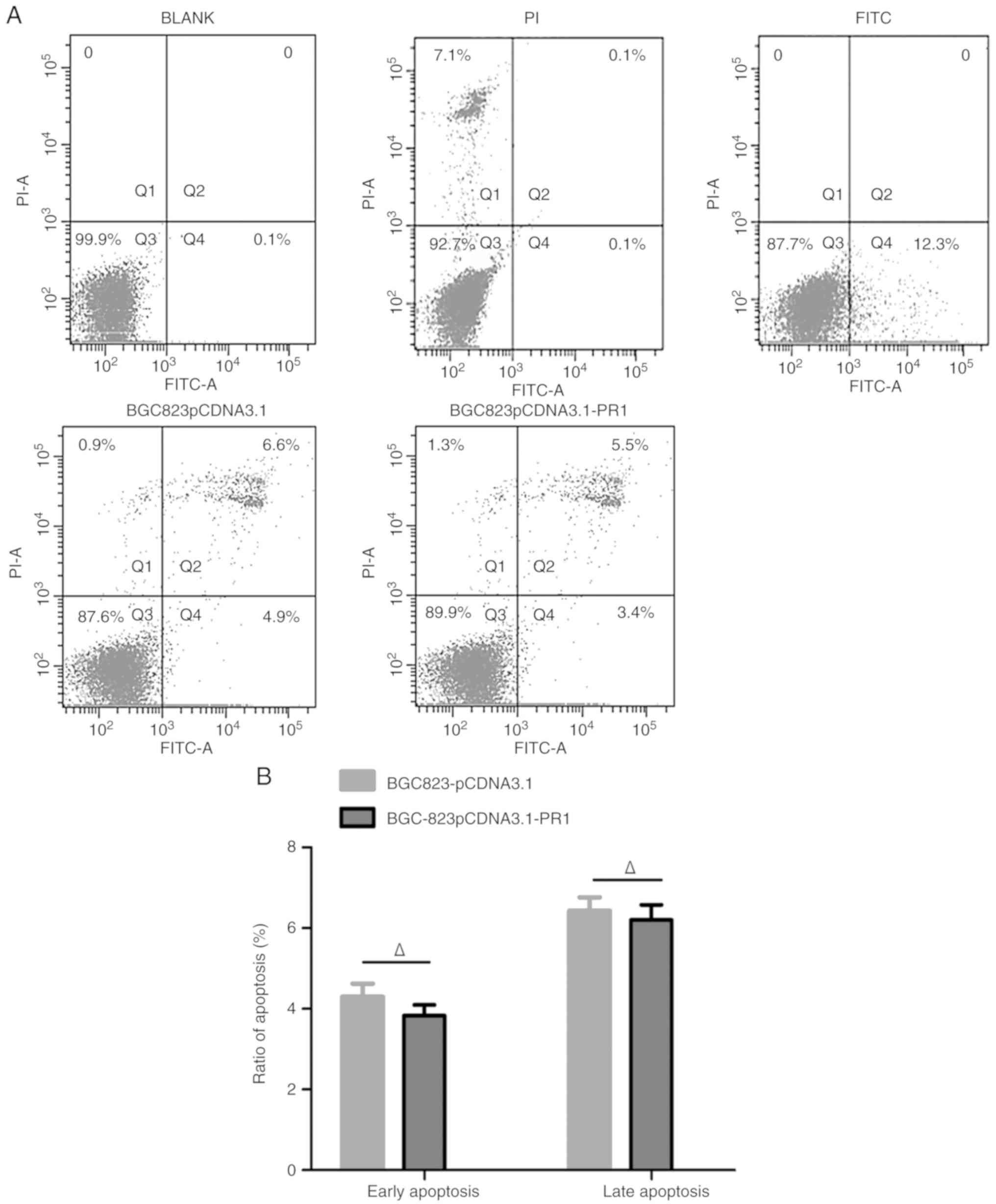
Effect of lncRNA RP1-163G9.1 on the
cell cycle and apoptosis of tumor cells
Apoptosis was detected using flow cytometry. The
results are presented in Fig. 6A.
The apoptotic level of BGC823-pCDNA3.1-PR1 did not differ
significantly from that of BGC823-pCDNA3.1 (P>0.05; Fig. 6B). This finding indicated that
lncRNA RP1-163G9.1 did not affect the apoptosis of tumor cells.
Notably, cell cycle analysis demonstrated that the proportion of
cells in the S-phase was significantly reduced in the
BGC823-pCDNA3.1-PR1 group compared with the BGC823-pCDNA3.1 group
(P<0.05; Fig. 6C and D).
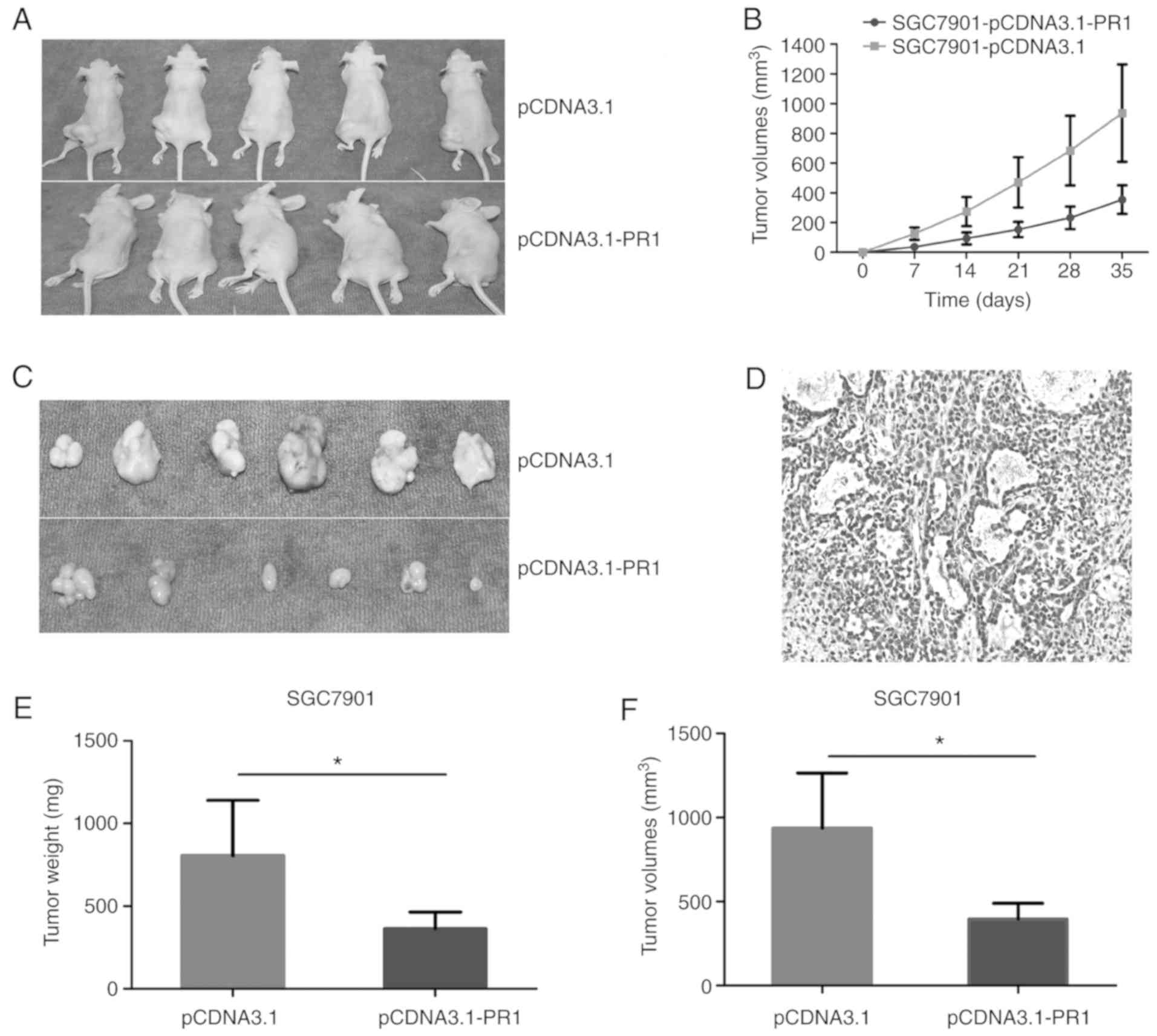
Tumor growth curve
Tumor cells were subcutaneously inoculated into nude
mice, and the animals were sacrificed at the end of the experiment.
Tumor growth in the two groups of nude mice was evaluated (Fig. 7A). Tumor volume was measured weekly
and a growth curve was plotted. The tumor volume of the
SGC7901-pCDNA3.1-PR1 group was smaller compared with that of the
SGC7901-pCDNA3.1 group, and the difference was statistically
significant (P<0.05; Fig.
7B).
lncRNARP1-163G9.1 inhibits tumor cell
proliferation in vivo
Tumor tissue was isolated, the tumor volume and size
were measured and representative tumor samples were subjected to
hematoxylin and eosin staining to determine the nature of the tumor
tissues (Fig. 7C and D).
Statistical analysis of tumor volume and size demonstrated that
there was a significant difference between the experimental and
control groups (P<0.05; Fig. 7E and
F). Therefore, these results revealed that lncRNARP1-163G9.1
overexpression can inhibit tumor growth in vivo.
Discussion
Based on the previous chip assay results, the
present study was the first to report that the expression of lncRNA
RP1-163G9.1 was downregulated in GA tissues. RT-qPCR results from
112 GA tissues indicated that the rate of downregulation was 75.89%
(85/112), and it was 23.94-fold lower in cancer samples compared
with paired non-cancerous tissues. The present investigation
further revealed that low expression of lncRNA RP1-163G9.1 was
associated with increased invasiveness of GA, lymph node
metastasis, larger tumor size and increased expression of the
immunohistochemical marker Ki-67. Furthermore, the overexpression
of lncRNA RP1-163G9.1 was shown to inhibit GA cell invasion and
colony formation, cause a decrease of the number of cells in the S
phase of the cycle in vitro, and reduce subcutaneous
tumorigenesis in nude mice in vivo. These data support that
lncRNA RP1-163G9.1 affects the proliferation ability of gastric
cancer cells. In addition, the results suggested that the low
expression of lncRNA RP1-163G9.1 in patients with GA is associated
with tumor proliferative capacity.
Cancer is a complex disease, which was caused by
multiple factors that are associated with changes in gene
expression. Research mostly focuses on protein-coding genes.
However, recently, emerging studies have indicated that lncRNAs are
not only involved in the occurrence and development of human tumors
(18), but may also be used as new
type of biomarker and therapeutic target (19,20).
More evidence has demonstrated that lncRNAs are closely associated
with human cancer, and play important roles in the regulation of
cell proliferation and apoptosis (21,22),
cell cycle, epigenetics and chromatin remodeling (23,24),
thereby participating in tumorigenesis and cancer progression.
Therefore, lncRNAs are becoming an area of interest in research.
lncRNAs are typically identified by the use of high-throughput chip
technology. In addition, other methods, using bioinformatics
techniques, have been used to predict the lncRNAs associated with
tumors, including Improved Random Walk with Restart for
lncRNA-Disease Association prediction (25) and KATZ measure for lncRNA-Disease
Association prediction (26). These
findings may be helpful in predicting gastric cancer-associated
lncRNAs by building a new computer model.
In the present study, low expression of lncRNA
RP1-163G9.1 was detected in GA tissues; however, its function in GA
remained unclear. An overexpression vector of this lncRNA was
therefore constructed and clones stably expressing lncRNA
RP1-163G9.1 were screened. lncRNA RP1-163G9.1 exerted an effect on
tumor proliferation and led to a cell cycle arrest in the
G1 phase. The results of the present study were similar
to the results obtained for other gastric cancer-associated-lncRNAs
to some degree. For example, the lncRNA taurine-upregulated 1 was
found to be overexpressed in GA through interacting with PRC2 to
affect cell cycle progression (27). The present results further confirm
that lncRNA dysregulation is closely associated with GA. Documented
literature has indicated that the function of lncRNAs that
participate in numerous signaling pathways is closely associated
with their location in the cell (28). The results of our FISH assay
revealed that lncRNA RP1-163G9.1 is located in the cytoplasm, and
so it may be hypothesized that lncRNA RP1-163G9.1 may be associated
with transcription regulation and protein translation.
Although the function of lncRNA RP1-163G9.1 was
investigated in the present study, the underlying molecular
mechanism requires further elucidation in future experiments. FISH
demonstrated that lncRNA RP1-163G9.1 is located in the cytoplasm.
According to current lncRNA research, it was hypothesized that
lncRNA RP1-163G9.1 may act as a ceRNA that combines with endogenous
miR and competes for the target gene (29). Alternatively, this lncRNA may act as
a bait molecule for target gene recruitment (30,31).
As regards the molecular mechanisms involved in lncRNA function, it
was previously demonstrated that lncRNAs may competitively combine
with miRs, leading to various effects associated with oncogenesis,
including cell proliferation, growth and metastasis (32). Thus, combined with bioinformatics
technology, coding-non-coding gene co-expression (CNC) and ceRNA
prediction analysis of lncRNA RP1-163G9.1 was performed in the
present study. A total of 54 mRNAs with the same expression
patterns were screened in CNC prediction (data not shown). Notably,
the expression of ~48 of these was negatively correlated with
lncRNA RP1-163G9.1 and the expression of 6 was positively
correlated. Furthermore, 41 miRs were predicted in ceRNA analysis
(data not shown), which have nucleotide binding sites for lncRNA
RP1-163G9.1. The results may be useful for further research on
identifying the possible mechanism of lncRNA RP1-163G9.1 in GA. We
hypothesized that lncRNA RP1-163G9.1 may not be related to
apoptosis-associated gene expression or participate in cell
apoptosis signaling pathway regulation. The exact role and
molecular mechanism involved in the association of lncRNA
RP1-163G9.1 with GA cell apoptosis requires further
investigation.
There were certain limitations to the present study.
These included an insufficient number of specimens and a lack of
study of the underlying molecular mechanism. Next, a larger number
of samples may be investigated to confirm its true clinical
application, and the expression of lncRNA RP1-163G9.1 may be
detected in collected blood samples (including whole blood, serum
and plasma). If its expression is consistent with that in the
tumor, a diagnostic cut-off value may be determined, which may
represent a good diagnostic marker for gastric cancer. According to
the results of bioinformatics prediction and the results of chip
assay conducted on GA cells stably expressing lncRNA RP1-163G9.1,
the high-score miRs and candidate target genes will be investigated
to determine their association with the expression of lncRNA
RP1-163G9.1. Furthermore, it was demonstrated that lncRNA
RP1-163G9.1 is a proliferation index of gastric cancer and, based
on this finding, the expression levels of genes that were related
to cell proliferation will be detected at the protein and mRNA
levels.
Taken together, the results of the present study
indicate that lncRNA RP1-163G9.1 acts as an important regulator of
tumor proliferation in GA and its expression is low in GA tissues.
The findings of the present study suggest that lncRNA RP1-163G9.1
may be a potential target for GA diagnosis and therapy, and may
provide a significant basis for understanding how lncRNAs are
involved in proliferation regulation and tumor progression and
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Innovation
Team Foundation of Fuzhou General Hospital (grant no.
2014CXTD04).
Availability of data and materials
All datasets generated and analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BS was involved in the design of the study, was
responsible for collecting the materials, conducting the literature
review, performing the relevant experiments and writing the
article. YD participated in the collection, preservation, quality
control and RNA extraction of some specimens. FZ, KL, XO and KW
were involved in the acquisition of specimens and the collection of
the clinical data of the patients. FZ participated in the cell
culture and animal tumor formation experiments. KL participated in
the design of some experiments. KW was involved in the RNA
extraction from some specimens and real-time RT-qPCR detection. XO
participated in H&E staining. QH was responsible for the design
of the whole study and revision of the manuscript. All the authors
have read and approved the final version of this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Fuzhou General
Hospital Ethics Committee. All patients provided permission for the
use of their tissue samples for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang K, Ruan J, Qian Q, Song H, Bao C,
Zhang X, Kong Y, Zhang C, Hu G, Ni J and Cui D: BRCAA1 monoclonal
antibody conjugated fluorescent magnetic nanoparticles for in vivo
targeted magnetofluorescent imaging of gastric cancer. J
Nanobiotechnology. 9:232011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu WK, Lee CW, Cho CH, Cho CH, Fan D, Wu
K, Yu J and Sung JJ: MicroRNA dysregulation in gastric cancer: A
new player enters the game. Oncogene. 29:5761–5771. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nammi D, Srimath-Tirumala-Peddinti RC and
Neelapu NR: Identification of drug targets in helicobacter pylori
by in silico analysis: Possible therapeutic implications for
gastric cancer. Curr Cancer Drug Targets. 16:79–98. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sierzega M, Kaczor M, Kolodziejczyk P,
Kulig J, Sanak M and Richter P: Evaluation of serum microRNA
biomarkers for gastric cancer based on blood and tissue pools
profiling: The importance of miR-21 and miR-331. Br J Cancer.
117:266–273. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiberg RA, Halligan DL, Ness RW, Necsulea
A, Kaessmann H and Keightley PD: Assessing recent selection and
functionality at long noncoding RNA loci in the Mouse genome.
Genome Biol Evol. 7:2432–2444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen X, Sun Y, Cai R, Wang G, Shu X and
Pang W: Long noncoding RNA: Multiple players in gene expression.
BMB Rep. 51:280–289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Terashima M, Tange S, Ishimura A and
Suzuki T: MEG3 long noncoding RNA contributes to the epigenetic
regulation of epithelial-mesenchymal transition in lung cancer cell
lines. J Biol Chem. 292:82–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma B, Wang J, Song Y, Gao P, Sun J, Chen
X, Yang Y and Wang Z: Upregulated long intergenic noncoding RNA
KRT18P55 acts as a novel biomarker for the progression of
intestinal-type gastric cancer. Onco Targets Ther. 9:445–453.
2016.PubMed/NCBI
|
|
11
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: LncRNA HOTAIR functions
as a competing endogenous RNA to regulate HER2 expression by
sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Y, Pan J, Zhang N, Wei W, Yu S and Ai
L: Knockdown of long non-coding RNA H19 inhibits multiple myeloma
cell growth via NF-kappaB pathway. Sci Rep. 7:180792017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang EB, Han L, Yin DD, Kong R, De W and
Chen J: c-Myc- induced, long, noncoding H19 affects cell
proliferation and predicts a poor prognosis in patients with
gastric cancer. Med Oncol. 31:9142014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhuang M, Gao W, Xu J, Wang P and Shu Y:
The long non-coding RNA H19-derived miR-675 modulates human gastric
cancer cell proliferation by targeting tumor suppressor RUNX1.
Biochem Biophys Res Commun. 448:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B
and Chen Y: Overexpression of long non-coding RNA HOTAIR predicts a
poor prognosis in patients with acute myeloid leukemia. Oncol Lett.
10:2410–2414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu
Y, Wang L, Wang Y and Huang Q: Expression and clinical significance
of long non-coding RNA HNF1A-AS1 in human gastric cancer.
World J Surg Oncol. 13:3022015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bai J, Yong HM, Chen FF, Mei PJ, Liu H, Li
C, Pan ZQ, Wu YP and Zheng JN: Cullin1 is a novel marker of poor
prognosis and a potential therapeutic target in human breast
cancer. Ann Oncol. 24:2016–2022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang B, Wei ZY, Wang BQ, Yang HC, Wang JY
and Bu XY: Down-regulation of the long noncoding RNA-HOX transcript
antisense intergenic RNA inhibits the occurrence and progression of
glioma. J Cell Biochem. 119:2278–2287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker
for diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ozes AR, Wang Y, Zong X, Fang F, Pilrose J
and Nephew KP: Therapeutic targeting using tumor specific peptides
inhibits long non-coding RNA HOTAIR activity in ovarian and breast
cancer. Sci Rep. 7:8942017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo JQ, Li SJ and Guo GX: Long noncoding
RNA AFAP1-AS1 promotes cell proliferation and apoptosis of gastric
cancer cells via PTEN/p-AKT pathway. Dig Dis Sci. 62:2004–2010.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu G, Xiang T, Wu QF and Wang WX: Long
noncoding RNA H19-derived miR-675 enhances proliferation and
invasion via RUNX1 in gastric cancer cells. Oncol Res. 23:99–107.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JJ, Kim M and Kim HP: Epigenetic
regulation of long noncoding RNA UCA1 by SATB1 in breast cancer.
BMB Rep. 49:578–583. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X and Yan GY: Novel human
lncRNA-disease association inference based on lncRNA expression
profiles. Bioinformatics. 29:2617–2624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Yan CC, Zhang X and You ZH: Long
non-coding RNAs and complex diseases: From experimental results to
computational models. Brief Bioinform. 18:558–576. 2017.PubMed/NCBI
|
|
27
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Atanasovska B, Rensen SS, van der Sijde
MR, Marsman G, Kumar V, Jonkers I, Withoff S, Shiri-Sverdlov R,
Greve JWM, Faber KN, et al: A liver-specific long noncoding RNA
with a role in cell viability is elevated in human nonalcoholic
steatohepatitis. Hepatology. 66:794–808. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He H, Wang N, Yi X, Tang C and Wang D:
Long non-coding RNA H19 regulates E2F1 expression by competitively
sponging endogenous miR-29a-3p in clear cell renal cell carcinoma.
Cell Biosci. 7:652017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Yu Y, Fan S and Luo L: Knockdown
of long noncoding RNA TUG1 inhibits the proliferation and cellular
invasion of osteosarcoma cells by sponging miR-153. Oncol Res.
26:665–673. 2018. View Article : Google Scholar : PubMed/NCBI
|