Introduction
Gastric cancer (GC) is a common malignancy
worldwide. The incidence of GC is highest in East Asian countries,
including Korea, Mongolia, Japan and China, with 40–60 cases per
100,000 individuals, followed by Eastern Europe (~35 cases per
100,000 people) (1,2). According to a statistical report, there
were ~679,000 new cases of GC and 498,000 associated mortalities in
China in 2015 (3). Current treatments
for GC include surgery, chemotherapy, radiation and immunotherapy,
all of which can be administered alone or in combination (4). Adjuvant treatment has been shown to be
beneficial for GC (4,5). In Japan, early diagnosis via endoscopy
and early tumor resection are used to improve the 5-year survival
rate of patients with GC (5,6). Those with GC often present symptoms only
in the later stages; however, the majority of patients do not
receive medical attention until symptoms present. At the time of
definitive diagnosis, many patients with GC are of an advanced
stage of disease, at which point treatment is less effective.
Despite advancements in treatment, no significant improvement in
the prognosis of patients with GC have been reported; the 5-year
overall survival rate was determined to be 30–35% (7). Thus, highly sensitive biomarkers to
increase the sensitivity of early diagnostic methods for GC are of
great interest for the development high-specificity drugs.
FK506 binding protein (FKBP65) is a 65-kDa protein
and highly conserved; almost all FKBP family members have peptide
precursor cis-trans isomerase activity (8). FKBP prolyl isomerase 10 (FKBP10) is a
gene encoding FKBP65, and is a member of a group of proteins termed
the immunophilins, belonging to the FKBP-type peptidylprolyl
cis/trans isomerase family (9,10). It is
located in the endoplasmic reticulum, and is a molecular chaperone
that interacts with collagen (11);
FKBP10 has been reported to directly interact with collagen I
(11). As an important intracellular
regulatory factor for extracellular matrix (ECM) reconstruction,
FKBP10 is an important potential target for the treatment of
idiopathic pulmonary fibrosis (12,13). In
addition, it is increasingly apparent that FKBP members serve a
very important role in the formation of tumors and may be
considered as novel biomarkers of cancer (14,15). For
example, FKBP10 is associated with ovarian cancer (16,17), lung
cancer (18), prostate cancer
(19), leukemia (20), renal cell carcinoma (21) and colorectal cancer (22).
In recent years, numerous studies have identified
genes related to the prognosis of GC (23–25). Some
of these genes can act as prognostic factors for GC, yet the
prognostic potential of these genes as biomarkers in GC remains
unknown. The importance of differentially expressed FKBP10 in GC
and its prognostic value in patients with GC require further
investigation.
In the present study, the differential expression
levels of FKBP10 mRNA in GC and normal tissues were compared using
the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas
(TCGA) databases; the removal of batch effects on same type
platforms was performed in our GEO analysis. In addition,
Kaplan-Meier survival and cox regression analyses were conducted to
explore the potential prognostic value of FKBP10 expression in
patients with GC.
Materials and methods
Data extraction of FKBP10 expression
from GEO and TCGA databases
The data were limited to microarray and RNA
sequencing uploaded before August 2018. Mesh-terms and free words
were used for increasing the search parameters in the GEO databases
(https://www.ncbi.nlm.nih.gov/geo/).
The search terms were: ‘Cancer’, ‘tumor’, ‘carcinoma’ or
‘neoplasm’, and ‘gastric’ or ‘stomach’. ‘Homo sapiens’ was
used to limit the search range. In total, 20 microarrays containing
957 samples of GC and 536 samples of paracancer tissues with FKBP10
expression information were downloaded (Table I). The normalized expression value and
the median expression value were obtained from multiple probes of
FKBP10. The data of FKBP10 expression from the TCGA database
(https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
were downloaded with University of Santa Cruz Xena (https://xena.ucsc.edu/), which provided a normalized
count of gene-level transcription.
 | Table I.Information of elected Gene
Expression Omnibus series dataset. |
Table I.
Information of elected Gene
Expression Omnibus series dataset.
|
|
|
| Case |
|---|
|
|
|
|
|
|---|
| ID | Type | Country | GC | non-GC |
|---|
| GSE29272 | GPL96 | USA | 134 | 134 |
| GSE37023 | GPL96 | Singapore | 112 | 39 |
| GSE54129 | GPL570 | China | 111 | 21 |
| GSE64951 | GPL570 | USA | 63 | 31 |
| GSE13911 | GPL570 | Italy | 38 | 31 |
| GSE19826 | GPL570 | China | 12 | 15 |
| GSE79973 | GPL570 | China | 10 | 10 |
| GSE51725 | GPL570 | Japan | 8 | 2 |
| GSE27342 | GPL5175 | USA | 80 | 80 |
| GSE63089 | GPL5175 | China | 45 | 45 |
| GSE13195 | GPL5175 | China | 25 | 25 |
| GSE33335 | GPL5175 | China | 25 | 25 |
| GSE56807 | GPL5175 | China | 5 | 5 |
| GSE26899 | GPL6947 | USA | 96 | 12 |
| GSE13861 | GPL6884 | USA | 65 | 19 |
| GSE65801 | GPL14550 | China | 32 | 32 |
| GSE103236 | GPL4133 | Romania | 10 | 9 |
| GSE84787 | GPL17077 | China | 10 | 10 |
Detection of FKBP10 mRNA expression
levels in gastric cancer
To investigate the expression of FKBP10, Gene
Expression Profiling Interactive Analysis (GEPIA) (26) was used to retrieve expression data of
GC tissues. In addition, data on FKBP10 expression in ~1,000 cell
lines were provided by The Cancer Cell Line Encyclopedia
(https://portals.broadinstitute.org/ccle). Except for
the TCGA data, FKBP10 expression was obtained from a microarray
series GEO dataset. After comparing FKBP10 expression in single
series datasets, same-type microarray platforms were combined to
expand sample capacity, in order to comprehensively analyze the
expression of FKBP10. The removal of batch effects across platforms
was performed using the ‘sva’ Bioconductor package of R (v3.5.0)
(26). The standardized mean
difference (SMD) method was used to assess the continuous variable,
FKBP10 expression. Data from 19 gene microarrays were combined with
a random effects model when heterogeneity (I2)>50%.
The results were presented as forest plots. Sensitivity analyses
and publication bias were used to evaluate the combined quality.
Continuous variables of FKBP10 expression were converted to true
positive, false positive, false negative and true negative counts.
Summary receiver operating characteristic (SROC) of 19 GEO
microarrays were used to comprehensively investigate the diagnostic
value of FKBP10. All analyses were conducted using STATA 12.0
software (StataCorp). The associations between FKBP10 expression
and certain clinicopathological parameters were analyzed using a
Student's t-test. Tumor and paracancerous samples from the same
patient were analyzed using a paired t-test, while an unpaired
t-test was used to analyze unpaired samples. Genetic alterations of
FKBP10 were determined by cBioPortal (https://www.cbioportal.org/). The DNA methylation
information of FKBP10 was obtained from the MethHC database
(http://methhc.mbc.nctu.edu.tw/)
(27).
Prognosis analysis of FKBP10 in
GC
We aimed to investigate the prognostic potential of
FKBP10 in GC in the TCGA and GEO databases independently. Overall
survival (OS) and disease-free survival (DFS) curves were drawn
based on data from TCGA and GEO (28)
via GEPIA (http://gepia.cancer-pku.cn/) and Kaplan-Meier Plotter
(http://kmplot.com/analysis/) (29). Univariate and multivariate cox
analyses were conducted with adjustments to age, sex, tumor stage,
histological grade and the clinical features of GC.
Detection of FKBP10 protein expression
by immunohistochemistry
The immunohistochemistry results of FKBP10 from the
Human Protein Atlas (HPA) were investigated, which contained
protein expression profiles as determined by immunohistochemistry
(https://www.proteinatlas.org/) (30). In addition, immunohistochemistry data
were verified using GC tissue and paired adjacent normal mucosa
tissue samples. In total, 40 cases of GC tissue and adjacent normal
tissue samples (20 male and 20 female, age range 25–79 years;
average age 56.8-yars old) were collected from patients with GC at
The First Affiliated Hospital of Guangxi Medical University
(Nanning, China), between January 2018 and August 2018. The present
study was approved by the Ethics Committee of The First Affiliated
Hospital of Guangxi Medical University and all patients provided
written informed consent. Antigen retrieval was conducted by
boiling tissue sections in sodium citrate buffer (pH 6.0) at
100–120°C for 5 min; endogenous peroxidase activity was blocked
with 3% hydrogen peroxide at room temperature for 10 min; sections
were then incubated with a rabbit anti-FKBP10 polyclonal antibody
(bs-13175R; 1:700; BIOSS) overnight at 4°C, followed by a
conjugated secondary antibody (cat. no. D-3004-15, Shanghai Long
Island Biotec, Co., Ltd.) at room temperature for 30 min; followed
by 3′,3′-diaminobenzidene staining at room temperature for 5 min.
The average score was calculated by randomly selecting five fields
under a light microscope (magnification ×200). The immunoreaction
score (IRS) was calculated according to the intensity of staining
and the percentage of positive cells. Intensity was scored as
follows: 0, negative; 1, weak; 2, moderate and 3, strong. The
percentage of positive cells was scored as follows: <5%, 0;
6–25%, 1; 26–50%, 2; 51–75%, 3; >76%, 4.
FKBP10 biological function
analysis
To further investigate the biological function of
FKBP10 in GC, we analyzed the possible interactions of FKBP10 using
a protein-protein interaction (PPI) network. An interaction score
of 0.4 was set as a cut-off value. In addition to PPI analysis, we
identified the genes associated with FKBP10 expression that may
also be involved in the regulation of GC development. FKBP10
co-expression networks were assessed using the GEPIA and Coexpedia
online tools (http://www.coexpedia.org/) (31). Pearson correlation analysis of FKBP10
was conducted using GEPIA. Then, Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were performed using the DAVID 6.8 (https://david.ncifcrf.gov/home.jsp).
Results
FKBP10 mRNA expression levels based on
TCGA and GEO databases
A flow chart of our study design was presented in
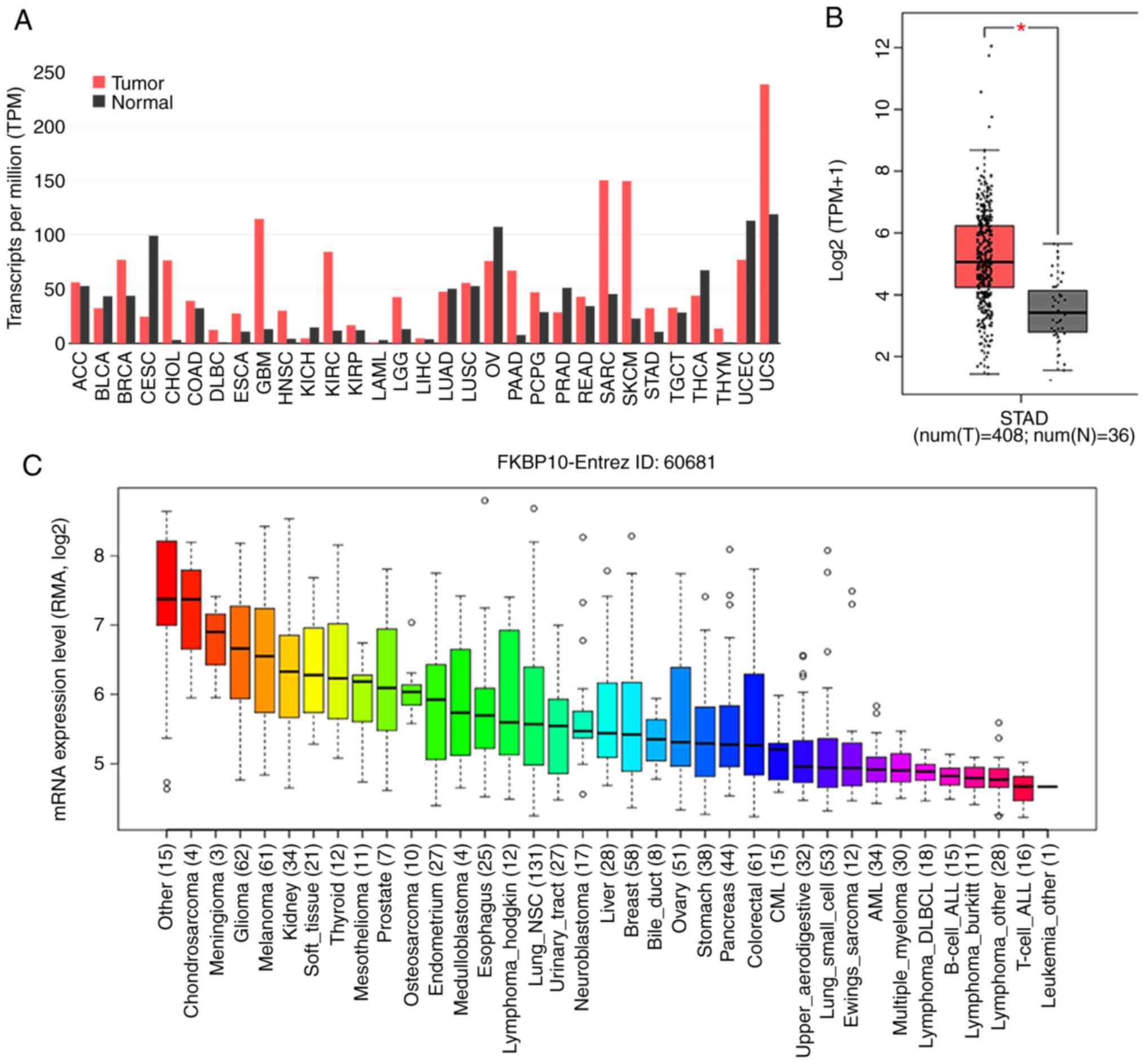
Fig. 1. FKBP10 is relatively
expressed at high levels in a variety of tumors, except
gynecological malignancies, including cervical squamous cell
carcinoma, uterine corpus endometrial carcinoma and ovarian cancer
(Fig. 2A). Additionally, FKBP10 was
significantly overexpressed in GC tissues {n=408,
log2[transcripts per million (TPM) + 1]=5.06} compared
with in adjacent gastric tissues in the TCGA database [n=36,
log2(TPM + 1)=3.53; P<0.001] (Fig.
2B). Expression levels in certain GC cell lines were consistent
with those in GC tissues, each exhibiting medium expression levels
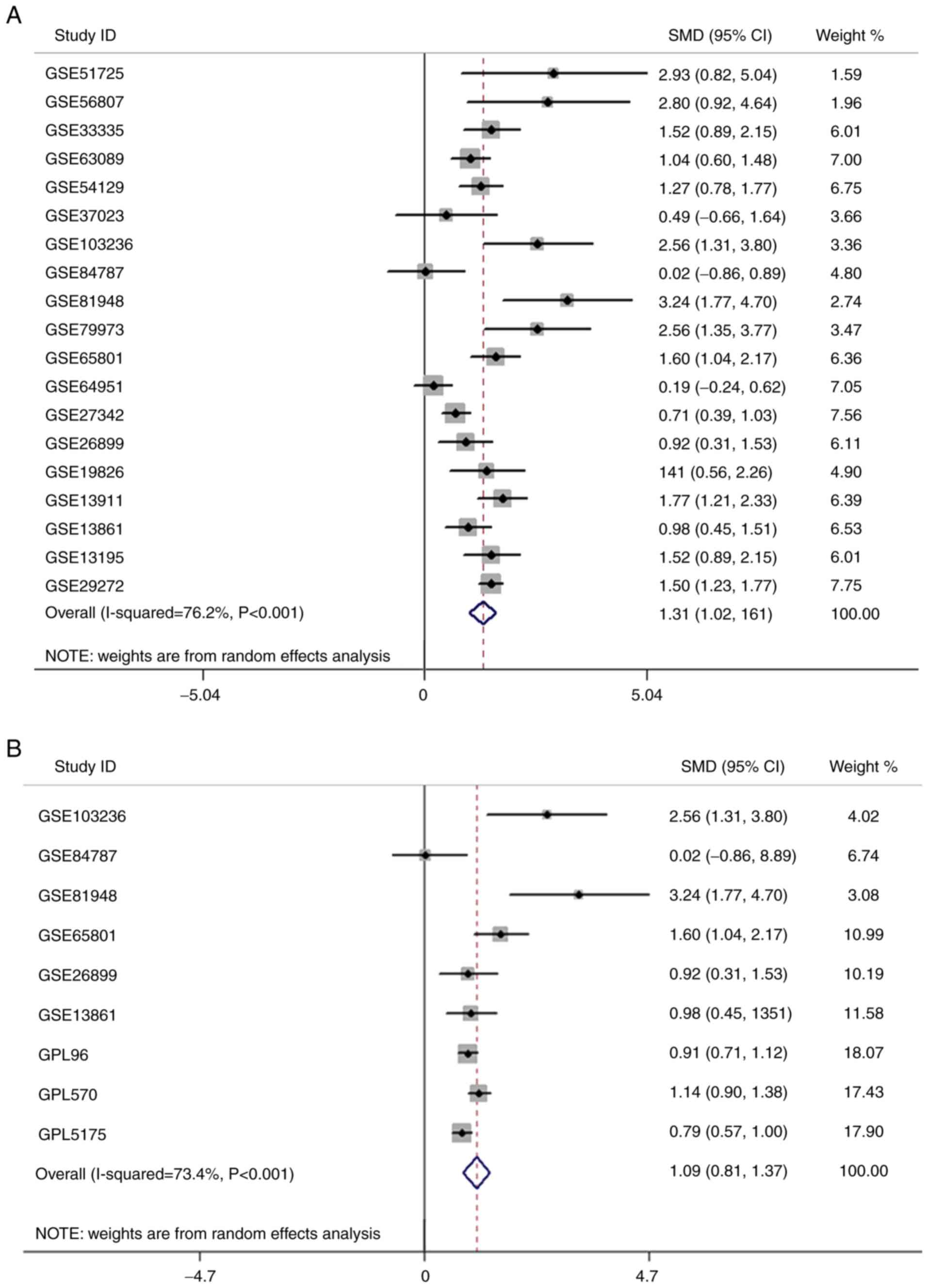
(Fig. 2C). Among 19 GEO microarray
analyses, FKBP10 expression levels in GC tissues were significantly
increased than in adjacent tissues (GSE29272, GSE54129, GSE26899,
GSE27342, GSE13861, GSE63089, GSE13911, GSE65801, GSE13195,
GSE33335, GSE89148, GSE19826, GSE79973, GSE103236, GSE51725,
GSE56807 and GSE2701) (Fig. 3). After
batch effects removal of platform GPL96, GPL570 and GPL5175, FKBP10
also exhibited significantly increased expression in the combined
GC samples compared with in normal samples (Fig. 3). Furthermore, a comprehensive
meta-analysis indicated that FKBP10 expression in GC tissues was
upregulated than in adjacent tissues [SMD=1.31, 95% confidence
interval (CI): 1.02–1.6; P<0.001] (Fig. 4A). Additionally, forest plots of
removal batch effects showed a consistent expression trend in GC
tissues (SMD=1.09, 95% CI: 0.81–1.37, P<0.001) (Fig. 4B). No significant publication bias was
determined in either funnel plot (P=0.125 and P=0.124) (data not
shown). The association between the differential expression of
FKBP10 and the clinicopathological features of patients with GC was
investigated. As for cancer status, FKBP10 expression in patients
with GC was significantly higher than in tumor-free patients, but
there were no significant differences between FKBP10 expression and
stage, grade, T stage, N stage and M stage (Table II). FKBP10 expression was
significantly associated with the clinicopathological factor of
person neoplasm cancer status. This indicated that tumor status
could be closely related to FKBP10 expression; however, FKBP10 was
not significantly linked to other clinical parameters. This may be
due an insufficient sample size. Histological types were classified
as gastrointestinal adenocarcinoma (tubular, papillary, not
otherwise specified and mucinous type) and stomach adenocarcinoma
(not otherwise specified, diffuse and signet ring type).
Unfortunately, FKBP10 expression did not significantly differ
between gastric and gastrointestinal adenocarcinoma. In addition,
there was no significant relationship between FKBP10 expression and
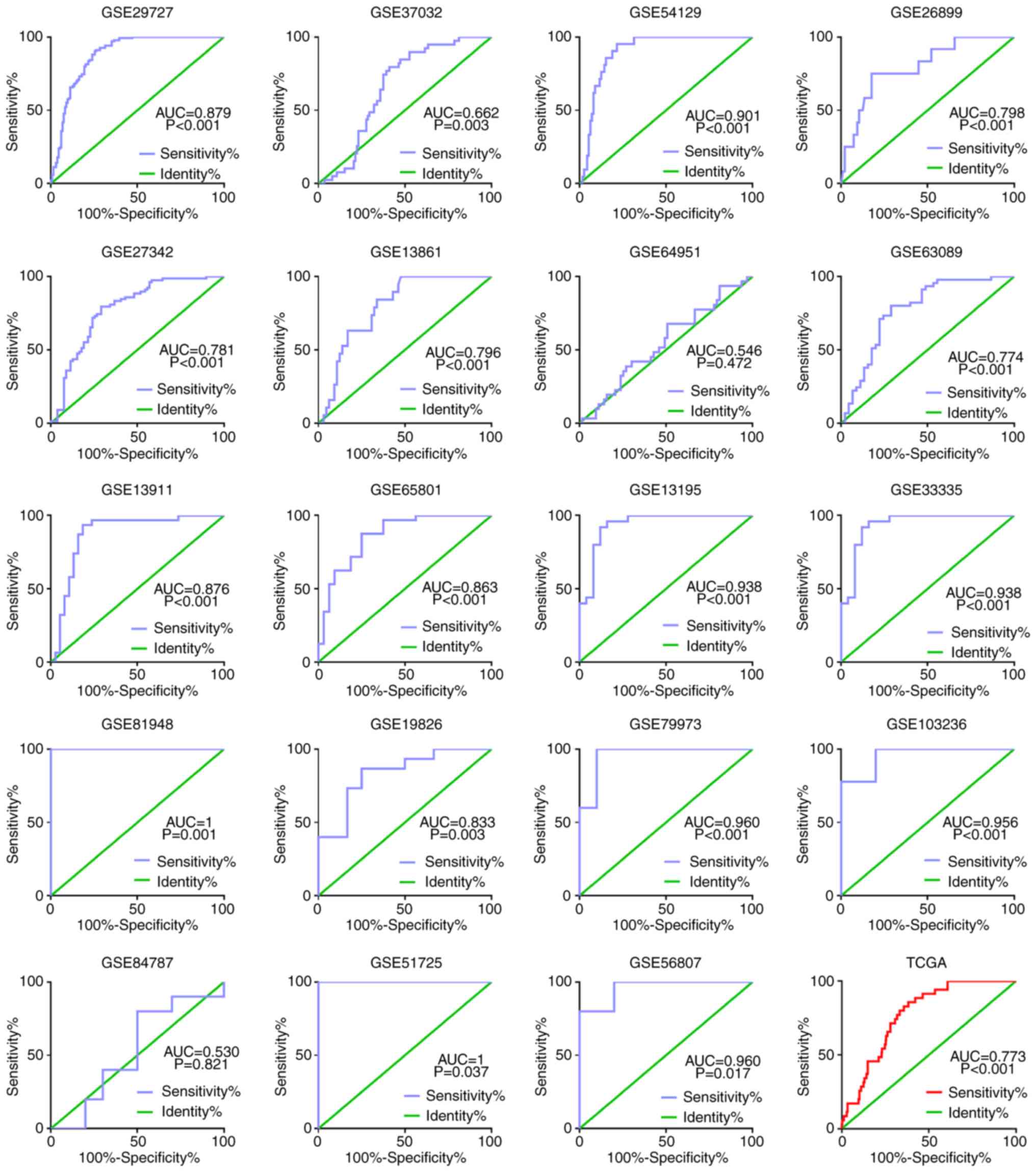
differentiated adenocarcinoma. The receiver operating
characteristic (ROC) curve for FKBP10 based on GEO was presented in
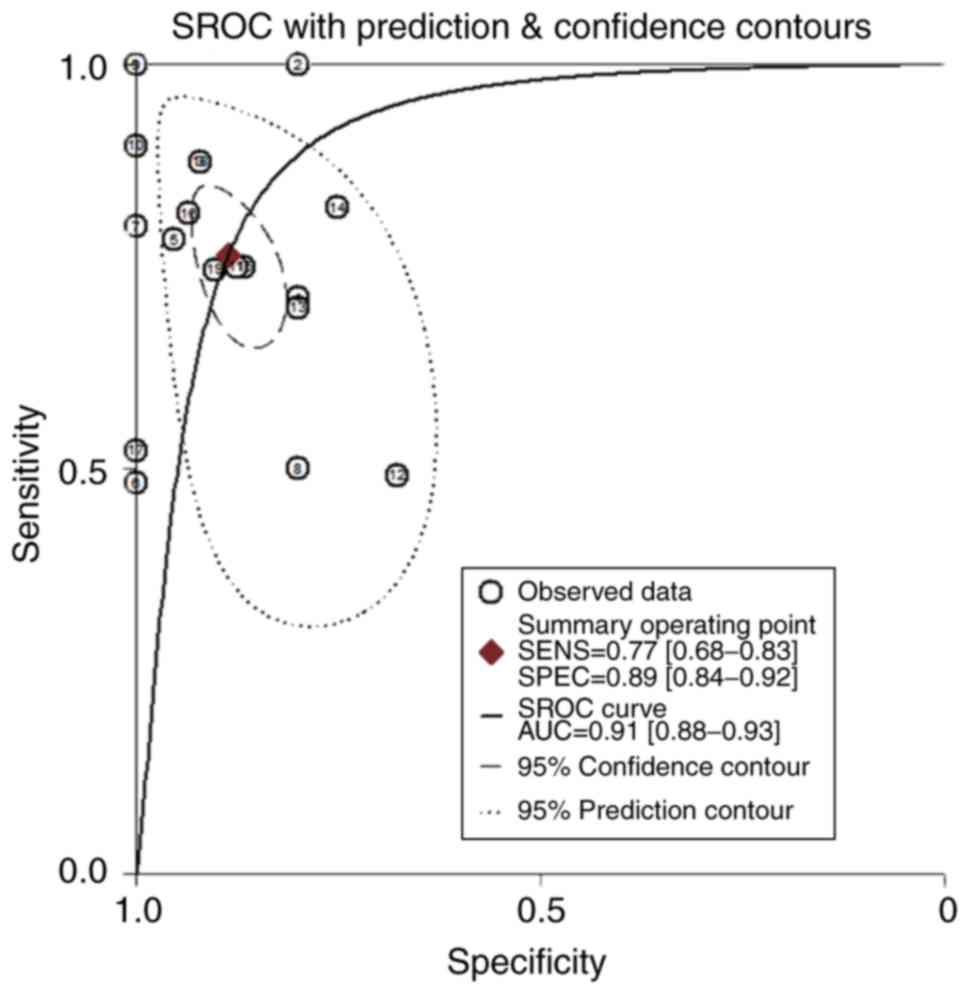
Fig. 5. The area under the curve was
0.774–1 (P=0.001). The ROC curve of FKBP10 was 0.773 in TCGA
(P<0.001; Fig. 5). The combined
microarray data had a sensitivity of 0.77 (0.64–0.86), a
specificity of 0.89 (0.83–0.93), and an area under the combined
SROC curve of 0.91 (0.89–0.94) (Fig.
6). No significant publication bias was observed (P=0.04). The
frequency of FKBP10 alterations in TCGA was 9% (35/393), with 24
amplifications, 6 missense mutations and 5 truncating mutations,
with no alterations in the remaining sections (Fig. 7A). Subsequently, DNA methylation
analysis revealed the methylation level across FKBP10 gene regions
[promoter, enhancer, TSS1500, TSS200, 5′untranslated region (UTR),
first exon, gene body and 3′UTR), as well as CpG islands/CPG island
regions, shelves and shores (Fig.
7B).
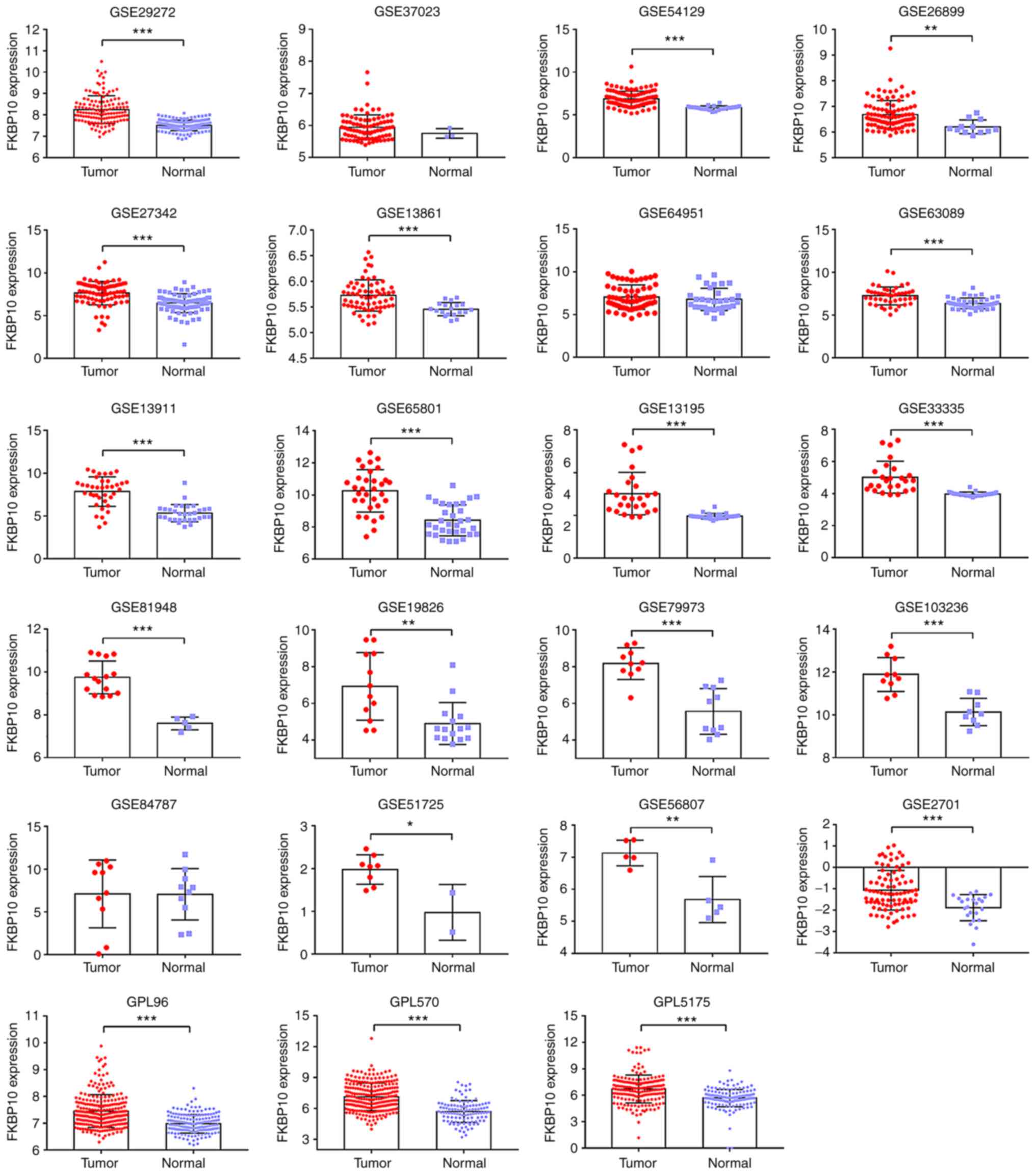 | Figure 3.FKBP10 expression based on Gene
Expression Omnibus microarray data. FKBP10 expression was assessed
by microarray analysis. A total of 17 series (GSE29272, GSE54129,
GSE26899, GSE27342, GSE13861, GSE63089, GSE13911, GSE65801,
GSE13195, GSE33335, GSE89148, GSE19826, GSE79973, GSE103236,
GSE51725, GSE56807, GSE4701) revealed FKBP10 to be significantly
overexpressed in gastric cancer relative to that in normal tissues.
The same platform microarrays underwent batch effects removal and
then processed into our workflow. GPL96 contained GSE29272 and
GSE37023, GPL570 contained GSE54129, GSE64951, GSE13911, GSE19826,
GSE79973 and GSE51725, while GPL5175 had GSE27342, GSE63089,
GSE13195, GSE33335 and GSE56807. *P<0.05; **P<0.01;
***P<0.001. A paired t-test was used to analyze the data of
paired samples, while unpaired t-test was used to analyze unpaired
samples. FKBP10, FK506 binding protein 10. |
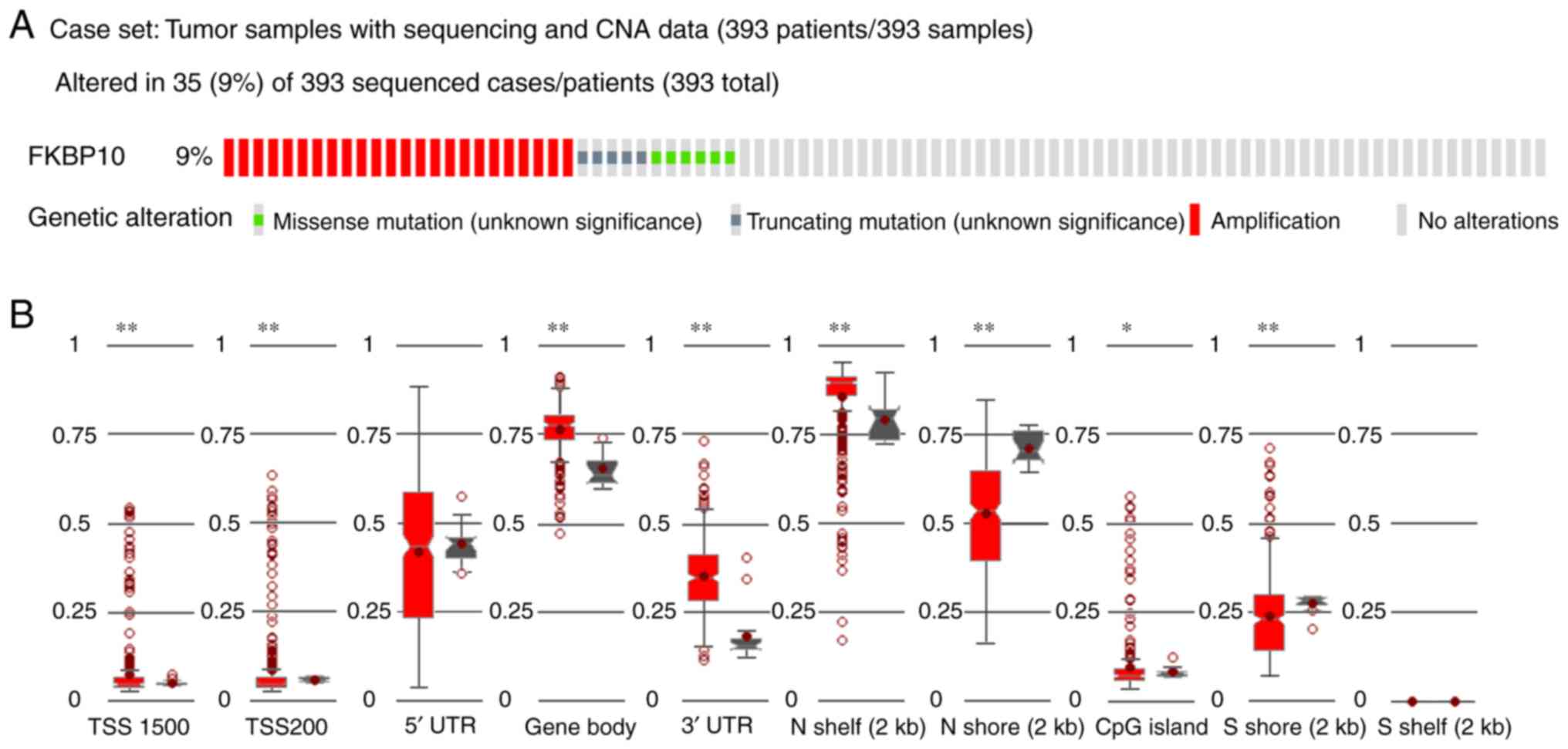 | Figure 7.Genetic alterations and methylation
of FKBP10. (A) FKBP10 was revealed to have more genetic alterations
in The Cancer Genome Atlas cohort. A total of 24 cases had FKBP10
amplifications, 5 had truncating mutations and 6 possessed missense
mutations. (B) The methylation levels of FKBP10 across gene regions
(TSS1500, TSS200, 5′UTR, gene body, 3′UTR) and CpG islands, as well
as shelves and shores in gastric cancer. *P<0.05, **P<0.005.
CNA, copy number alteration; FKBP10, FK506 binding protein 10; UTR,
untranslated region. |
 | Table II.Association between gastric cancer
and FKBP10 expression and clinicopathological features in The
Cancer Genome Atlas. |
Table II.
Association between gastric cancer
and FKBP10 expression and clinicopathological features in The
Cancer Genome Atlas.
| Clinicopathological
parameters | Cases (n) | FKBP10 expression
levels | T | P-value |
|---|
| Age |
| <60
years | 122 | 10.612 | 1.089 | 0.277 |
| ≥60
years | 288 | 10.439 |
|
|
| Sex |
|
Male | 268 | 10.553 | 1.216 | 0.225 |
|
Female | 147 | 10.369 |
|
|
| Stage |
|
I+II | 181 | 10.385 | −1.043 | 0.298 |
|
III+IV | 211 | 10.543 |
|
|
| Grade |
|
G1+G2 | 160 | 10.626 | 1.519 | 0.129 |
| G3 | 255 | 10.400 |
|
|
| T stage |
|
T1+T2 | 110 | 10.499 | 0.177 | 0.860 |
|
T3+T4 | 296 | 10.469 |
|
|
| N stage |
| N1 | 123 | 10.251 | −1.862 | 0.063 |
|
N2+N3 | 273 | 10.550 |
|
|
| M stage |
| M0 | 367 | 10.460 | −1.113 | 0.266 |
| M1 | 27 | 10.786 |
|
|
| Person neoplasm
cancer status |
| Tumor
free | 237 | 10.365 | −2.107 | 0.036a |
| With
tumor | 135 | 10.704 |
|
|
| Recurrence |
| No | 313 | 10.416 | −1.728 | 0.085 |
|
Yes | 102 | 10.706 |
|
|
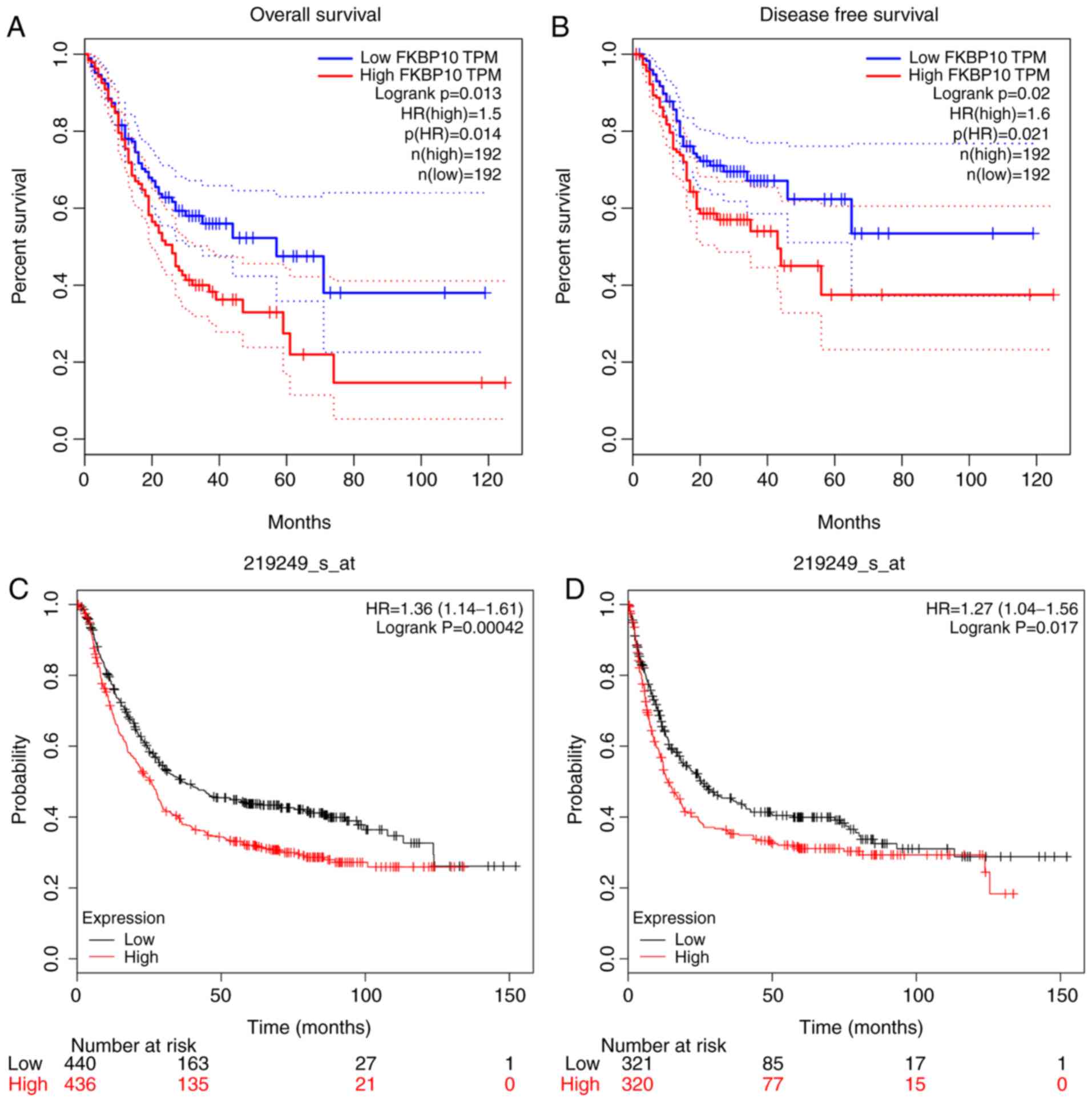
Prognostic value of FKBP10
FKBP10 has potential for predicting the prognosis of
patients with GC. Patients with high FKBP10 expression had a
significantly shorter OS time relative to patients with low
expression FKBP10 (hazard ratio (HR)=1.5; P=0.014) (Fig. 8A). In addition, patients with high
expression of FKBP10 had shorter durations of DFS than those with
low expression (HR=1.6, P=0.021; Fig.
8B). We also verified in the GEO database that patients with
high expression of FKBP10 had significantly shorter OS and DFS
times than patients with low expression (HR=1.36, P<0.001;
HR=1.27, P=0.017) (Fig. 8C and D).
Using univariate cox analysis, we found that FKBP10 expression
levels, age, tumor, node and metastasis (TNM) stage, grade, T stage
and N stage were closely associated with prognosis. Subsequently,
multivariate analysis indicated that FKBP10 expression, age and TNM
staging could be independent prognostic factors for patients with
GC (Table III).
 | Table III.Cox regression model analysis of
overall survival in patients with gastric cancer. |
Table III.
Cox regression model analysis of
overall survival in patients with gastric cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Covariates | HR | 95% confidence
interval | P-value | HR | 95% CI | P-value |
|---|
| FKBP10 expression
level | 1.192 | 1.076–1.319 | 0.001a | 1.159 | 1.042–1.289 | 0.006a |
| Age (<60 vs. ≥60
years) | 1.482 | 1.028–2.136 | 0.035 | 1.627 | 1.117–2.370 | 0.011 |
| Sex (male vs.
female) | 0.772 | 0.546–1.091 | 0.143 |
|
|
|
| T stage (T1-2 vs.
T3-4) | 1.799 | 1.195–2.708 | 0.005a |
|
|
|
| N stage (N0 vs.
N1-3) | 2.096 | 1.389–3.164 |
<0.001a | 2.059 | 1.357–3.122 | 0.001a |
| M stage (M0 vs.
M1) | 1.137 | 0.614–2.103 | 0.683 |
|
|
|
| Histological grade
(G1-2 vs. G3) | 1.398 | 0.997–1.961 | 0.052 |
|
|
|
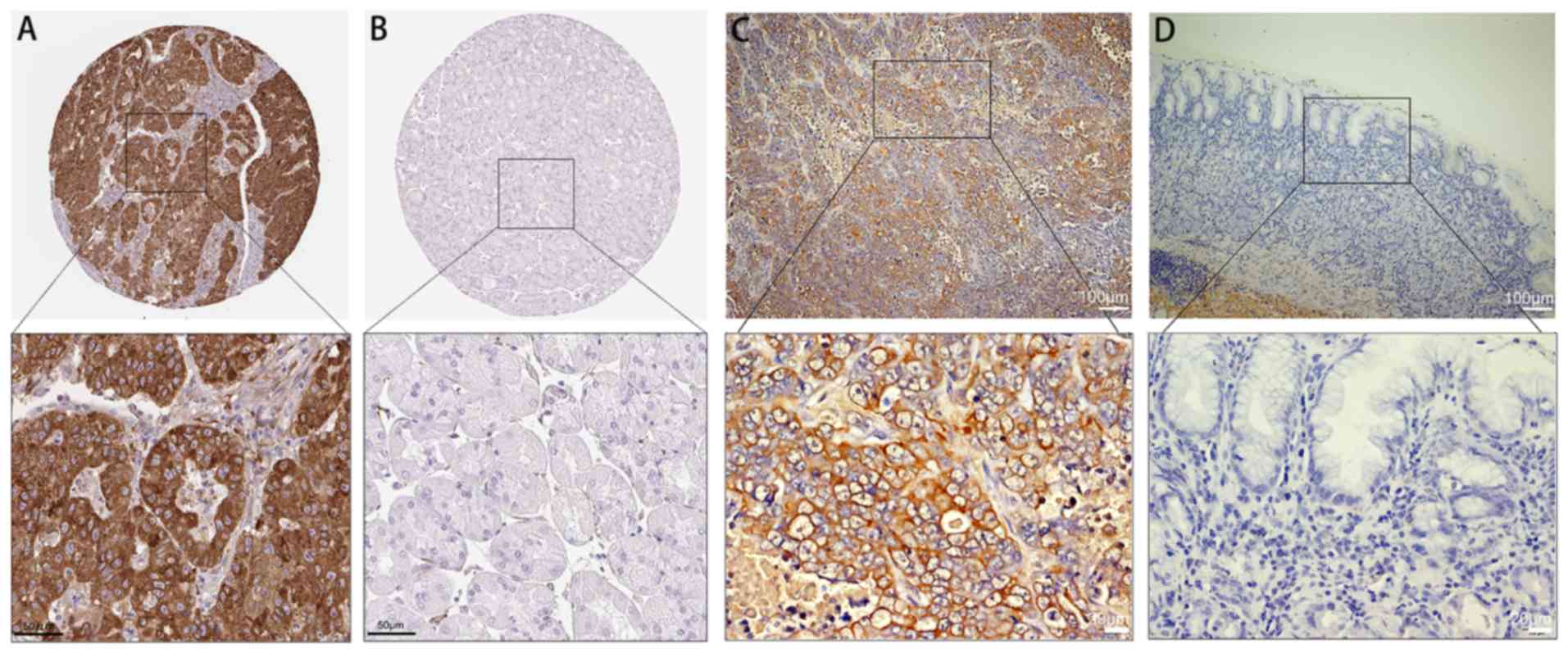
FKBP10 protein expression in GC
We downloaded FKBP10 protein expression data
pertaining to GC from the HPA. We reported that 4/10 GC tissue
samples exhibited positive staining with HPA057021 antibody
(Fig. 9A). Compared with cancer
tissues, upregulated FKBP10 protein expression was not detected in
normal tissues (Fig. 9B). In
addition, we validated 40 pairs of GC tissues and corresponding
adjacent tissues by immunohistochemistry and calculated the IRS
scores. The expression of FKBP10 in GC tissues (IRS=5.6) was
significantly increased than in adjacent tissues (IRS=0.002,
P<0.001; Fig. 9C and D).
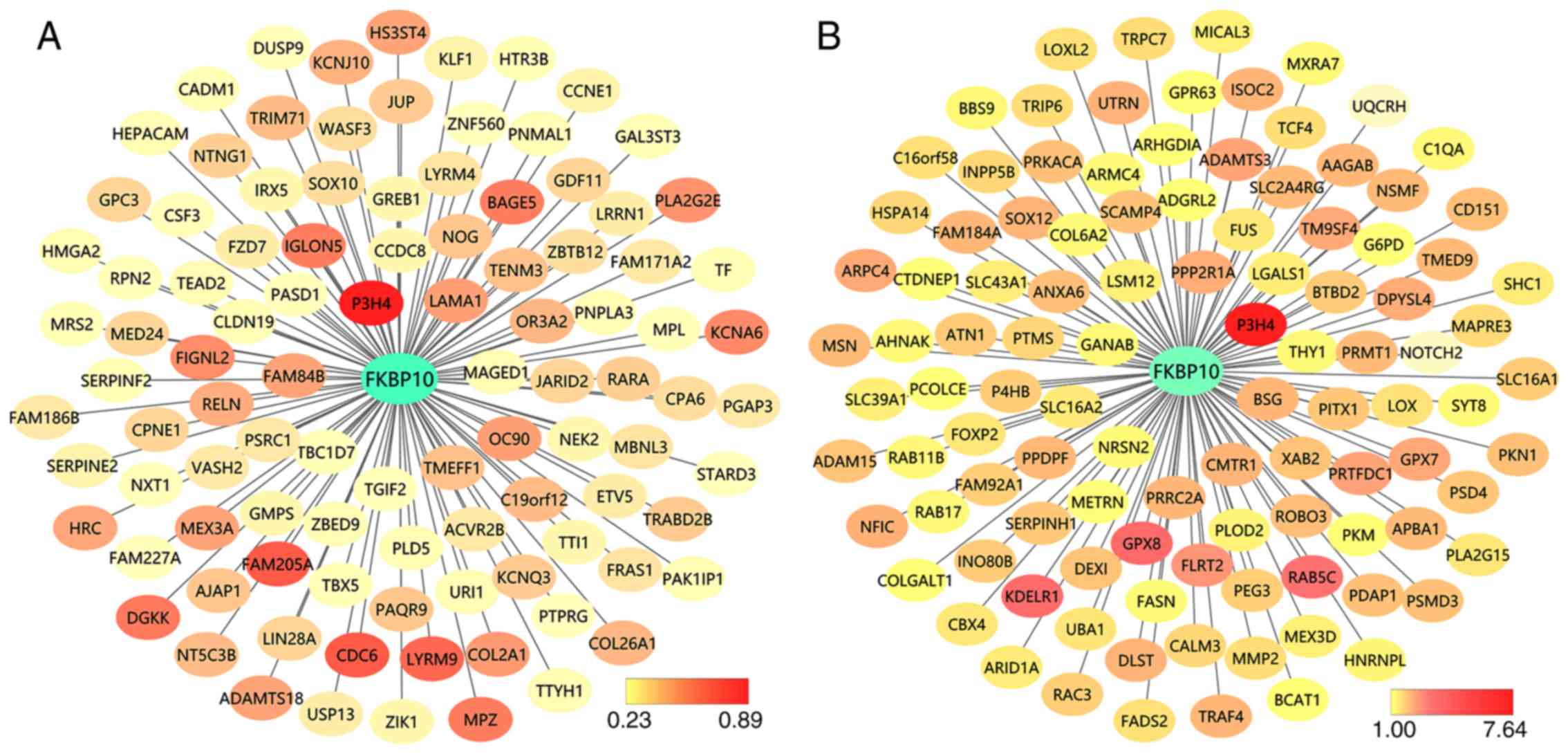
Biological function analysis
To explore the biological function of FKBP10, we
identified and analyzed the proteins that interact with FKBP10 via
PPI analysis. We found that dystonin, leucine- and proline-enriched
proteoglycan 1 (also known as P3H1), keratin 14 and transmembrane
protein 38B could interact with FKBP10 (genes combined score
>0.7) (Fig. 10). We also searched
for genes closely related to FKBP10 expression; a FKBP10
co-expression network of TCGA and GEO data was created and analyzed
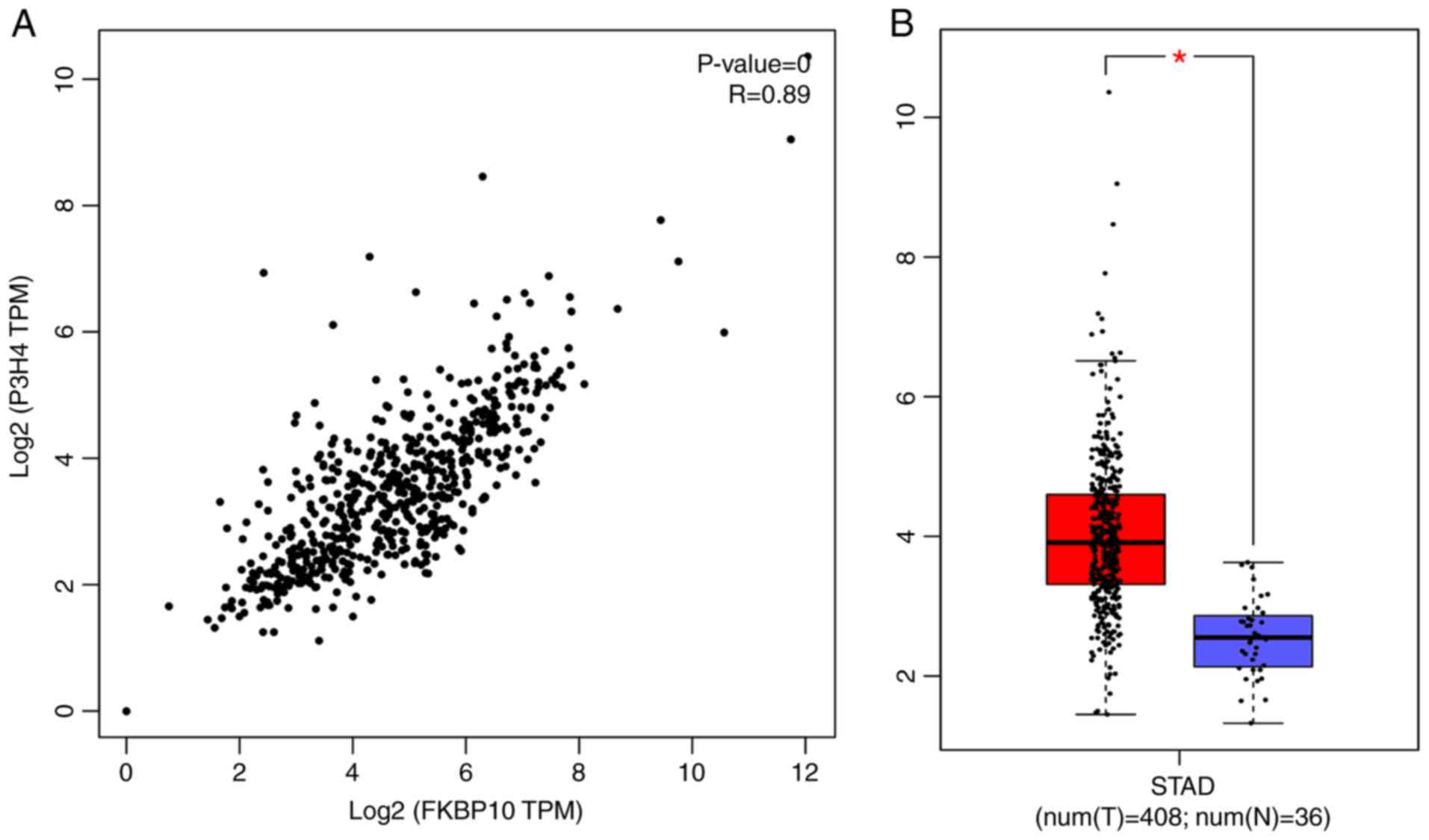
by GEPIA and Coexpedia, respectively (Fig. 11). Additionally, prolyl 3-hydroxylase
family member 4 (P3H4, also known as LEPREL4) was identified as the
most closely related gene in from TCGA and GEO network analyses
(R=0.89, P<0.001; Fig. 12). We
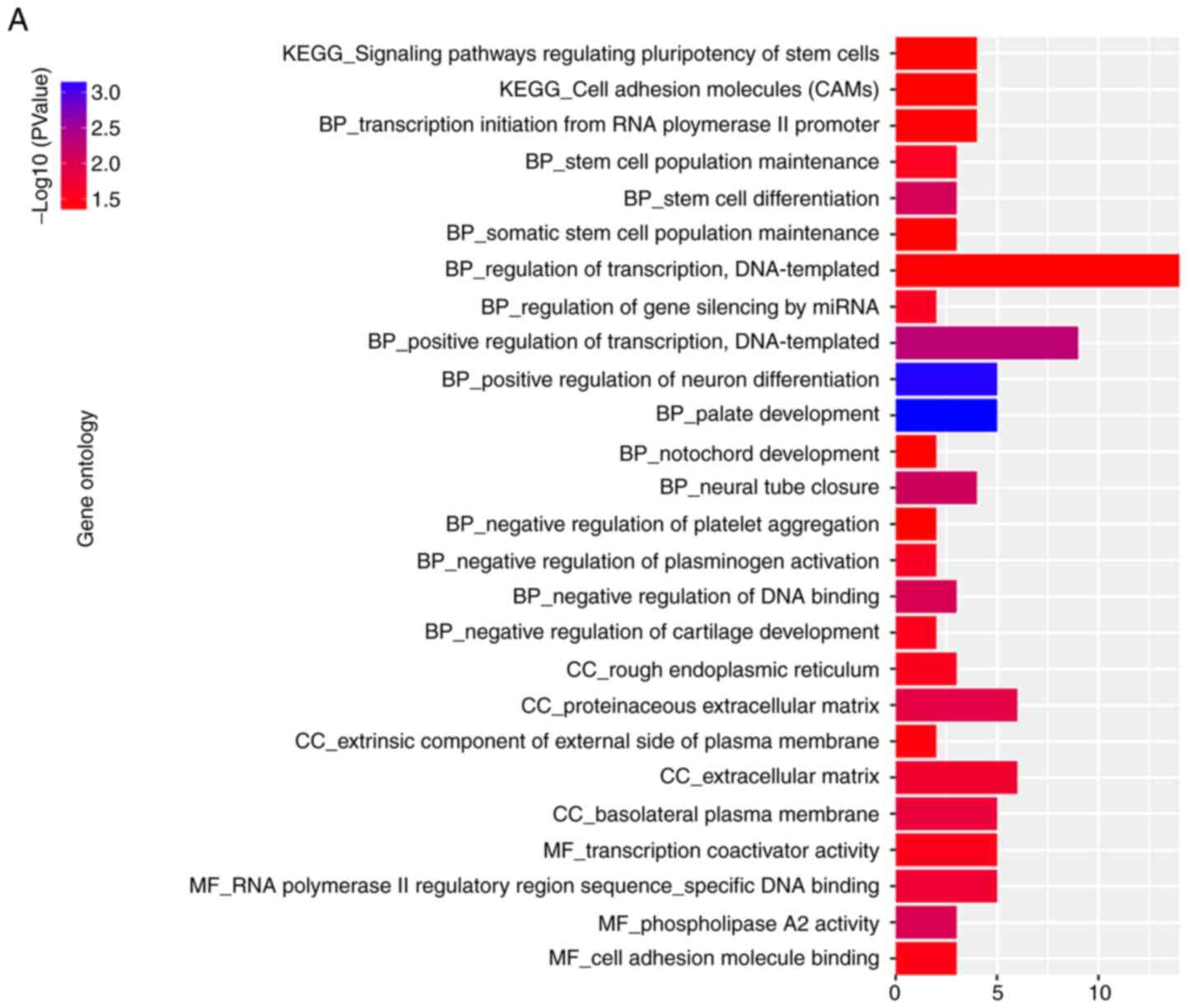
also ran GO and KEGG pathway analyses of co-expressed genes. The
results of KEGG enrichment showed that the most significantly
enriched pathways were regulating the ‘pluripotency of stem cells’,
‘cell adhesion molecules’, ‘vasopressin-regulated water
reabsorption’, ‘ras signaling’, ‘lysine degradation’, ‘insulin
signaling’, ‘glutathione metabolism’, ‘glucagon signaling’ and
‘estrogen signaling pathway’ (Fig.
13).
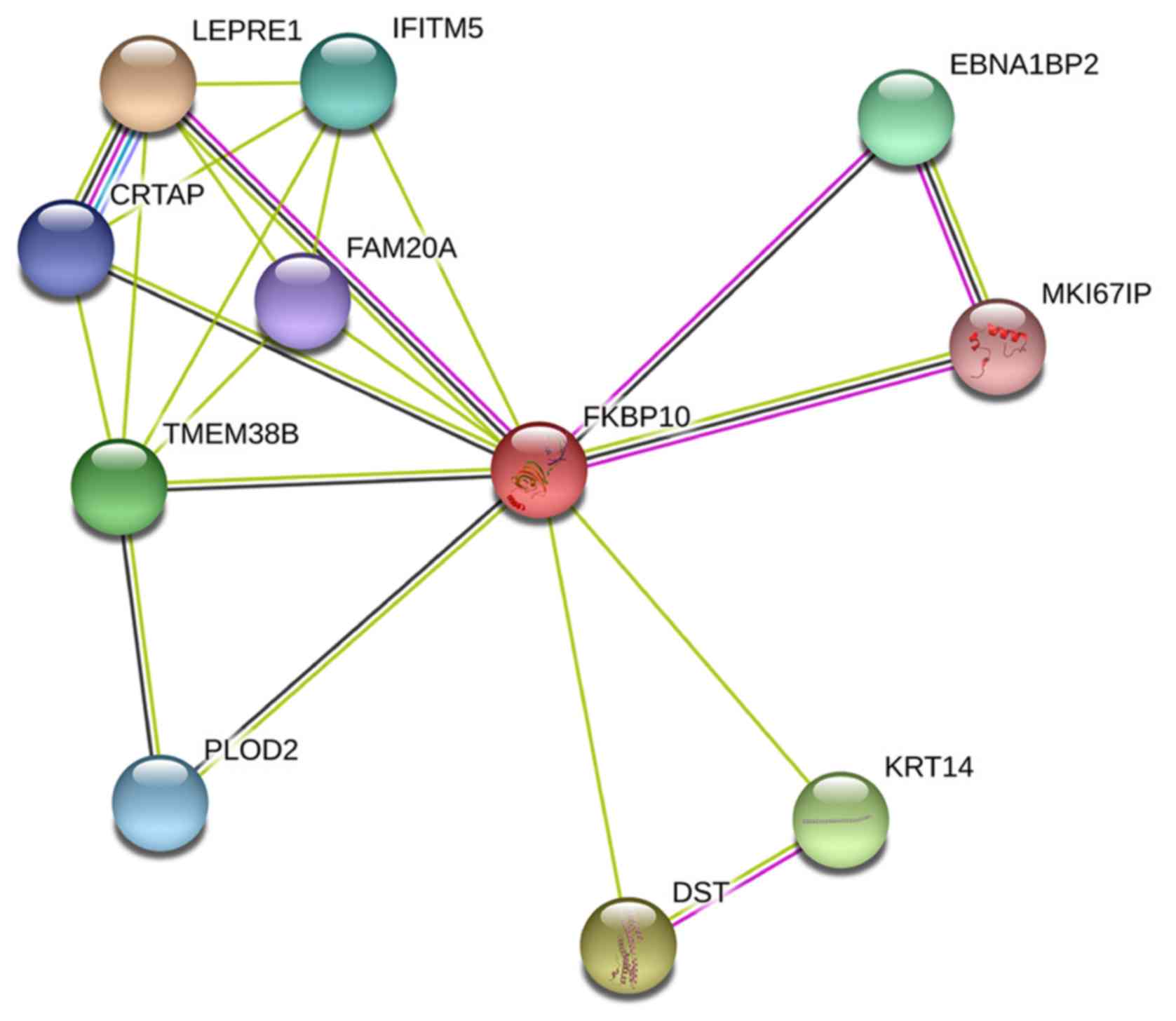 | Figure 10.Protein-protein interaction analysis
of FKBP10. FKBP10 is likely to associate with DST, CRTAP (P3H5),
LEPRE1 (P3H1), KRT14 and TMEM38B. An interaction score of 0.4 was
set as cut-off value. DST, dystonin; CRTAP, cartilage associated
protein; EBNA1BP2, EBNA1 binding protein 2; FAM20A, family with
sequence similarity 20, member A; FKBP10, FK506 binding protein 10;
IFITM5, interferon-induced transmembrane protein 5; LEPRE1,
leprecan; KRT14, keratin 14; PLOD2, procollagen-lysine,
2-oxoglutarate 5-dioxygenase 2; TMEM38B, transmembrane protein
38B. |
Discussion
To the best of our knowledge, the present study is
the first to comprehensively investigate the characterization of
FKBP10 in GC. We initially found that FKBP10 is highly expressed in
GC using the GEO and TCGA databases. Subsequently, we verified that
the protein expression levels of FKBP10 were consistent with the
data of GEO and TCGA by immunohistochemistry. In addition, the
expression of FKBP10 was determined to be closely associated with
clinical prognosis. It was found in the TCGA and GEO datasets that
patients with GC and high expression of FKBP10 had lower DFS and OS
times than those with low expression. In addition, multivariate COX
analysis demonstrated that FKBP10 was an independent prognostic
factor for GC. Collectively, the results of the present study
suggests that FKBP10 may be a key target gene involved in the
development of GC.
As an endoplasmic reticulum localization protein,
FKBP65 binds to tropoelastin throughout the secretory process
(32). Investigations into FKBP10
have primarily focused on pulmonary fibrosis and osteochondrosis;
FKBP10 mutations has been linked to the onset of many diseases
(33,34). As a connective tissue disease, Bruck
syndrome is mainly characterized by the loss of endopeptide lysine
hydroxylation at the molecular level, leading to the reduction of
collagen pyrimidine cross-linking (35). The literature indicates that FKBP65
crosslinks with pyridine by mediating the dimerization of LH2
(35–37). FKBP10 expression was determined to be
upregulated in idiopathic pulmonary fibrosis and bleomycin-induced
lung fibrosis (12). Importantly, the
loss of FKBP10 expression significantly suppressed collagen
secretion by primary human lung fibroblasts (12). Downregulated expression of FKBP10
leads to decreased collagen type I a 1chain mRNA levels, resulting
in liver fibrosis and collagen accumulation (38).
In recent years, the relationship between FKBP10 and
cancer has been investigated. The main function of FKBP10 is to
control folding, trafficking and secretion of protein during the
production of extracellular matrix proteins (32). FKBP10 is highly expressed in melanoma
and colorectal cancer (22,39). Downregulated FKBP10 can suppress tumor
growth in KRAS-driven lung tumors (18). Decreasing the expression of FKBP10 can
inhibit the proliferation and migration of renal cancer cells,
affecting the cell cycle (21). The
transcription factor ETS variant 1 targets FKBP10 to regulate the
invasion and migration of prostate cancer cells (19,40).
However, in ovarian cancer (16,17), the
expression profile of FKBP10 opposed that to other tumors; the
mechanism of action by which FKBP10 may be involved differs, yet
further investigation is required. In addition, the expression and
function of FKBP10 has not yet been reported in GC.
According to GO analysis, FKBP10 significantly
correlated with protein and collagen production. KEGG pathway
enrichment analysis showed that FKBP10 may be related to cell
migration, particularly via cell adhesion molecules and
ECM-receptor interaction pathways. These results are similar to
those of Romano et al (32).
FKBP10 protein localizes to the endoplasmic reticulum (ER) and acts
as a molecular chaperone (18). We
reported that P3H4 was the most commonly expressed gene with
FKBP10. The data indicated that P3H4 acts as part of an ER complex
with prolyl 3-hydroxylase 3, which affects collagen lysine
hydroxylation (41). This suggests
that FKBP10 may interact with proline 3-hydroxylase to affect
collagen synthesis. Although our study found that the expression of
FKBP10 could be related to the prognosis of patients with GC, how
FKBP10 regulates the progression of GC and how this can be applied
for the targeted treatment of GC requires further
investigation.
Of note, there were limitations to the present
study. First, survival analysis was conducted using GEPIA and
Kaplan Meier Plotter tools. The survival time of DFS should be
shorter than OS. However, the result of our analysis showed the
reverse. Among the TCGA and GEO data, some patients are still being
followed up, so the data are incomplete and still required further
sample verification. We aim to verify the relationship between
FKBP10 and patient prognosis using our own samples in the future.
At the protein level, we only verified this relationship using
immunohistochemistry; however, further validation using PCR and
western blotting will be conducted in the future. In addition, the
biological role of FKBP10 in gastric cancer cells should be
investigated. Regarding the biological mechanism of FKBP10 in
cancer, we have only proposed some options, yet the oncogenic
function of FKBP10 should be further confirmed by in vitro
and in vivo experiments.
In summary, the present study reported that FKBP10
is significantly elevated in GC; thus; FKBP10 could be considered
as a potential novel therapeutic target for the treatment of this
disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Research
and Development Program of Science and Technology Department of
Guangxi (grant no. 2017AA45153), the Scientific Research and
Technology-development Program of Guangxi (grant no. 1598011-4) and
Innovation Project of Guangxi Graduate Education (grant no.
YCBZ2019043).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JQC and GC designed and directed the project. LL and
KZ processed and analyzed data; LL wrote and revised the
manuscript. XGQ and JHZ performed the immunohistochemistry
experiment. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Guangxi Medical
University and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Correa P: Gastric cancer: Overview.
Gastroenterol Clin North Am. 42:211–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsukanov VV, Butorin NN, Maady AS,
Shtygasheva OV, Amelchugova OS, Tonkikh JL, Fassan M and Rugge M:
Helicobacter pylori infection, intestinal metaplasia, and
gastric cancer risk in Eastern Siberia. Helicobacter. 16:107–112.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39:10104283177146262017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sitarz R, Skierucha M, Mielko J, Offerhaus
GJA, Maciejewski R and Polkowski WP: Gastric cancer: Epidemiology,
prevention, classification, and treatment. Cancer Manag Res.
10:239–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Japanese Gastric Cancer Association:
Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric
Cancer. 20:1–19. 2017. View Article : Google Scholar
|
|
7
|
Chon SH, Berlth F, Plum PS, Herbold T,
Alakus H, Kleinert R, Moenig SP, Bruns CJ, Hoelscher AH and Meyer
HJ: Gastric cancer treatment in the world: Germany. Transl
Gastroenterol Hepatol. 2:532017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang CB, Hong Y, Dhe-Paganon S and Yoon
HS: FKBP family proteins: Immunophilins with versatile biological
functions. Neurosignals. 16:318–325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coss MC, Winterstein D, Sowder RC II and
Simek SL: Molecular cloning, DNA sequence analysis, and biochemical
characterization of a novel 65-kDa FK506-binding protein (FKBP65).
J Biol Chem. 270:29336–29341. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patterson CE, Schaub T, Coleman EJ and
Davis EC: Developmental regulation of FKBP65. An ER-localized
extracellular matrix binding-protein. Mol Biol Cell. 11:3925–3935.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishikawa Y, Vranka J, Wirz J, Nagata K and
Bachinger HP: The rough endoplasmic reticulum-resident
FK506-binding protein FKBP65 is a molecular chaperone that
interacts with collagens. J Biol Chem. 283:31584–31590. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Staab-Weijnitz CA, Fernandez IE, Knüppel
L, Maul J, Heinzelmann K, Juan-Guardela BM, Hennen E, Preissler G,
Winter H, Neurohr C, et al: FK506-binding protein 10, a potential
novel drug target for idiopathic pulmonary fibrosis. Am J Respir
Crit Care Med. 192:455–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knüppel L, Heinzelmann K, Lindner M, Hatz
R, Behr J, Eickelberg O and Staab-Weijnitz CA: FK506-binding
protein 10 (FKBP10) regulates lung fibroblast migration via
collagen VI synthesis. Respir Res. 19:672018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Solassol J, Mange A and Maudelonde T: FKBP
family proteins as promising new biomarkers for cancer. Curr Opin
Pharmacol. 11:320–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yao YL, Liang YC, Huang HH and Yang WM:
FKBPs in chromatin modification and cancer. Curr Opin Pharmacol.
11:301–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Quinn MC, Wojnarowicz PM, Pickett A,
Provencher DM, Mes-Masson AM, Davis EC and Tonin PN: FKBP10/FKBP65
expression in high-grade ovarian serous carcinoma and its
association with patient outcome. Int J Oncol. 42:912–920. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Henriksen R, Sørensen FB, Ørntoft TF and
Birkenkamp-Demtroder K: Expression of FK506 binding protein 65
(FKBP65) is decreased in epithelial ovarian cancer cells compared
to benign tumor cells and to ovarian epithelium. Tumour Biol.
32:671–676. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramadori G, Konstantinidou G,
Venkateswaran N, Biscotti T, Morlock L, Galié M, Williams NS,
Luchetti M, Santinelli A, Scaglioni PP and Coppari R: Diet-induced
unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell
Metab. 21:117–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paulo P, Ribeiro FR, Santos J, Mesquita D,
Almeida M, Barros-Silva JD, Itkonen H, Henrique R, Jerónimo C,
Sveen A, et al: Molecular subtyping of primary prostate cancer
reveals specific and shared target genes of different ETS
rearrangements. Neoplasia. 14:600–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun Z, Dong J, Zhang S, Hu Z, Cheng K, Li
K, Xu B, Ye M, Nie Y, Fan D and Zou H: Identification of
chemoresistance-related cell-surface glycoproteins in leukemia
cells and functional validation of candidate glycoproteins. J
Proteome Res. 13:1593–1601. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ge Y, Xu A, Zhang M, Xiong H, Fang L,
Zhang X, Liu C and Wu S: FK506 binding protein 10 is overexpressed
and promotes renal cell carcinoma. Urol Int. 98:169–176. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Olesen SH, Christensen LL, Sørensen FB,
Cabezón T, Laurberg S, Orntoft TF and Birkenkamp-Demtröder K: Human
FK506 binding protein 65 is associated with colorectal cancer. Mol
Cell Proteomics. 4:534–544. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu JG, Wang JJ, Jiang X, Lan JP, He XJ,
Wang HJ, Ma YY, Xia YJ, Ru GQ, Ma J, et al: MiR-125b promotes cell
migration and invasion by targeting PPP1CA-Rb signal pathways in
gastric cancer, resulting in a poor prognosis. Gastric Cancer.
18:729–739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Imaoka H, Toiyama Y, Okigami M, Yasuda H,
Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y and Kusunoki M:
Circulating microRNA-203 predicts metastases, early recurrence, and
poor prognosis in human gastric cancer. Gastric Cancer. 19:744–753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang M, Shi B, Liu J, He L, Yi G, Zhou L,
Yu G and Zhou X: Downregulation of miR203 induces overexpression of
PIK3CA and predicts poor prognosis of gastric cancer patients. Drug
Des Devel Ther. 9:3607–3616. 2015.PubMed/NCBI
|
|
26
|
Leek JT, Johnson WE, Parker HS, Fertig EJ,
Jaffe AE, Storey JD, Zhang Y and Torres LC: sva: Surrogate variable
analysis. R package version 3.32.1. 2019.
|
|
27
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:D856–D861.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang S, Kim CY, Hwang S, Kim E, Kim H,
Shim H and Lee I: COEXPEDIA: Exploring biomedical hypotheses via
co-expressions associated with medical subject headings (MeSH).
Nucleic Acids Res. 45:D389–D396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Romano S, D'Angelillo A and Romano MF:
Pleiotropic roles in cancer biology for multifaceted proteins
FKBPs. Biochim Biophys Acta. 1850:2061–2068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Christiansen HE, Schwarze U, Pyott SM,
AlSwaid A, Al Balwi M, Alrasheed S, Pepin MG, Weis MA, Eyre DR and
Byers PH: Homozygosity for a missense mutation in SERPINH1, which
encodes the collagen chaperone protein HSP47, results in severe
recessive osteogenesis imperfecta. Am J Hum Genet. 86:389–398.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Essawi O, Symoens S, Fannana M, Darwish M,
Farraj M, Willaert A, Essawi T, Callewaert B, De Paepe A, Malfait F
and Coucke PJ: Genetic analysis of osteogenesis imperfecta in the
Palestinian population: Molecular screening of 49 affected
families. Mol Genet Genomic Med. 6:15–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Y, Terajima M, Banerjee P, Guo H, Liu
X, Yu J, Yamauchi M and Kurie JM: FKBP65-dependent peptidyl-prolyl
isomerase activity potentiates the lysyl hydroxylase 2-driven
collagen cross-link switch. Sci Rep. 7:460212017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gjaltema RA, van der Stoel MM, Boersema M
and Bank RA: Disentangling mechanisms involved in collagen
pyridinoline cross-linking: The immunophilin FKBP65 is critical for
dimerization of lysyl hydroxylase 2. Proc Natl Acad Sci USA.
113:7142–7147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Duran I, Martin JH, Weis MA, Krejci P,
Konik P, Li B, Alanay Y, Lietman C, Lee B, Eyre D, et al: A
chaperone complex formed by HSP47, FKBP65, and BiP modulates
telopeptide Lysyl hydroxylation of type I procollagen. J Bone Miner
Res. 32:1309–1319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vollmann EH, Cao L, Amatucci A, Reynolds
T, Hamann S, Dalkilic-Liddle I, Cameron TO, Hossbach M, Kauffman
KJ, Mir FF, et al: Identification of novel fibrosis modifiers by in
vivo siRNA silencing. Mol Ther Nucleic Acids. 7:314–323. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hagedorn M, Siegfried G, Hooks KB and
Khatib AM: Integration of zebrafish fin regeneration genes with
expression data of human tumors in silico uncovers potential novel
melanoma markers. Oncotarget. 7:71567–71579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rahim S, Minas T, Hong SH, Justvig S,
Celik H, Kont YS, Han J, Kallarakal AT, Kong Y, Rudek MA, et al: A
small molecule inhibitor of ETV1, YK-4-279, prevents prostate
cancer growth and metastasis in a mouse xenograft model. PLoS One.
9:e1142602014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heard ME, Besio R, Weis M, Rai J, Hudson
DM, Dimori M, Zimmerman SM, Kamykowski JA, Hogue WR, Swain FL, et
al: Sc65-Null mice provide evidence for a novel endoplasmic
reticulum complex regulating collagen Lysyl hydroxylation. PLoS
Genet. 12:e10060022016. View Article : Google Scholar : PubMed/NCBI
|