Introduction
Osteoporosis, which is characterized by low bone
mass and microarchitectural deterioration of the bone tissue, is a
significant public health issue (1-3).
Therefore, it has received extensive attention. With the global
trend of population aging, the incidence of osteoporosis is
increasing (1,2). The prevalence of osteoporosis in the
elderly population in China is ~36% (4). Fractures, which can lead to chronic
pain, disability, depression and other associated diseases, are the
most serious complication of osteoporosis (3,5).
Therefore, identifying novel biomarkers for predicting individuals
at risk of osteoporosis is of great importance for the early
intervention to prevent fractures.
Human bone metabolism is a complex process that
involves bone absorption mediated by osteoclasts and
osteoblast-mediated bone formation (6). Bone absorption and formation are
stable under physiological conditions. However, disruption of this
balance, which can lead to impaired osteoblast and osteoclast
activities, can result in osteoporosis (7).
Bone marrow mesenchymal stem cells (BMSCs), which
were first isolated from bone marrow and named by Friedenstein
et al (8) in 1976, are
closely associated with the occurrence and development of
osteoporosis. Abnormal BMSC differentiation, proliferation and
senescence are the most common causes of osteoporosis (9). BMSCs are normally differentiate into
osteoblasts, chondrocytes and adipocytes, while their
differentiation can be affected by several factors, including age
(10) and intracellular biological
factors (11). When only a few
BMSCs differentiate into osteoblasts, which in turn results in
reduced bone formation, osteoporosis can occur (11,12).
Furthermore, it has been reported that BMSC senescence is regulated
by several factors, such as DNA damage, oxidative stress and
mitogenic signals (13,14). Therefore, emerging evidence has
suggested that BMSC senescence plays a significant role in the
occurrence of osteoporosis (15,16).
Although circular RNAs (circRNAs/circ) are
classified as non-coding RNAs, several of them exert protein-coding
potential (17,18). Owing to their loop structure,
without 3' and 5' ends, circRNAs are more stable compared with
linear RNAs and less prone to degradation by exonucleases (4,19). In
addition, previous studies indicated that circRNAs, possessing
numerous miRNA binding sites, could act as competing endogenous
RNAs (ceRNAs) via sponging microRNAs (miRNAs), thus playing an
essential regulatory role in the progression of several diseases,
including osteoporosis (19-21).
For example, a study revealed that circ_0001052 was involved in the
regulation of BMSC differentiation and the pathogenesis of
osteoporosis via acting as ceRNA through sponging miR-124-3p
(22). In addition, circRNA_0048211
could protect against postmenopausal osteoporosis via targeting
miRNA-93-5p (23). Furthermore, an
increasing number of evidence has indicated that circRNA-miRNA-mRNA
networks serve a regulatory role in osteoporosis (21,23). A
previous study from our laboratory demonstrated that
hsa_circ_0122913 was upregulated in the peripheral blood of
patients with senile osteoporotic vertebral compression fracture
(24). However, the effect of
hsa_circ_0122913 on the proliferation and osteogenic
differentiation of BMSCs has not been previously investigated.
Furthermore, a previous study also showed that icariin exerted
beneficial effects on the skeletal system in vitro,
including the enhancement of the osteogenic differentiation and
proliferation of BMSCs (25).
However, whether circRNAs are involved in the mechanisms underlying
the protective effects of icariin remains elusive. Therefore, the
present study aimed to explore the role of hsa_circ_0122913 in the
proliferation and osteogenic differentiation of BMSCs, and whether
it could be involved in the protective effects of icariin on the
skeletal system. Overall, the results of the current study could
provide novel insights into the pathophysiological mechanisms
underlying the onset of osteoporosis and could potentially pave the
way for the development of therapeutic interventions.
Materials and methods
Cell culture and treatment
The human BMSC cell line (cat. no. HUXMA-01001,
passage 2 upon receipt) was obtained from Cyagen Biosciences Inc.
and cultured in BMSCs growth medium (Cyagen Biosciences, Inc.) at
37˚C in an incubator with 5% CO2. Cells were then
treated with 1x10-6 mol/l icariin (Shanghai Yuanye
Bio-Technology, Co., Ltd.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Following culture and treatment with the indicated
compounds, BMSCs were collected, and total RNA was then isolated
using the Total RNA Rapid Extraction Kit (cat. no. GK3016; Generay
Biotech Co., Ltd.). Subsequently, total RNA was reverse transcribed
into cDNA using the HiScript II Q RT SuperMix (cat. no. R222-01;
Vazyme Biotech Co., Ltd.) strictly according to the manufacturer's
protocols. qPCR was performed using the ChamQ SYBR Color qPCR
Master Mix Kit (cat. no. Q411-02; Vazyme Biotech Co., Ltd.) on the
CFX Connect Real-Time PCR System (Bio-Rad Laboratories, Inc.). The
thermocycling protocol included an initial denaturation step at
95.0˚C for 30 sec followed by 40 cycles consisting of 95.0˚C for 10
sec and 60.0˚C for 30 sec, with a final melt curve analysis
performed from 70˚C to 95˚C using 0.5˚C increments and 5-sec holds
at each temperature step. The expression levels of target genes
were normalized to β-actin as the housekeeping gene, and relative
gene expression was calculated using the 2-ΔΔCq method
(26). The primer sequences used
for PCR are listed in Table I.
 | Table ISequences of primers used for reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene name | Primer sequence
(5'-3') |
|---|
| Actin | F:
TGACGTGGACATCCGCAAAG |
| | R:
CTGGAAGGTGGACAGCGAG |
| Osteopontin | F:
GAAGTTTCGCAGACCTGACAT |
| | R:
GTATGCACCATTCAACTCCTCG |
| Osterix | F:
GTGGAACAGGAGTGGAGCTG |
| | R:
AGGCAGATGGAGAGAGCTGG |
| Runt-related
transcription factor 2 | F:
CGCCTCACAAACAACCACAG |
| | R:
ACTGCTTGCAGCCTTAAATGAC |
| Defective in cullin
neddylation 1 domain containing 1 | F:
TGCCTACTGGAACTTAGTGCT |
| | R:
CTGCAATCATCGTACTGAAGTCT |
Alizarin red staining
BMSCs at a density of 6x104 cells/well
were seeded into 12-well culture plates and cultured for 7 days.
Subsequently, the medium was discarded and BMSCs were washed twice
with PBS. Cells were then fixed with 2 ml 4% formaldehyde at room
temperature for 30 min, followed by washing twice with PBS.
Subsequently, BMSCs were stained with alizarin red solution (cat.
no. G3280; Beijing Solarbio Science & Technology Co., Ltd.) for
5 min. Following washing with PBS for two times, images of the
stained cells were captured under a fluorescent inverted microscope
(cat. no. IX73; Olympus Corporation) at a magnification of x200 and
a scale bar of 50 µm.
Osteogenic differentiation assay
The differentiation potential of osteocytes was
verified using a differentiation assay. Briefly, BMSCs at a density
of 6x104 cells/well were seeded into 12-well culture
plates and differentiated in osteogenic medium containing alizarin
red staining solution (Cyagen Biosciences, Inc.), according to the
manufacturer's instructions.
Western blot analysis
Total proteins were extracted from BMSCs using a
Cell Lysis Buffer for Western and IP (Beyotime Institute of
Biotechnology) supplemented with 1 mM phenyl-methyl-sulfonyl
fluoride (Beyotime Institute of Biotechnology). Subsequently, the
protein concentration was determined using a bicinchoninic protein
assay kit (Beyotime Institute of Biotechnology). The protein
extracts were then separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, with 30 µg of protein
loaded per lane, followed by transferring onto polyvinylidene
fluoride membranes (MilliporeSigma). Following blocking with 5%
fat-free milk at room temperature for 1 h, the membranes were
incubated with primary antibodies at 4˚C overnight, followed by
washing with Tris-buffer saline containing 0.05% Tween-20 (TBST)
for three times for 10 min each. Subsequently, the membranes were
incubated with the corresponding secondary antibodies at room
temperature for 1 h and washed again with TBST. The protein bands
were visualized using an enhanced chemiluminescence kit (Beyotime
Institute of Biotechnology) and images were captured using an
automatic chemiluminescence imaging analysis system (Bio-Rad
Laboratories, Inc.). The densitometric analysis of the protein
bands was performed using ImageJ software (version 1.4.3.67;
National Institutes of Health). The primary antibodies used against
defective in cullin neddylation 1 domain containing 1 (DCUN1D1;
cat. no. ab181233; 1:5,000), osterix (OSX; cat. no. ab209484;
1:1,000), runt-related transcription factor 2 (Runx2; cat. no.
ab236639; 1:1,000) and osteopontin (OPN; cat. no. ab214050;
1:1,000) were all purchased from Abcam. The anti-β-actin primary
antibody (cat. no. ab008; 1:5,000) was obtained from Hangzhou
MultiSciences (Lianke) Biotech Co., Ltd. The corresponding
peroxidase-conjugated secondary antibodies, including goat
anti-mouse IgG (cat. no. GAM007, 1:5,000) and goat anti-rabbit IgG
(cat. no. GAR0072, 1:5,000), were also obtained from Hangzhou
MultiSciences (Lianke) Biotech Co., Ltd.
Annexin V-APC/7-AAD assay
Cell apoptosis was assessed using an Annexin
V-APC/7-AAD apoptosis kit [Hangzhou MultiSciences (Lianke) Biotech
Co., Ltd.]. Briefly, cells were collected after trypsinization
without EDTA at 48 and 72 h, and 7 days of cell proliferation,
followed by washing twice with PBS. The cells were resuspended in
binding buffer with Annexin V-APC (5 µl/tube) and 7-AAD (10
µl/tube) and incubated in the dark at 4˚C for 5 min. Finally, cells
were analyzed using a BD Accuri™ C6 Plus flow cytometer
(BD Biosciences) and the corresponding supporting BD Accuri C6
software.
Cell transfection
For silencing experiments, viral particles
encompassing short hairpin (sh)RNAs targeting hsa_circ_0122913 and
DCUN1D1, and their corresponding negative control (NC) shRNAs were
purchased from Hanbio Biotechnology Co., Ltd. For transfection,
BMSCs at a density of 4x104 cells/well were seeded into
12-well plates. When the cell confluence reached 50%, the culture
medium was replaced with fresh medium containing viral particles at
multiplicities of infection (MOI) of 50 or 100. The viral titer for
all constructs was 2x108 TU/ml, with volumes of 10 µl
(MOI=50) or 20 µl (MOI=100) added per well. Polybrene was included
at a 1:1,000 dilution to enhance transduction efficiency. The viral
particles were incubated with the cells for 6 h at 37˚C and 5%
CO2. Following the incubation, the transfection medium
was replaced with fresh complete culture medium and the cells were
cultured for 48 h before collection for further analysis. The shRNA
sequences are listed in Table
II.
 | Table IISequences of shRNAs. |
Table II
Sequences of shRNAs.
| Name of shRNA | Strand (5'-3') |
|---|
| DCUN1D1 | Sense:
GATCCGTACGATGATTGCAGATGACTCGAGTCATCTGCAATCATCGTACTTTTTTG |
| | Antisense:
AATTCAAAAAAGTACGATGATTGCAGATGACTCGAGTCATCTGCAATCATC
GTACG |
|
hsa_circ-0122913 | Sense:
GATCCGATGAAGAAGAACAAGTTGCTCGAGCAACTTGTTCTTCTTCATCTTTTTTG |
| | Antisense:
AATTCAAAAAAGATGAAGAAGAACAAGTTGCTCGAGCAACTTGTTCTTCTTCATCG |
| Negative
control | Sense:
GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTC |
| | Antisense:
AATTGAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAACG |
Cell counting kit 8 (CCK-8) assay
The cultured BMSCs were reaped and seeded into
96-well plates (density, 8x103 cells/well), followed by
incubation in an incubator at 37˚C with 5% CO2. Cell
proliferation at 24, 48 and 72 h, as well as 7 days after treatment
was assessed using a CCK-8 Assay kit [Hangzhou MultiSciences
(Lianke) Biotech Co., Ltd.]. Briefly, after removing the
supernatants, each well was supplemented with 10 µl CCK-8 solution
and cells were incubated at 37˚C in the dark for 1 h. The
absorbance in each well at wavelengths of 450 and 650 nm was
measured using a microplate reader (SpectraMAX Plus; Molecular
Devices, LLC.).
Dual-luciferase reporter assay
Bioinformatics analysis using the CircBank database
(http://www.circbank.cn/) was carried out to
predict whether hsa_circ_0122913 could encompass a biding site for
microRNA (miR)-501-5p. MiR-501-5p mimic and the corresponding NC
mimic were synthesized by Guangzhou RiboBio Co., Ltd. The sense
sequence of the miR-501-5p mimic is: 5'-AAUCCUUUGUCCCUGGGUGAGA-3'
and the antisense sequence is: 5'-UCUACCCAGGGACAAAGGAUU-3'. The
sense sequence of the NC mimic is: 5'-UUUGUACUACACAAAAGUACUG-3',
and the antisense sequence is: 5'-AAACAUGAUGUGUUUUCAUGAC-3'. The
reporter plasmids used in the experiment were the
pmirGLO-hsa-circ-0122913-WT and pmirGLO-hsa-circ-0122913-MUT
plasmids, synthesized by Beijing Tsingke Biotech Co., Ltd. For the
transfection experiments, 293T cells, which were obtained from
Chinese National Collection of Authenticated Cell Cultures, at ~80%
confluency in 24-well plates were co-transfected with 20 pmol of
miRNA mimics and 500 ng of reporter plasmids using Lipofectamine
3000 transfection reagent (cat. no. L3000150; Thermo Fisher
Scientific, Inc.). The interaction between miR-501-5p and
hsa_circ_0122913 was verified using the Dual-Luciferase Reporter
Assay System (Promega Corporation), with Firefly luciferase
activity normalized to Renilla luciferase activity,
according to the manufacturer's instructions.
Statistical analysis
All quantitative data were obtained from at least
three biological replicates and are expressed as the mean ±
standard deviation. All analyses were performed using GraphPad
Prism 9.0 (GraphPad Software, Inc.; Dotmatics). For comparisons
between two groups, independent unpaired t-tests were used. For
multiple group comparisons, one-way ANOVA was performed, followed
by Tukey's post-hoc test for pairwise comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
Hsa_circ_0122913 suppresses the
proliferation and osteogenic differentiation of BMSCs in vitro
To investigate the effect of hsa_circ_0122913 on the
proliferation and osteogenic differentiation of BMSCs,
hsa_circ_0122913 was silenced in BMSCs using the corresponding
shRNAs. Therefore, the RT-qPCR results showed that
sh-circ_0122913-1 significantly decreased the expression levels of
hsa_circ_0122913 in BMSCs. Therefore, hsa_circ_0122913-sh1 was used
in the subsequent experiments (Fig.
1A). Additionally, CCK-8 assays (Fig. 1B) and flow cytometric analysis
(Figs. 1C and S1) demonstrated that hsa_circ_0122913
silencing improved the viability and reduced the apoptosis of BMSCs
at 72 h and 7 days compared with the NC group. However, no
difference was observed at 48 h. Furthermore, the role of
hsa_circ_0122913 in the osteogenic differentiation of BMSCs was
assessed using alizarin red S staining, RT-qPCR and western blot
analysis. Alizarin staining demonstrated that hsa_circ_0122913
knockdown could significantly enhance the osteogenic
differentiation ability of BMSCs on day 7 compared with the control
group (Fig. 1D). In addition, the
RT-qPCR and western blot results revealed that hsa_circ_0122913
silencing could increase the expression levels of several
significant osteoblast differentiation-related markers, including
those of OSX, RUNX2 and OPN, at both the mRNA and protein levels
(Fig. 1E and F).
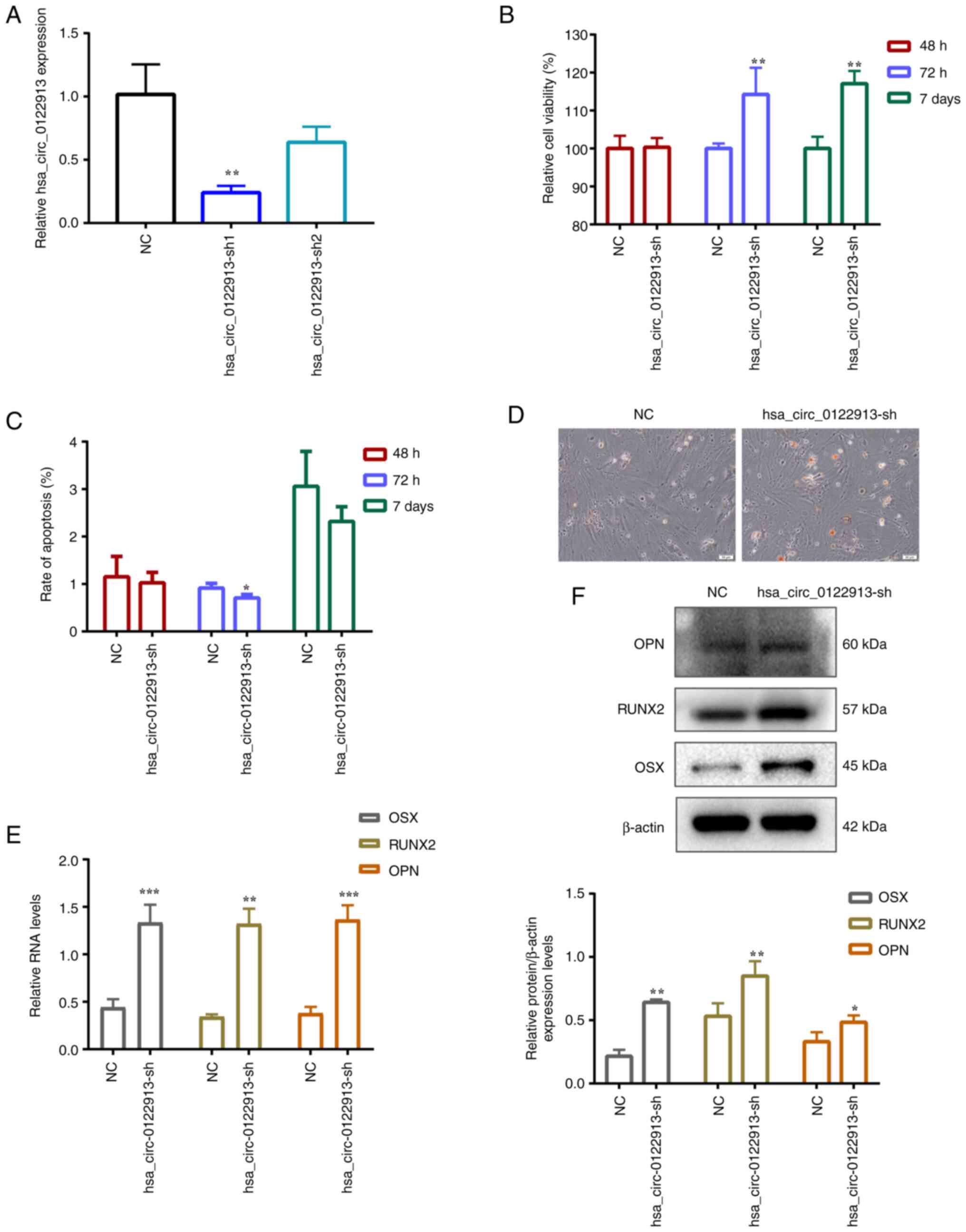 | Figure 1Hsa_circ_0122913 suppresses the
proliferation and osteogenic differentiation of BMSCs in
vitro. (A) The expression of knocked down hsa_circ_0122913 in
BMSCs determined by RT-qPCR. (B) The cell viability of
hsa_circ_0122913 knocking-down in BMSCs after 48 h, 72 h and 7 d
determined by Cell Counting Kit-8 assay. (C) The apoptotic rate of
hsa_circ_0122913 knocking-down in BMSCs group after 48, 72 h and 7
d determined by flow cytometric analysis. (D) The osteogenic
differentiation ability after 7 days of osteogenic induction
detected by alizarin staining. (E) The mRNA expression of OSX,
RUNX2 and OPN in BMSCs after knockdown of hsa_circ_0122913 detected
by RT-qPCR. (F) The protein expression of OSX, RUNX2 and OPN in
BMSCs after knockdown of hsa_circ_0122913 detected by western blot
analysis. *P<0.05, **P<0.01 and
***P<0.001 vs. NC. BMSCs, bone marrow mesenchymal
stem cells; RT-qPCR, reverse transcription-quantitative PCR; OSX,
osterix; RUNX2, runt-related transcription factor 2; OPN,
osteopontin; NC, negative control. |
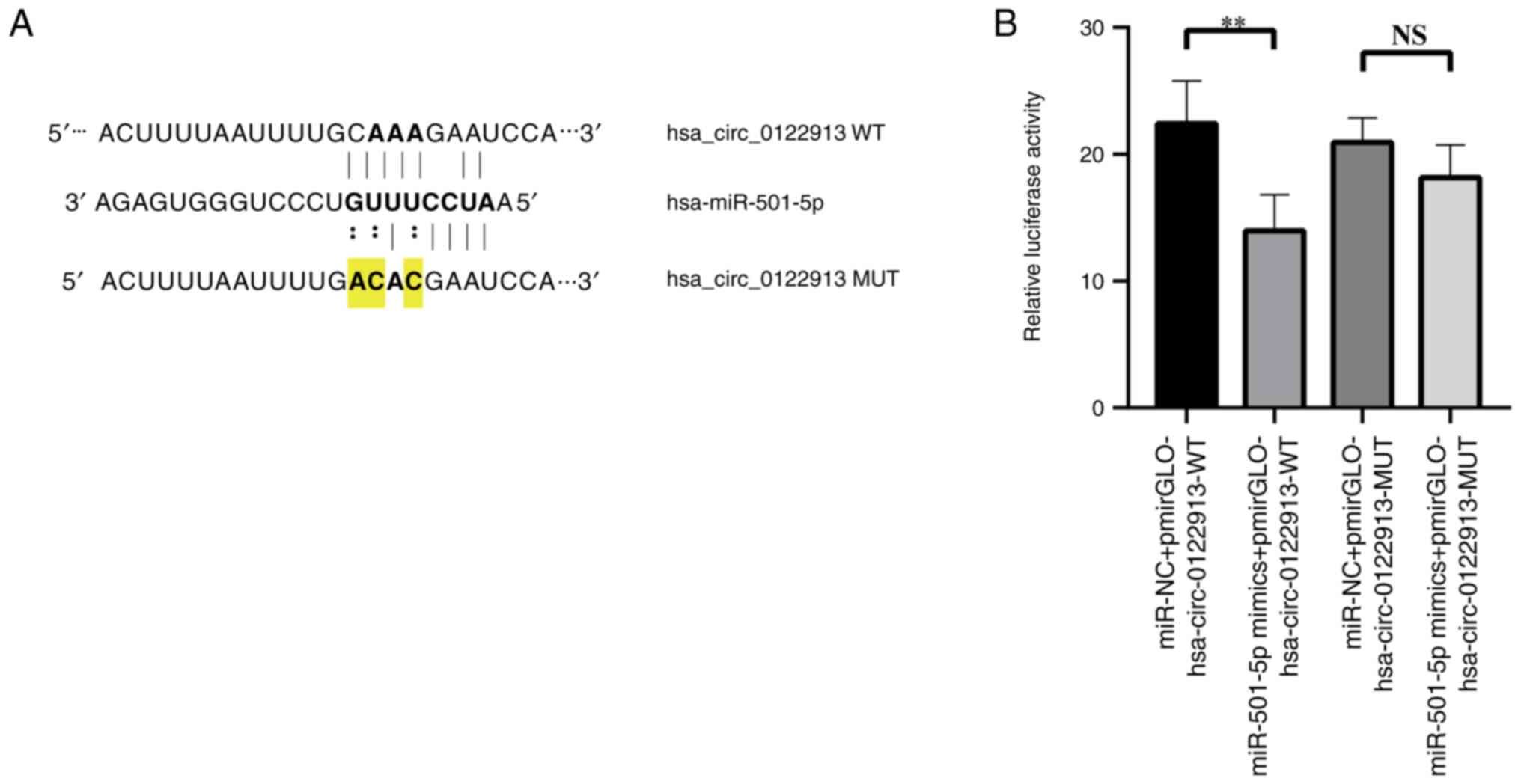
Hsa_circ_0122913 acts a ceRNA for
miR-501-5p
The aforementioned results suggested that
hsa_circ_0122913 could play a key role in the occurrence and
development of osteoporosis. Therefore, it was hypothesized that
hsa_circ_0122913 could regulate the expression levels of miRNAs via
a ceRNA network. Bioinformatics analysis predicted that
hsa_circ_0122913 encompassed a binding site for miR-501-5p
(Fig. 2A). Subsequently, a
dual-luciferase reporter assay was carried out to verify whether
hsa_circ_0122913 could sponge miR-501-5p. Therefore, wild-type (WT)
and binding motif-mutant (MUT) luciferase reporter plasmids were
constructed, namely hsa_circ_0122913-WT/MUT. Then, BMSCs were
co-transfected with miR-501-5p mimic and hsa_circ_0122913-WT/MUT
plasmids. The results indicated that compared with the vector
control group, the luciferase activity was reduced upon
co-transfection of the miR-501-5p mimic. However, transfection of
the miR-501-5p mimic had no effect on the luciferase activity of
hsa_circ_0122913-MUT, thus indicating that hsa_circ_0122913 could
sponge miR-501-5p (Fig. 2B).
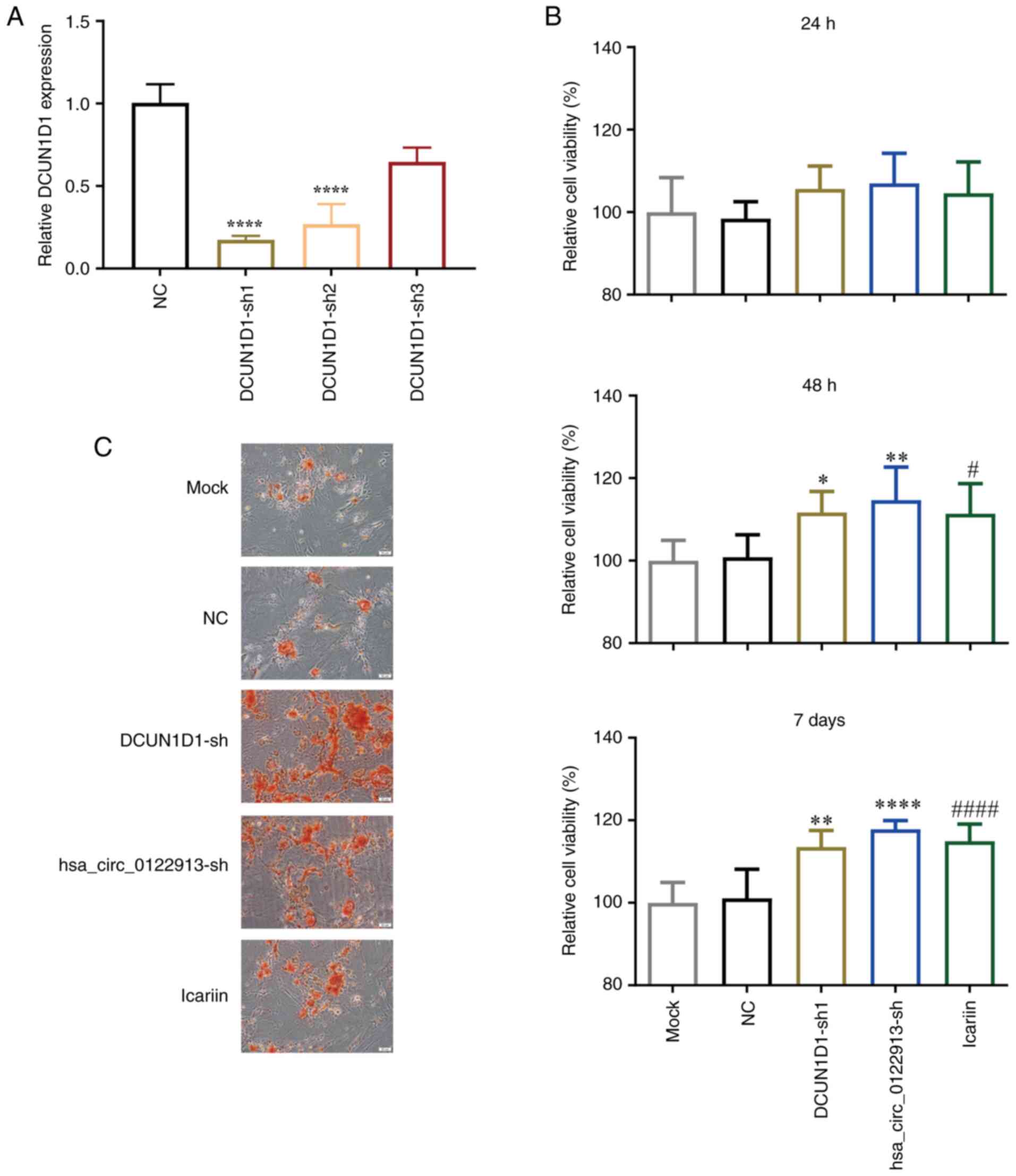
Icariin treatment and DCUN1D1
knockdown can promote the proliferation and osteogenic
differentiation of BMSCs in vitro
Bioinformatics analysis using the CircBank database
predicted that DCUN1D1 could be a host gene of hsa_circ_0122913.
Therefore, shRNAs were used to knockdown DCUN1D1 in BMSCs and the
RT-qPCR results showed that DCUN1D1 expression was most
significantly decreased in BMSCs transfected with DCUN1D1-sh1
(Fig. 3A). Therefore, this
construct was used for DCUN1D1 knockdown in subsequent experiments.
The CCK-8 assay results demonstrated that both hsa_circ_0122913 and
DCUN1D1 knockdown increased cell viability compared with NC
controls, with non-significant effects at 24 h, but statistically
significant enhancement at 48 h and 7 days. Similarly, icariin
treatment showed a progressive improvement in viability compared
with mock controls, with marked increases observed at 48 h and 7
days (Fig. 3B). Furthermore, the
alizarin red staining results showed that hsa_circ_0122913 and
DCUN1D1 silencing promoted the osteogenic differentiation of BMSCs
to a similar extent as icariin treatment (Fig. 3C).
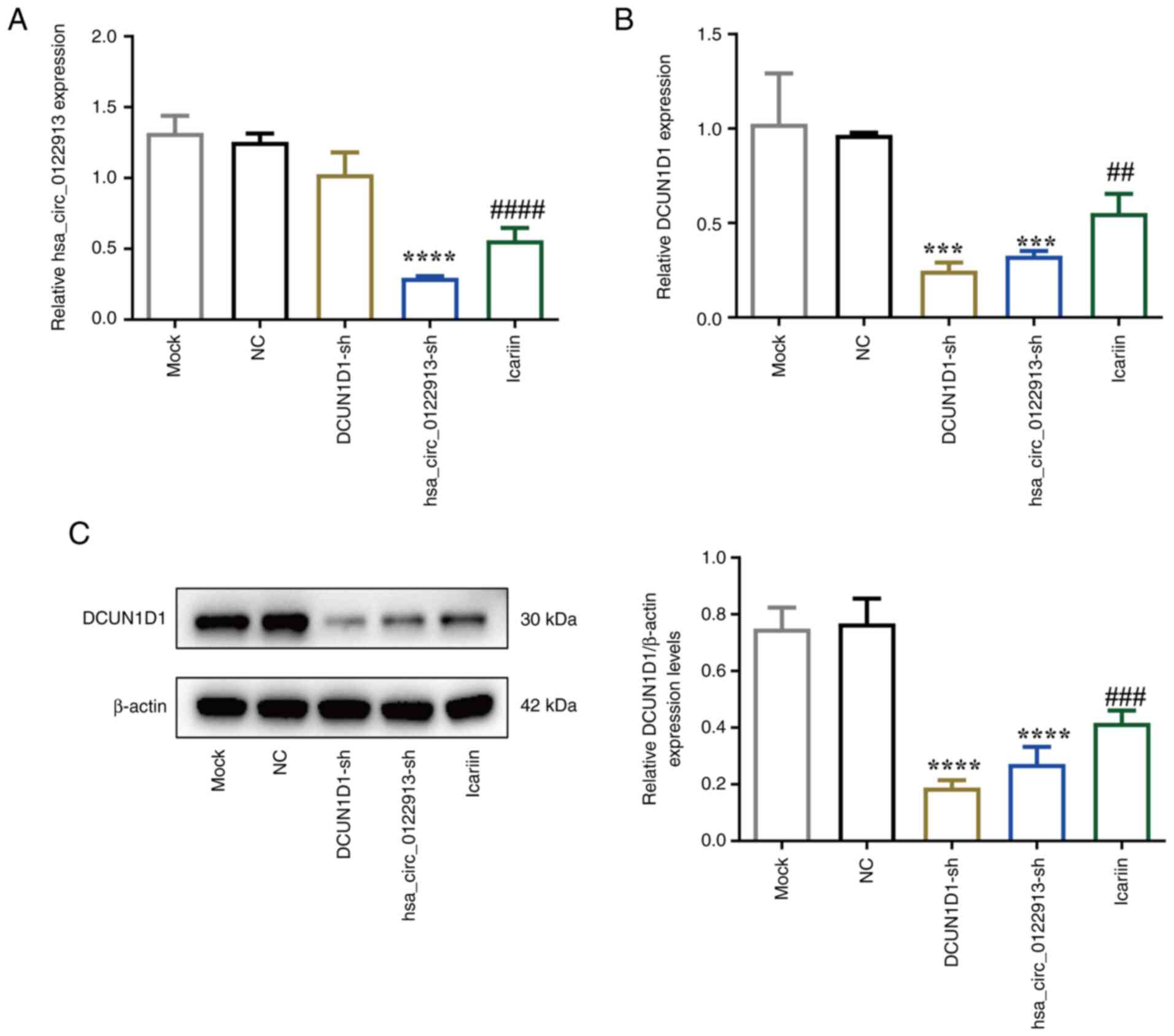
Hsa_circ_0122913/DCUN1D1 axis is
involved in the effects of icariin treatment on BMSCs
The aforementioned results suggested that the
effects of icariin on the proliferation and differentiation ability
of BMSCs were similar to those observed in circ_0122913- or
DCUN1D1-depleted BMSCs. However, no association between icariin and
the hsa_circ_0122913/DCUN1D1 pathway was obtained. Therefore,
RT-qPCR and western blot analyses were performed to verify that
icariin could suppress the mRNA expression levels of
hsa_circ_0122913 (Fig. 4A) and
those of DCUN1D1 at both the mRNA and protein levels (Fig. 4B and C). Furthermore, hsa_circ_0122913 knockdown
significantly decreased the mRNA and protein expression levels of
DCUN1D1 in BMSCs (Fig. 4B and
C). However, DCUN1D1 silencing had
no effect on the expression levels of hsa_circ_0122913 (Fig. 4A).
Discussion
CircRNAs are a relatively new class of non-coding
RNAs that play significant roles in several biological processes.
Therefore, circRNAs are considered as potential therapeutic
biomarkers in various diseases (27,28),
such as cancer (29), Crohn's
disease (30), cardiovascular
diseases (31) and renal diseases
(32). The present study mainly
focused on the effects of hsa_circ_0122913 on the proliferation and
osteogenic differentiation capacities of BMSCs, since this circRNA
was found to be upregulated in patients with senile osteoporotic
vertebral compression fracture (24). The results showed that
hsa_circ_0122913 could reduce the viability and osteogenic
differentiation ability of BMSCs in vitro. Consistent with
previous studies, the aforementioned finding suggested that
hsa_circ_0122913 was closely associated with the occurrence and
development of osteoporosis.
CeRNA networks, where competitive non-coding RNAs
and miRNAs regulate target gene expression, are considered as a
typical mechanism of action of non-coding RNAs (28). A previous study predicted the top
five miRNAs that could regulate hsa_circ_0122913, including
miR-501-5p (24). Other studies
demonstrated that miR-501-5p could play a significant regulatory
role in several types of cancer, such as neck squamous cell
carcinoma (33) and hepatocellular
carcinoma (34). However, its role
in osteogenic differentiation has not been previously investigated.
In the present study, the results indicated that hsa_circ_0122913
could act as a sponge for miR-501-5p. However, how this ceRNA
network acts in BMSCs remains poorly understood and requires
further investigation.
The regulation of circRNA expression and its
interplay with host genes is complex and depending on context.
Indeed, emerging evidence suggests that circRNAs can regulate the
expression of host genes (35). For
instance, knockdown experiments targeting circRNAs such as
circAMOTL1(36) and
circ-ENO1(37) have demonstrated
lower expression levels of their respective host genes, paralleling
our observation that the knockdown of hsa_circ_0122913 coincided
with a downregulation of the host gene DCUN1D1. However, it is
crucial to recognize that circRNA expression can be influenced by
various factors, including cell type and tissue specificity. In
some cases, circRNA expression may not directly correlate with the
expression levels of the linear host gene, as supported by previous
research (38,39). This is consistent with our results,
where the expression level of hsa_circ_0122913 was not
significantly affected by DCUN1D1 knockdown.
Furthermore, DCUN1D1 knockdown and icariin treatment
were found to enhance the viability and osteogenic differentiation
potential of BMSCs. Additionally, the RT-qPCR and western blot
analysis results showed that icariin treatment could regulate the
expression of hsa_circ_0122913 and DCUN1D1, thus indicating that
the hsa_circ_0122913/DCUN1D1 axis could be involved in the effect
of icariin treatment on BMSCs. Previous studies demonstrated that
DCUN1D1 could serve a key role in tumor progression (40,41),
and was involved in several biological functions, including
activation of matrix metalloproteinase 2(42), regulation of CD8+ T-cell
infiltration and depigmentation (43). Notably, Shao et al (44) found that DCUN1D1 may be one of the
genes regulated by upregulated miRNAs in osteoporosis. Combining
the aforementioned previous findings with those of the current
study, it was hypothesized that the hsa_circ_0122913/DCUN1D1
signaling pathway could play a significant role in the occurrence
and development of osteoporosis.
Although the results of the present study could be
helpful in the investigation of the molecular mechanisms associated
with osteoporosis, the present study has some limitations. Firstly,
while the experimental design used in previous studies was
referenced (45,46), where scrambled shRNA was used as a
negative control, it is acknowledged that the inclusion of both
blank and positive controls will be essential in future, as well as
more in-depth studies to strengthen the experimental design and
enhance the reliability of the results. Secondly, more studies are
needed on ceRNA networks to fully explain how hsa_circ_0122913
could regulate the proliferation and differentiation of BMSCs.
Lastly, although the results demonstrated that icariin could
regulate the expression of hsa_circ_0122913 and DCUN1D1, more
studies are needed to uncover the association between the
underlying molecular mechanisms of icariin and those of the
hsa_circ_0122913/DCUN1D1 axis.
In conclusion, the results of the current study
showed that hsa_circ_0122913 could suppress the proliferation and
osteogenic differentiation of BMSCs via sponging miR-501-5p.
Furthermore, hsa_circ_0122913 and DCUN1D1 could be involved in
icariin-regulated biological events in BMSCs. Overall, the
aforementioned results could provide novel insights into the
development of new therapies for osteoporosis.
Supplementary Material
Flow cytometric analysis of apoptosis
in the hsa_circ_0122913-knockdown BMSC group at 48, 72 h and 7
days. BMSC, bone marrow mesenchymal stem cell.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by major scientific and
technological plan projects for social development in Xiaoshan
(grant nos. 2019212 and 2020304) and Zhejiang Traditional Chinese
medicine science and technology project (grant no. 2024ZL801).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HH designed the study. YW, ML and CL performed the
experiments and analyzed the results. YW and ML confirm the
authenticity of all the raw data. YW and HH wrote the manuscript.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lane NE: Epidemiology, etiology, and
diagnosis of osteoporosis. Am J Obstet Gynecol. 194 (Suppl
2):S3–S11. 2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang Y, Tao Y, Hyman ME, Li J and Chen Y:
Osteoporosis in China. Osteoporos Int. 20:1651–1662.
2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Johnston CB and Dagar M: Osteoporosis in
older adults. Med Clin North Am. 104:873–884. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ji H, Cui X, Yang Y and Zhou X: CircRNA
hsa_circ_0006215 promotes osteogenic differentiation of BMSCs and
enhances osteogenesis-angiogenesis coupling by competitively
binding to miR-942-5p and regulating RUNX2 and VEGF. Aging (Albany
NY). 13:10275–10288. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Anthamatten A and Parish A: Clinical
update on osteoporosis. J Midwifery Womens Health. 64:265–275.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi
G, Zhichen W, Zirui W and Shengwang W: The roles of miRNA, lncRNA
and circRNA in the development of osteoporosis. Biol Res.
53(40)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xiao L, Zhong M, Huang Y, Zhu J, Tang W,
Li D, Shi J, Lu A, Yang H, Geng D, et al: Puerarin alleviates
osteoporosis in the ovariectomy-induced mice by suppressing
osteoclastogenesis via inhibition of TRAF6/ROS-dependent MAPK/NF-κB
signaling pathways. Aging (Albany NY). 12:21706–21729.
2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Friedenstein AJ, Gorskaja JF and Kulagina
NN: . Fibroblast precursors in normal and irradiated mouse
hematopoietic organs. Exp Hematol. 4:267–274. 1976.PubMed/NCBI
|
|
9
|
Hu L, Yin C, Zhao F, Ali A, Ma J and Qian
A: Mesenchymal stem cells: cell fate decision to osteoblast or
adipocyte and application in osteoporosis treatment. Int J Mol Sci.
19(360)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang C, Meng H, Wang X, Zhao C, Peng J and
Wang Y: Differentiation of bone marrow mesenchymal stem cells in
osteoblasts and adipocytes and its role in treatment of
osteoporosis. Med Sci Monit. 22:226–233. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Qadir A, Liang S, Wu Z, Chen Z, Hu L and
Qian A: Senile osteoporosis: The involvement of differentiation and
senescence of bone marrow stromal cells. Int J Mol Sci.
21(349)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Infante A and Rodríguez CI: Osteogenesis
and aging: Lessons from mesenchymal stem cells. Stem Cell Res Ther.
9(244)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mattiucci D, Maurizi G, Leoni P and Poloni
A: Aging- and senescence-associated changes of mesenchymal stromal
cells in myelodysplastic syndromes. Cell Transplant. 27:754–764.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fathi E, Charoudeh HN, Sanaat Z and
Farahzadi R: Telomere shortening as a hallmark of stem cell
senescence. Stem Cell Investig. 6(7)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Guo Y, Jia X, Cui Y, Song Y, Wang S, Geng
Y, Li R, Gao W and Fu D: Sirt3-mediated mitophagy regulates
AGEs-induced BMSCs senescence and senile osteoporosis. Redox Biol.
41(101915)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang P, Zhang H, Lin J, Xiao T, Xu R, Fu
Y, Zhang Y, Du Y, Cheng J and Jiang H: Insulin impedes osteogenesis
of BMSCs by inhibiting autophagy and promoting premature senescence
via the TGF-β1 pathway. Aging (Albany NY). 12:2084–2100.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen L, Wang C, Sun H, Wang J, Liang Y,
Wang Y and Wong G: The bioinformatics toolbox for circRNA discovery
and analysis. Brief Bioinform. 22:1706–1728. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shafabakhsh R, Mirhosseini N, Chaichian S,
Moazzami B, Mahdizadeh Z and Asemi Z: Could circRNA be a new
biomarker for pre-eclampsia? Mol Reprod Dev. 86:1773–1780.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gao M, Zhang Z, Sun J, Li B and Li Y: The
roles of circRNA-miRNA-mRNA networks in the development and
treatment of osteoporosis. Front Endocrinol (Lausanne).
13(945310)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu N, Lu W, Qu X and Zhu C: LLLI promotes
BMSC proliferation through circRNA_0001052/miR-124-3p. Lasers Med
Sci. 37:849–856. 2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Qiao L, Li CG and Liu D: CircRNA_0048211
protects postmenopausal osteoporosis through targeting miRNA-93-5p
to regulate BMP2. Eur Rev Med Pharmacol Sci. 24:3459–3466.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yao X, Liu M, Jin F and Zhu Z:
Comprehensive analysis of differentially expressed circular RNAs in
patients with senile osteoporotic vertebral compression fracture.
Biomed Res Int. 2020(4951251)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu Y, Xia L, Zhou Y, Xu Y and Jiang X:
Icariin induces osteogenic differentiation of bone mesenchymal stem
cells in a MAPK-dependent manner. Cell Prolif. 48:375–384.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yu L and Liu Y: circRNA_0016624 could
sponge miR-98 to regulate BMP2 expression in postmenopausal
osteoporosis. Biochem Biophys Res Commun. 516:546–550.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chia W, Liu J, Huang YG and Zhang C: A
circular RNA derived from DAB1 promotes cell proliferation and
osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death
Dis. 11(372)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gao Y, Shang S, Guo S, Li X, Zhou H, Liu
H, Sun Y, Wang J, Wang P, Zhi H, et al: Lnc2Cancer 3.0: An updated
resource for experimentally supported lncRNA/circRNA cancer
associations and web tools based on RNA-seq and scRNA-seq data.
Nucleic Acids Res. 49:D1251–D1258. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ye Y, Zhang L, Hu T, Yin J, Xu L, Pang Z
and Chen W: CircRNA_103765 acts as a proinflammatory factor via
sponging miR-30 family in Crohn's disease. Sci Rep.
11(565)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ju J, Song YN, Chen XZ, Wang T, Liu CY and
Wang K: circRNA is a potential target for cardiovascular diseases
treatment. Mol Cell Biochem. 477:417–430. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Jin J, Sun H, Shi C, Yang H, Wu Y, Li W,
Dong YH, Cai L and Meng XM: Circular RNA in renal diseases. J Cell
Mol Med. 24:6523–6533. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Giordano L, Porta GD, Peretti GM and
Maffulli N: Therapeutic potential of microRNA in tendon injuries.
Br Med Bull. 133:79–94. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC
and Jiang N: MiR-501-5p regulates CYLD expression and promotes cell
proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol.
45:738–744. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen
D, Li M and Xiao L: Exosome-transmitted circ_MMP2 promotes
hepatocellular carcinoma metastasis by upregulating MMP2. Mol
Oncol. 14:1365–1380. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang
F, Li X, Li T, Zhao L, Ren Y and Xu Y: circAMOTL1 motivates AMOTL1
expression to facilitate cervical cancer growth. Mol Ther Nucleic
Acids. 19:50–60. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhou J, Zhang S, Chen Z, He Z, Xu Y and Li
Z: CircRNA-ENO1 promoted glycolysis and tumor progression in lung
adenocarcinoma through upregulating its host gene ENO1. Cell Death
Dis. 10(885)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li J, Yu T, Yan M, Zhang X, Liao L, Zhu M,
Lin H, Pan H and Yao M: DCUN1D1 facilitates tumor metastasis by
activating FAK signaling and up-regulates PD-L1 in non-small-cell
lung cancer. Exp Cell Res. 374:304–314. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Huang S, Liu Z, Jiang F, He H and Zhong W:
DCUN1D1 promotes tumour progress in prostate cancer and its effect
on DU145 in vitro. J Pak Med Assoc. 71:473–478. 2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
O-charoenrat P, Sarkaria I, Talbot SG,
Reddy P, Dao S, Ngai I, Shaha A, Kraus D, Shah J, Rusch V, et al:
SCCRO (DCUN1D1) induces extracellular matrix invasion by activating
matrix metalloproteinase 2. Clin Cancer Res. 14:6780–6789.
2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhao Y, Hu Y, Shen Q, Xiong R, Song X and
Guan C: DCUN1D1, a new molecule involved in depigmentation via
upregulating CXCL10. Exp Dermatol. 32:457–468. 2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Shao JL, Li H, Zhang XR, Zhang X, Li ZZ,
Jiao GL and Sun GD: Identification of serum exosomal MicroRNA
expression profiling in menopausal females with osteoporosis by
high-throughput sequencing. Curr Med Sci. 40:1161–1169.
2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen SC, Jiang T, Liu QY, Liu ZT, Su YF
and Su HT: Hsa_circ_0001485 promoted osteogenic differentiation by
targeting BMPR2 to activate the TGFβ-BMP pathway. Stem Cell Res
Ther. 13(453)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Fan L, Yang K, Yu R, Hui H and Wu W:
circ-Iqsec1 induces bone marrow-derived mesenchymal stem cell
(BMSC) osteogenic differentiation through the miR-187-3p/Satb2
signaling pathway. Arthritis Res Ther. 24(273)2022.PubMed/NCBI View Article : Google Scholar
|