Introduction
The coronavirus disease 2019 (COVID-19) pandemic,
caused by severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2), had a profound impact on global health and society.
The World Health Organization (WHO) officially declared COVID-19 a
pandemic on March 11, 2020, acknowledging the widespread and severe
nature of the disease (1,2). Morbidity and mortality due to COVID-19
markedly increase with age and pre-existing health conditions, such
as cancer and cardiovascular diseases. While most patients recover
from the illness, even the youngest and healthiest can unexpectedly
succumb to COVID-19(3). These
observations raise questions about how much these variations in
disease severity are due to the genetic susceptibility of the
patient: Genetic factors may contribute both to increased
transmissibility of the virus and to the worsening of the disease,
as observed in a small fraction of those affected (4).
The SARS-CoV-2 virus enters host cells through a
specific mechanism involving the angiotensin-converting enzyme 2
(ACE2) receptor located on the host cell membrane and the
viral spike (S) protein (5-7).
The spike protein (S) is initially synthesized in an inactive
precursor form and is converted by host cell proteases into its
active form in a process known as activation (8). After the binding of the spike protein
to the ACE2 receptor, transmembrane serine protease 2
(TMPRSS2) cleaves the spike protein (9), inducing the fusion of the viral
envelope's plasma membrane and the direct entry of SARS-CoV into
the cells and its activation (10).
Research indicates that the expression of the
ACE2 receptor may increase with age, which accounts for the
greater susceptibility of older individuals to COVID-19 (11,12).
Furthermore, increased expression of TMPRSS2 is associated
with worse prognosis in infections by the H1N1 virus and
susceptibility to infections by the H7N9 virus (13). Understanding these molecular
interactions between the virus and host cell receptors is crucial
for developing targeted therapies and interventions to disrupt
viral entry and replication (14).
In fact, identifying host factors involved in viral entry, such as
TMPRSS2, has emerged as a potential therapeutic approach to
prevent COVID-19 infection (15).
By unraveling the mechanisms of viral entry and pathogenesis,
effective antiviral strategies and treatments can be developed to
combat the spread and impact of COVID-19.
The aim of the present study was to evaluate changes
in the expression of ACE2 and TMPRSS2 genes in
nasopharyngeal cells from patients with varying degrees of
COVID-19, with the aim of determining their prognostic
potential.
Patients and methods
Patient samples and
classification
The present study collected nasopharyngeal cell
samples from individuals attended at hospitals and emergency
services within the public health network of the municipalities of
Santo André, São Bernardo do Campo, São Caetano and São Mateus (São
Paulo, Brazil) during 2020. The inclusion criteria were as follows:
Participants of both sexes, aged between 18 and 80 years, with
symptoms of respiratory diseases and who underwent SARS-CoV-2
detection tests, with or without the need for intensive care unit
(ICU) admission, from April to December 2020. Samples were
collected prior to the initiation of the National COVID-19
Vaccination Operationalization Plan.
The exclusion criteria were as follows: Patients
under the age of 18 years and those who were hospitalized for
reasons unrelated to respiratory diseases.
Disease severity classification in this study
followed the clinical criteria established by the World Health
Organization (WHO), which define mild cases as those with symptoms
such as fever, cough, sore throat, malaise, headache, muscle pain,
nausea, or anosmia, but without dyspnea, abnormal imaging, or the
need for hospitalization (16).
Moderate cases present with clinical or radiological signs of lower
respiratory disease and maintain SpO2 ≥90% on room air.
Severe cases are defined by respiratory distress, SpO2
<90%, respiratory rate >30 breaths per minute, or the need
for intensive care. In the present study, these WHO clinical
definitions were combined with the level of care required (such as
outpatient, hospital ward, or ICU admission) to classify patients
accordingly. In cases of discrepancy between the WHO-defined
clinical severity and the level of care received, for example, a
patient with SpO2 ≥90% admitted to the ICU due to
comorbidities or precautionary monitoring, the clinical criteria
were prioritized whenever sufficient data were available. However,
if clinical data were insufficient to confidently determine
severity, the level of care served as a surrogate marker for
disease severity.
Although symptom information was documented during
routine triage in 2020, a standardized and complete symptom survey
was not systematically applied across all patients, as data
collection occurred during routine clinical care under overwhelming
operational constraints at the peak of the COVID-19 pandemic.
The study was approved by the Ethics Committee of
FMABC University Center (protocol number 5.610.755; São Paulo,
Brazil) on August 29, 2022. This is a retrospective analysis of
nasopharyngeal samples that were obtained during the COVID-19
pandemic as part of routine diagnostics. The samples were promptly
processed and stored under appropriate conditions according to
established guidelines, including storage at -80˚C for long-term
storage, ensuring the integrity and stability of the specimens for
subsequent research analysis. Patients registered at public health
units in the region, or their families, were subsequently
approached by the research team to obtain written informed consent
for the use of their samples and participation in the study.
Total RNA isolation
Total RNA was isolated from the nasal swab samples
using the PureLink™ Total RNA Blood Kit (cat no.
K156001), following the manufacturer's protocol, and quantified
using a Qubit 4 fluorometer (cat no. Q33238) with the Qubit RNA HS
Assay Kit (cat no. Q32852; all from ThermoFisher Scientific, Inc.).
RNA samples (100 ng) were converted into cDNA using the QuantiTect
Reverse Transcription Kit (cat no. 205311; QIAGEN), following the
manufacturer's instructions.
Gene expression
The expression of the ACE2 and TMPRSS2
genes in nasopharyngeal cells was evaluated by quantitave
polymerase chain reaction (qPCR). Specific primers were designed
using the Primer3 Input 0.4.0 software (https://bioinfo.ut.ee/primer3-0.4.0/) and validated
for specificity using the Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The sequences of the primers and characteristics of the amplicons
are described in Table I.
 | Table ICharacteristics of specific primers
and their amplicons. |
Table I
Characteristics of specific primers
and their amplicons.
| Gene | Sequence
(5'-3') | Amplicon |
|---|
| ACE2 | F:
AGGGCGACTTCAGGATCCTT | |
| | R:
TGTGGCTGCAGAAAGTGACA | 185 bp |
| TMPRSS2 | F:
GAACACAAGTGCCGGCAATG | |
| | R:
CTGGACGTGCAGGCTGAC | 193 bp |
| RPL13a | F:
TTGAGGACCTCTGTGTATTTGTCAA | |
| | R:
CCTGGAGGAGAAGAGGAAAGAGA | 126 bp |
The relative expression of the target genes was
normalized by the reference RPL13α gene expression (17-19).
RPL13α was selected as the reference gene because its Ct
values consistently remained below 35 and its amplification showed
greater stability compared with other candidate reference genes
tested (including GUSB, TFRC, GAPDH, and
β-actin). Amplifications were performed in an ABI 7500
thermocycler (Applied Biosystems; ThermoFisher Scientific, Inc.) in
a final volume of 15 µl containing 1X SYBR Green
(QuantiFast® SYBR Green PCR kit; cat. no. 204054;
QIAGEN), 10 pmol of each specific primer, and 2 µl of cDNA. The
amplification parameters consisted of an initial hot start at 95˚C
for 10 min, followed by 40 cycles at 95˚C for 15 sec and 60˚C for
25 sec. Differential expression was determined using the formula
2-ΔΔCq (20).
Statistical analysis
An initial descriptive statistical analysis was
performed, expressing categorical variables as absolute and
relative frequencies. Quantitative variables were summarized using
medians, interquartile ranges (IQR), minimum, and maximum values.
The assumption of normality for quantitative variables was formally
assessed through the Shapiro-Wilk test. Based on the distribution,
comparisons between two groups were performed using the
Mann-Whitney U test for non-normal data, and comparisons among more
than two groups were performed using the Kruskal-Wallis test.
Graphical representations were standardized according to data
distribution: Variables with a normal distribution are presented as
the means ± standard deviation, whereas non-normally distributed
variables are shown as medians and interquartile ranges (IQR).
To evaluate the expression levels of the ACE2
and TMPRSS2 genes across different COVID-19 severity groups,
the non-parametric Kruskal-Wallis test was used (version 10.4.1
GraphPad Prism; Dotmatics). When significant differences were
detected, Dunn's post hoc test was applied for pairwise
comparisons. Correlation analyses were performed using Spearman's
rank correlation. All statistical tests were two-tailed, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
The study included samples from 491 individuals, of
whom 158 tested negative and 333 tested positive for SARS-CoV-2 via
molecular testing. The demographic and clinical characteristics of
the patients are summarized in Table
II. Among the 158 individuals with a negative test, only 3
required ICU admission for reasons unrelated to COVID-19. By
contrast, among the 333 individuals who tested positive, 208
experienced mild to moderate symptoms and did not require ICU
admission, while 125 developed severe symptoms and required
intensive care. Although a standardized symptom survey was not
employed in the present study, the symptoms most frequently
documented in patients classified as mild cases included fever,
cough, sore throat, and fatigue. For moderate cases, in addition to
these symptoms, patients often presented dyspnea and tachypnea,
consistent with clinical signs of pneumonia. Severe cases
predominantly exhibited respiratory distress, hypoxia (oxygen
saturation <90%), and altered mental status, which aligned with
the WHO criteria for severe COVID-19 and frequently justified ICU
admission (16). While exact
frequencies could not be uniformly quantified due to variability in
record completeness, these symptom patterns were consistent with
the clinical stratification applied throughout the cohort.
 | Table IIClinical characteristics of the
patients. |
Table II
Clinical characteristics of the
patients.
|
Characteristics | n (%) |
|---|
| Sex | |
|
Female | 250 (50.9) |
|
Male | 241 (49.1) |
| Age (years) | |
|
18 to
40 | 227 (46.2) |
|
41 to
60 | 154 (31.4) |
|
61 to
80 | 110 (22.4) |
| SARS-CoV-2-negative
(n=158) | |
|
No ICU
admission | 155 (98.1) |
|
With ICU
admission | 3 (1.9) |
| SARS-CoV-2-positive
(n=333) | |
|
No ICU
admission | 208 (62.5) |
|
With ICU
admission | 125 (37.5) |
| Sex distribution
(SARS CoV-2-positive) (n=333) | |
|
Male | 176 (52.9) |
|
Female | 157 (47.1) |
| Sex distribution
(SARS CoV-2-negative) (n=158) | |
|
Male | 65 (41.1) |
|
Female | 93 (58.9) |
| Mean age (±SD)
(n=491) | |
|
SARS
CoV-2-positive | 47.4±19 |
|
SARS
CoV-2-negative | 41.8±18 |
Men represented the majority of participants
infected with the virus: 176 (52.9%) compared with 157 (47.1%)
women; among participants without SARS-CoV-2 infection, 93 (58.9%)
were female and 65 (41.1%) were male. Additionally, infected
participants had a higher mean age (47.4±19) than those without the
virus (41.8±18) (P=0.001), as shown in Table II. Regarding severity, as expected,
there was a higher proportion of severe cases (37.5%) and a lower
proportion of moderate cases (14.4%) among those with SARS-CoV-2
infection compared with the group without the infection (Table III).
 | Table IIIContingency by severity compared with
SARS-CoV-2 diagnosis. |
Table III
Contingency by severity compared with
SARS-CoV-2 diagnosis.
| Severity |
|---|
| Without SARS-CoV-2
infection, n (%) | With SARS-CoV-2
infection, n (%) |
|---|
| Mild | 88 (55.7) | Mild | 160 (48.0) |
| Moderate | 67 (42.4) | Moderate | 48 (14.4) |
| Severe | 3 (1.9) | Severe | 125 (37.5) |
The analysis of gene expression data for ACE2
and TMPRSS2 among patients diagnosed with COVID-19 was
conducted to investigate possible associations with sex, age and
disease severity.
Both ACE2 [men (n=125), 0.3021±0.5056 vs.
women (n=104), 0.3686±0.6313] and TMPRSS2 [men (n=102),
0.0463±0.0896 vs. women (n=89) 0.0745±0.1192] expression levels
were slightly higher in women than in men; however, the difference
was not statistically significant for ACE2 (P=0.24), while
TMPRSS2 expression showed a trend toward significance
(P=0.07) (Table IV). These values
refer only to the subset of samples in which the respective gene
expression was detected.
 | Table IVComparison of ACE2 and
TMPRSS2 levels and age between men and women. |
Table IV
Comparison of ACE2 and
TMPRSS2 levels and age between men and women.
| Variable | Group | P-value | Statistical
significance |
|---|
| ACE2
expression | | | |
|
Men
(n=125) | 0.3021±0.5056 | 0.24 | NS |
|
Women
(n=104) | 0.3686±0.6313 | | |
| TMPRSS2
expression | | | |
|
Men
(n=102) | 0.0463±0.0896 | 0.07 | Trend towards
significance |
|
Women
(n=89) | 0.0745±0.1192 | | |
| Age (years) | | | |
|
Men | 48±19 | 0.42 | |
|
Women | 46±18 | NS | |
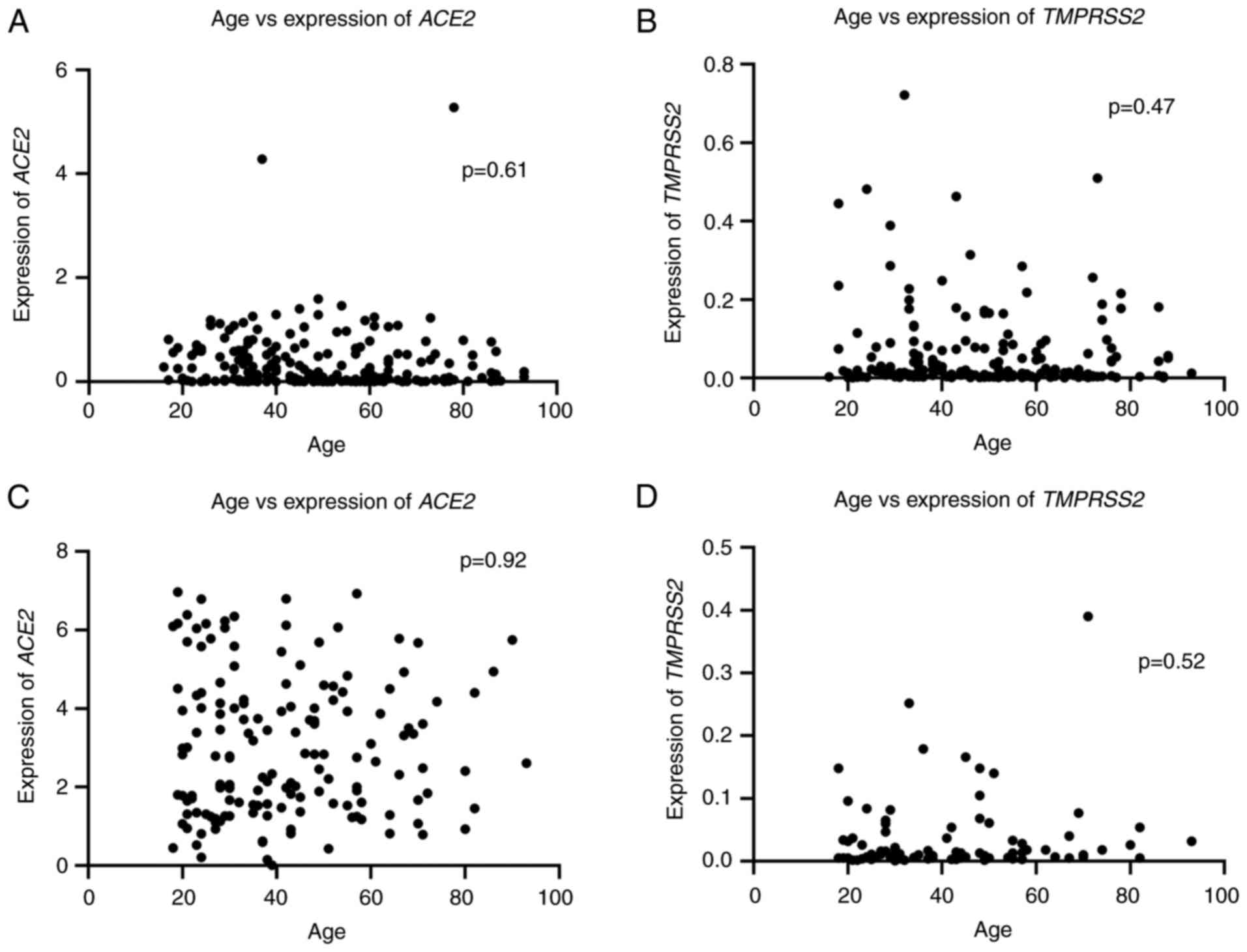
The analysis of the age of patients revealed no
significant difference between sexes (P=0.42), with a mean age of
48±19 years for men and 46±18 years for women, as shown on Table IV. Similarly, no significant
correlation was found between age and the expression of ACE2
or TMPRSS2 in either SARS-CoV-2-positive or -negative groups
(Fig. 1). These findings indicated
that gene expression in the nasal epithelium is not directly
associated with age or SARS-CoV-2 infection status.
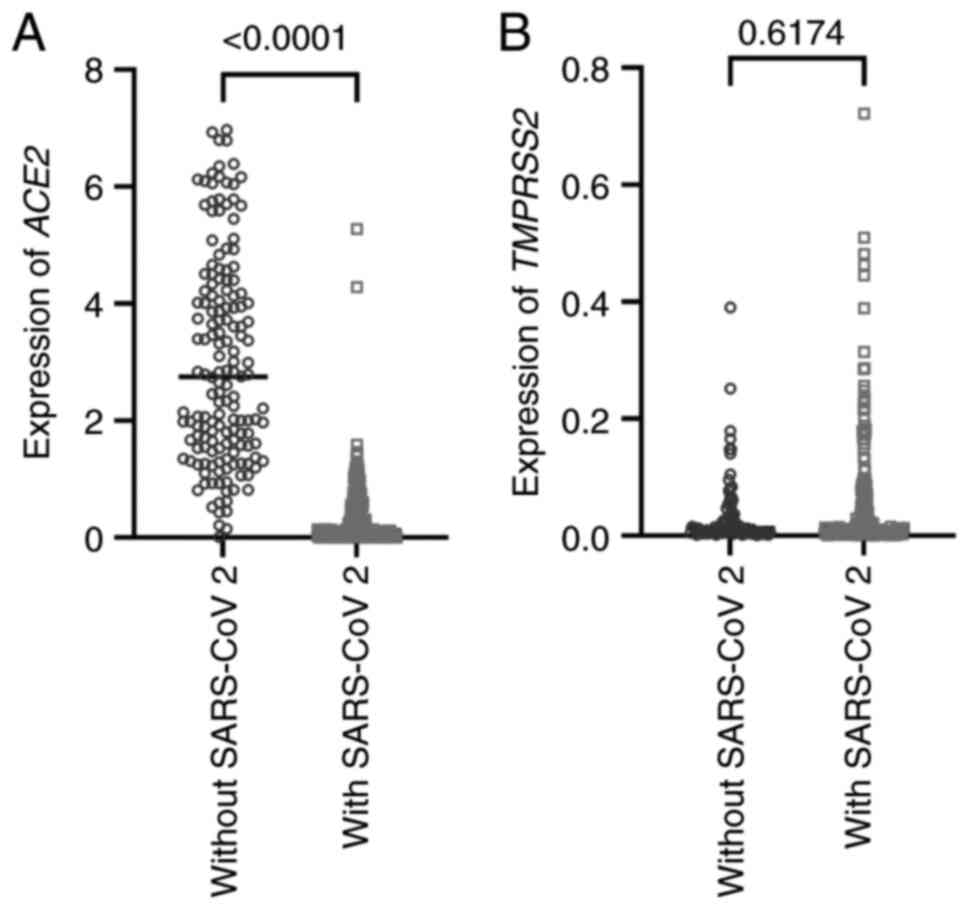
ACE2 expression was revealed to be
significantly higher in uninfected individuals (2.750±1.775)
compared with those with SARS-CoV-2 infection (0.122±0.528)
(Fig. 2A). While reduced
ACE2 expression in SARS-CoV-2-infected individuals may
initially appear counterintuitive given the receptor's role in
viral entry, an alternative explanation is that this reduction
reflects a host-mediated defense mechanism aimed at limiting viral
spread. By downregulating ACE2, host tissues may decrease
the number of entry points available to the virus, thereby
restricting replication and dissemination (21).
Analysis of TMPRSS2 expression revealed
greater variability among individuals, with no significant
difference between SARS-CoV-2-infected and uninfected groups
(Fig. 2B). This suggests that
TMPRSS2 levels in the nasopharyngeal epithelium are not directly
affected by infection, although the presence of TMPRSS2 may still
facilitate viral entry when ACE2 is available.
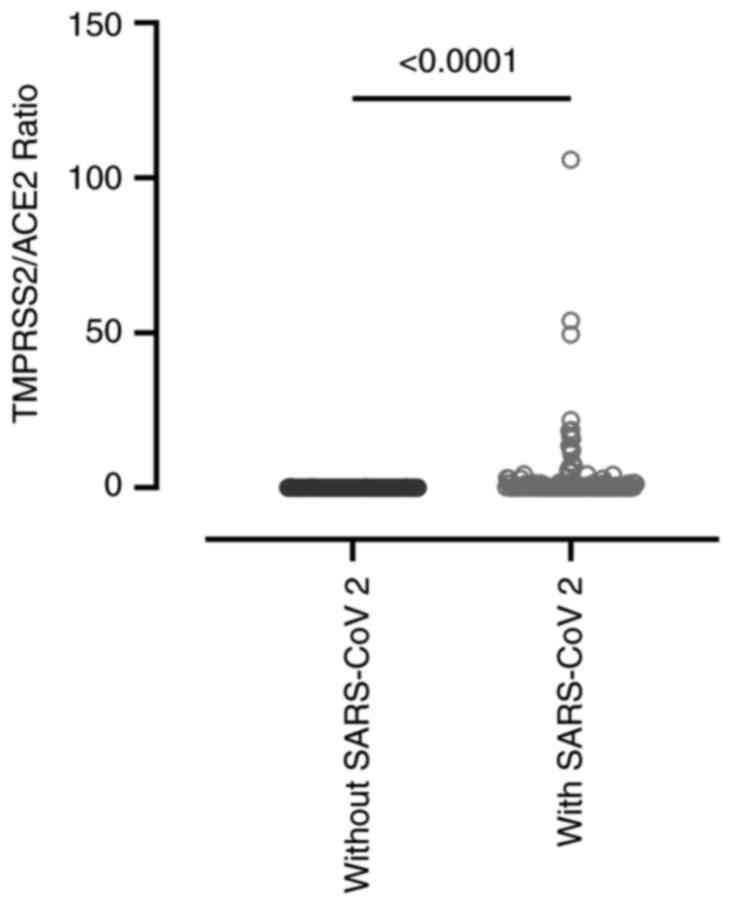
The present study analyzed the expression dynamics
of the ACE2 and TMPRSS2 receptors in the
nasopharyngeal epithelium and their relationship with COVID-19
severity. While no direct association was observed between
ACE2 expression and disease severity, the
TMPRSS2/ACE2 ratio more effectively distinguished
individuals with SARS-CoV-2 infection from those without it (with
SARS-CoV-2, 3.094±10.920 vs. without SARS-CoV-2, 0.024±0.047;
P<0.0001), indicating a possible synergistic interaction between
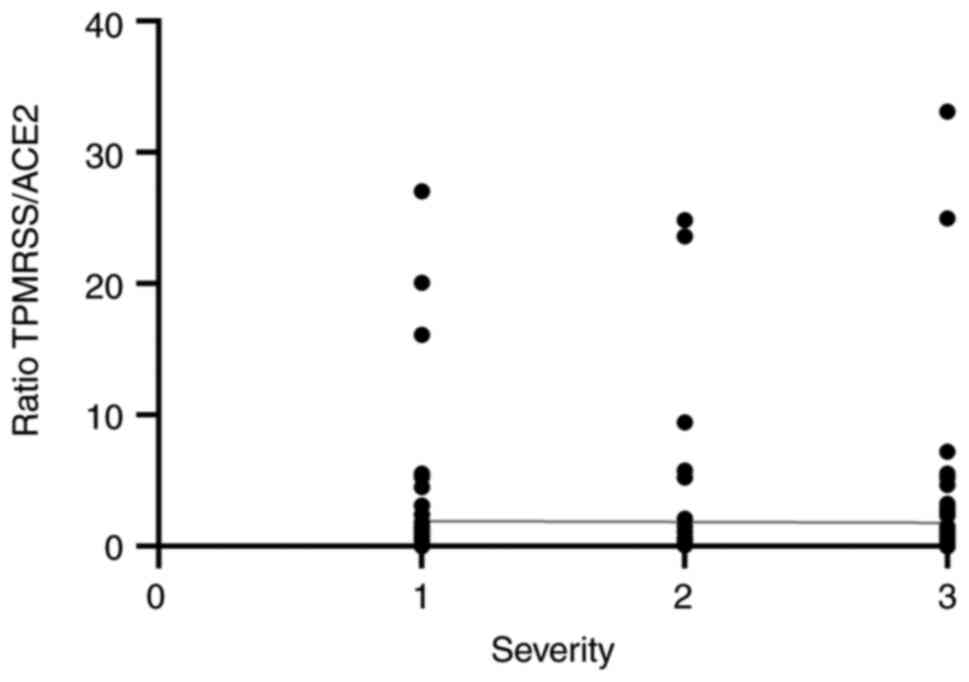
these receptors in viral pathogenesis (Fig. 3). On the other hand, when evaluating
whether this ratio varied according to disease severity within the
COVID-19-positive group, it appeared to be independent of severity
(r=-0.033; P=0.67) (Fig. 4).
To further explore the association between
ACE2 and TMPRSS2 expression levels and COVID-19
severity, patients were categorized according to clinical
presentation. Among individuals with respiratory symptoms caused by
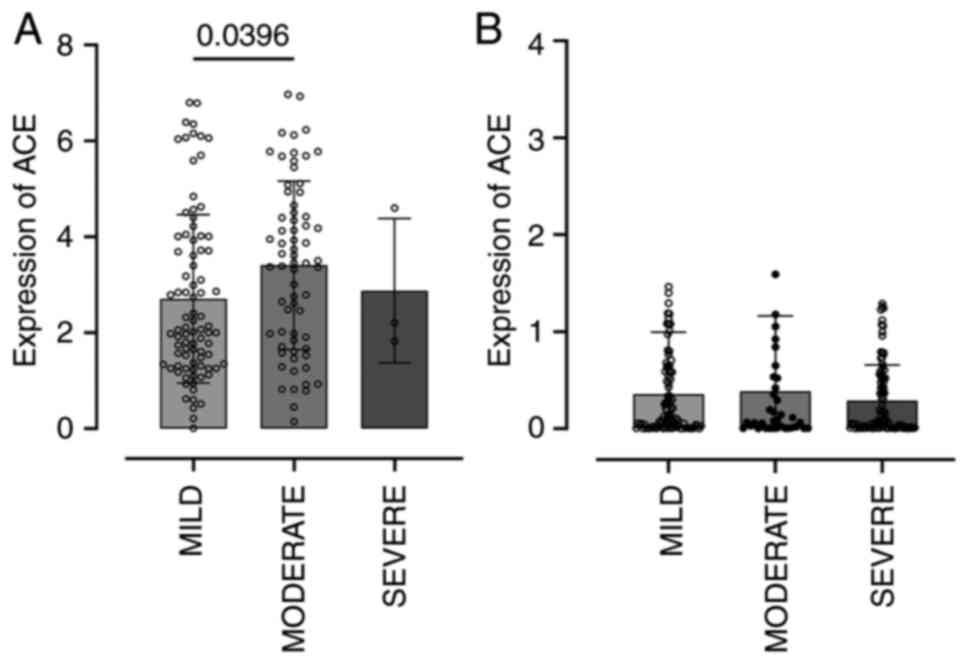
agents other than SARS-CoV-2, ACE2 expression was significantly
increased in nasopharyngeal cells from patients with moderate
symptoms (Fig. 5A). By contrast,
this pattern was not observed among SARS-CoV-2-infected
individuals: ACE2 expression remained downregulated
following infection, regardless of clinical severity (Fig. 5B).
Gene expression patterns differed between COVID-19
patients and those with other respiratory infections. In non-COVID
infections, ACE2 expression increased during moderate
symptoms, whereas in COVID-19 patients, ACE2 remained
suppressed even after infection, suggesting a potential viral
strategy to evade host defenses. Additionally, both ACE2 and
TMPRSS2 showed age-related increases, with males over 65
having significantly higher ACE2 levels, which may help
explain the higher mortality in this group.
In SARS-CoV-2-infected individuals, opposing
patterns were observed: Symptomatic patients had lower baseline
ACE2, which increased during active infection, while
TMPRSS2 levels, initially elevated, decreased after
infection. As previously shown in Fig.
4, the TMPRSS2/ACE2 ratio did not show a significant
correlation with clinical severity in COVID-19 cases (Spearman's
rs=-0.033; P=0.67), suggesting that this parameter is
not associated with disease progression. Despite limitations, such
as the small number of severe cases in the non-COVID cohort,
sensitivity analyses confirmed the robustness of these
findings.
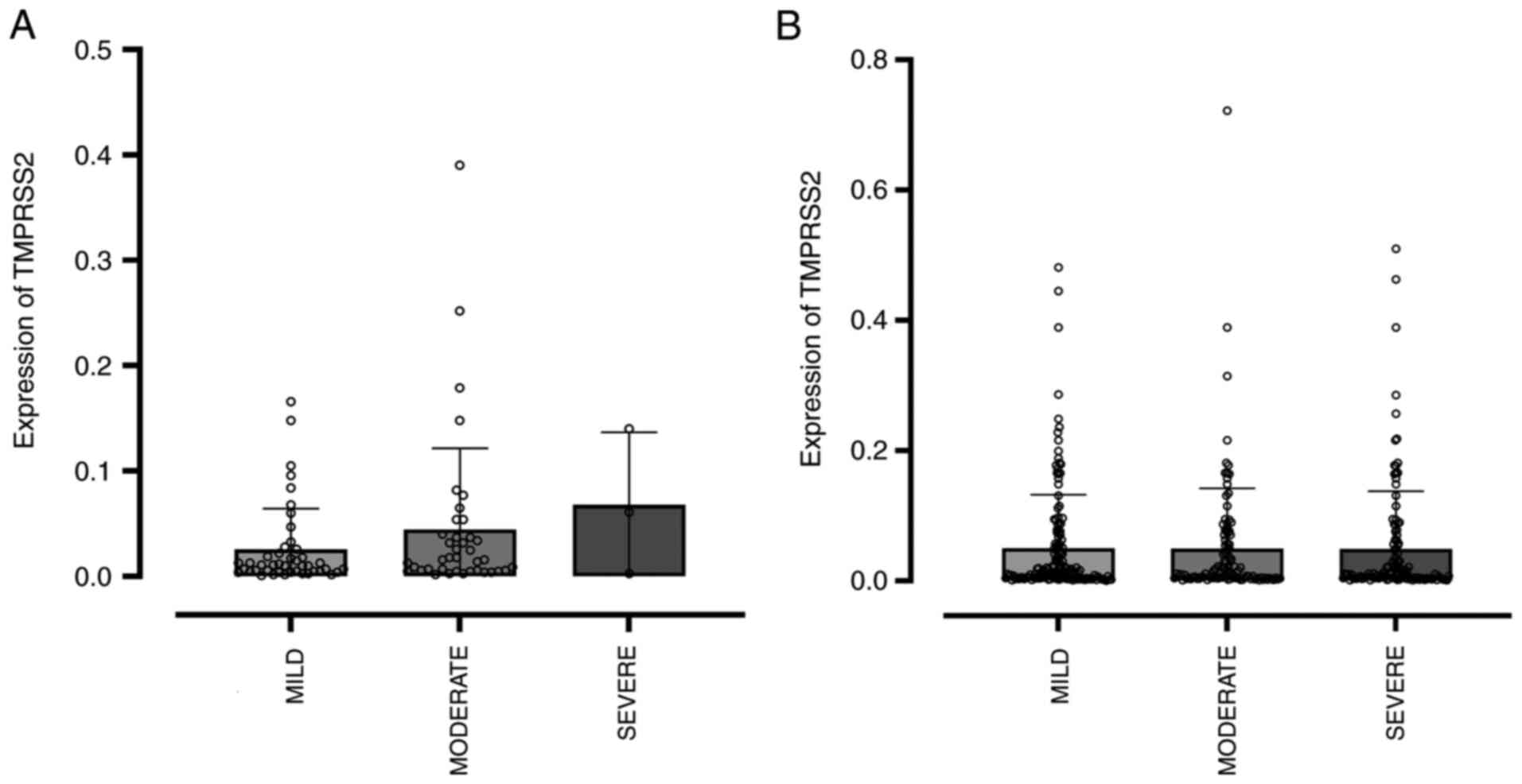
Similarly, an evaluation of TMPRSS2 gene
expression was conducted according to symptom severity in
individuals without and with SARS-CoV-2 infection (Fig. 6). In both scenarios, no difference
in the expression of this gene in the nasopharyngeal cells was
associated with disease severity. There was no significant
correlation between TMPRSS2 expression and disease severity
in patients infected with COVID-19 (Spearman's
rs=-0.103; P=0.154; n=191), indicating that
TMPRSS2 expression does not appear to be associated with
disease severity in this cohort. Likewise, no significant
correlation was observed between ACE expression and disease
severity in COVID-19 patients (Spearman's rs=-0.030;
P=0.649; n=229), as shown in Table
V, suggesting that ACE expression is not associated with
disease severity in this cohort. However, the decrease in
expression observed among infected individuals may indicate a
condition of infection that could potentially render the patient
more vulnerable, but does not directly determine the severity of
the disease.
 | Table VCorrelation between gene expression
and COVID-19 severity. |
Table V
Correlation between gene expression
and COVID-19 severity.
| Gene | No. of
patients | rs | P-value |
|---|
| TMPRSS2 | 191 | -0.103 | 0.154 |
| ACE2 | 229 | -0.030 | 0.649 |
This observation supports the notion that receptor
expression dynamics in COVID-19 may be more reflective of viral
evasion mechanisms and host-pathogen interactions, rather than
directly predicting disease outcomes. Further investigation into
how these changes in receptor expression interact with other
factors, such as immune response or co-morbidities, will be
essential to fully understand the complex determinants of COVID-19
severity.
Discussion
The presence of COVID-19 is associated with greater
severity in patient hospitalizations. Patients diagnosed with
COVID-19 are more likely to present severe cases, which can
influence clinical decisions and treatment strategies. These
findings may support the need for more stringent measures for the
prevention and treatment of COVID-19, aiming to reduce the severity
of hospitalizations. Therefore, there is strong evidence that the
detection of COVID-19 is significantly associated with greater
severity in patient hospitalizations. This study aimed to
understand how the ACE2 and TMPRSS2 receptors vary in
patients with different levels of COVID-19, seeking to determine
their potential as prognostic markers. By comparing their
expression in respiratory cells of patients with COVID-19 and
individuals without the virus, the goal was to identify which
receptor is more useful for predicting clinical outcomes.
ACE2 and TMPRSS2 are essential proteins for the entry
of the SARS-CoV-2 virus into cells and play a crucial role in the
pathogenesis of COVID-19(22). The
expression of these genes can be influenced by factors such as
genetic polymorphisms, cytokines associated with asthma, hormones
including testosterone, and even obesity (23,24).
Additionally, the expression of these genes can vary across
different tissues, such as the lungs, upper respiratory tract, and
intestine, which are potential sites of SARS-CoV-2 replication
(25).
It is known that ACE2 expression in the
airways is high in the nasal epithelium but progressively decreases
in the bronchial and alveolar regions, correlating with the levels
of SARS-CoV-2 infection in different compartments of the airways
(26). However, the results of the
present study indicated that ACE2 expression was
significantly lower in the nasal epithelial cells of patients
infected with SARS-CoV-2 compared with individuals without the
virus, suggesting a possible negative regulation of this gene
during infection. Research has indicated that SARS-CoV-2 infection
can modulate ACE2 function and subsequent inflammatory
responses, leading to a decrease in the expression of this gene
overtime (22). In fact, there is
evidence suggesting that the virus leads to a reduction in
ACE2 expression, occurring through various mechanisms,
including the release of ACE2 from tissues, the reduction of
ACE2 levels in infected cells, the induction of clathrin-
and AP2-dependent endocytosis, and subsequent lysosomal degradation
(27-29).
This downregulation of ACE2 by the virus has been associated
with lung injury and inflammation, further exacerbating
pathological processes in infected individuals (29). In the intestines, on the other hand,
the absence of a positive correlation between susceptibility to
infection and ACE2 expression has been described in a
subpopulation of enterocytes considered the primary target of the
virusAccording to the aforementioned study, infected cells
activated strong pro-inflammatory programs and produced interferon,
while the expression of interferon-stimulated genes was limited to
uninfected cells due to the virus's suppression of interferon
(30).
The decrease in ACE2 expression by SARS-CoV-2
may play a role in disease progression and its severity, including
the development of acute respiratory distress syndrome (31), as it disrupts the physiological
balance between ACE/ACE2 and Ang-II/angiotensin-(1-7),
potentially causing severe organ damage (32). Understanding the mechanisms
underlying ACE2 downregulation by the virus is essential for
the development of targeted therapeutic interventions to mitigate
the effects of SARS-CoV-2 infection.
In order to assess whether there is an association
between ACE2 expression in the nasal epithelium of
individuals with various clinical manifestations of COVID-19, the
patients were grouped according to the severity of the disease
(mild, moderate, or severe). No significant changes in ACE2
expression associated with COVID-19 severity were observed. In
fact, studies indicate that the presence of ACE2 in nasal
epithelial cells does not necessarily correlate with an increase in
the severity of SARS-CoV-2 infection (33,34).
It appears that greater vulnerability to severe outcomes in
infected individuals may be attributed to other factors, such as
the immune response and the presence of different ACE2
variants, which could influence the severity of the infection
(35,36). Therefore, while ACE2
expression is crucial for viral entry into cells, it may not serve
as a reliable predictor of disease severity (33).
TMPRSS2, a transmembrane protease, is crucial
for the activation of SARS-CoV-2 within human airway cells
(35). This protease is expressed
in various tissues, including nasal epithelial cells, where it
cleaves and activates the viral S protein, facilitating viral entry
and infection (36). The
co-expression of TMPRSS2 and ACE2 in nasal epithelial
cells has been associated with increased SARS-CoV-2 infectivity and
transmissibility, suggesting a potential role of these cells in the
early stages of infection (37).
However, despite the decreased ACE2 expression in infected
individuals, the results did not reveal any alteration in
TMPRSS2 gene expression between individuals with and without
the virus, indicating that the regulation of this gene may not be
influenced by its presence. Research conducted on SARS-CoV, a
member of the coronavirus family, has revealed that TMPRSS2
expression is indeed not affected during viral production (23). According to this study, TMPRSS2
influences viral entry but not other phases of viral replication,
suggesting that the spatial orientation of TMPRSS2 in relation to
the S protein is a key mechanism underlying this phenomenon
(23).
In the present study, it was observed that the
expression of the ACE2 and TMPRSS2 genes does not
vary between men and women with COVID-19, nor with the age of
infected patients. A previous study suggests that ACE2 expression
may vary with age (38), while
another study did not find a consistent correlation (39).
Although a relationship between age, the expression
of these genes, and susceptibility or severity of SARS-CoV-2
infection was anticipated, other factors beyond this variable
appear to have a greater influence on the modulation of the
expression of these genes (23,40).
Factors such as genetic variations, pre-existing health conditions,
environmental exposures, and other biological and social
determinants may play a significant role in regulating ACE2
and TMPRSS2 expression, thus influencing SARS-CoV-2
susceptibility and severity.
Additionally, the expression of these genes may be
modulated by factors such as inflammation, smoking exposure, and
diseases such as asthma and chronic obstructive pulmonary disease
(40). Therefore, it is crucial to
consider a broad range of factors beyond age when investigating the
expression of these genes and their relationship with COVID-19. A
potential limitation of the study is that it did not account for
pre-infection factors, such as those aforementioned, which may
influence gene expression. However, it is important to emphasize
that the present study was conducted during the peak of the
pandemic in Brazil, a time when strict social isolation rules and
regulations imposed significant limitations on data collection and
analysis. These constraints may have impacted the ability to fully
explore the role of these additional factors in the context of
COVID-19 severity. Furthermore, it is important to highlight that,
as this classification relied exclusively on RT-qPCR results, the
possibility of false-negative results cannot be ruled out.
Therefore, some individuals classified as SARS-CoV-2-negative might
have actually been infected, especially in cases of improper sample
collection or low viral load at the time of testing.
The downregulation of ACE2 observed during
SARS-CoV-2 infection, in contrast to the stable expression of
TMPRSS2, highlights the importance of considering multiple
influences on gene modulation, such as environmental exposures and
other external factors. These insights underscore the need for
multifactorial approaches in COVID-19 research and treatment to
better understand and mitigate susceptibility and disease
severity.
In conclusion, the present study demonstrated that
ACE2 gene expression is significantly reduced in individuals
infected with SARS-CoV-2, regardless of symptom severity, age, or
sex, suggesting that this downregulation may be a direct
consequence of viral infection. By contrast, TMPRSS2
expression exhibited no significant variation between infected and
non-infected individuals, nor across different levels of disease
severity. Although both proteins are essential for viral entry,
their expression levels in nasopharyngeal cells do not serve as
reliable biomarkers for predicting COVID-19 severity. These
findings highlight the complexity of COVID-19 pathogenesis and
underscore the need for multifactorial approaches, integrating
genetic, immunological, and clinical factors, for improved
understanding and management of disease progression.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
All authors (SSC, ACBCS, MYSG, MCP, RSS, IDRT, GLV,
FLAF, BCAA) contributed equally to the conception, design, data
acquisition, analysis, and writing of the manuscript. SSC and BCAA
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of FMABC University Center (protocol number 5.610.755;
São Paulo, Brazil) and adheres to Resolution 466/12 of the National
Health Council, and all patients received information concerning
their participation in the study and provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Riou J and Althaus CL: Pattern of early
human-to-human transmission of Wuhan 2019 novel coronavirus
(2019-nCoV), December 2019 to January 2020. Euro Surveill.
25(2000058)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
World Health Organization (WHO):
Coronavirus disease 2019 (COVID-19) situation report-52. WHO,
Geneva, 2020.
|
|
3
|
Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z
and Tong S: Epidemiology of COVID19 among children in China.
Pediatrics. 145(e20200702)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hou Y, Zhao J, Martin W, Kallianpur A,
Chung MK, Jehi L, Sharifi N, Erzurum S, Eng C and Cheng F: New
insights into genetic susceptibility of COVID-19: An ACE2 and
TMPRSS2 polymorphism analysis. BMC Med. 18(216)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ejaz H, Alsrhani A, Zafar A, Javed H,
Junaid K, Abdalla AE, Abosalif KOA, Ahmed Z and Younas S: COVID-19
and comorbidities: Deleterious impact on infected patients. J
Infect Public Health. 13:1833–1839. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hassanpour M, Rezaie J, Nouri M and Panahi
Y: The role of extracellular vesicles in COVID-19 virus infection.
Infect Genet Evol. 85(104422)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chaudhry F, Lavandero S, Xie X, Sabharwal
B, Zheng YY, Correa A, Narula J and Levy P: Manipulation of ACE2
expression in COVID-19. Open Heart. 7(e001424)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zmora P, Moldenhauer AS, Hofmann-Winkler H
and Pöhlmann S: TMPRSS2 isoform 1 activates respiratory viruses and
is expressed in viral target cells. PLoS One.
10(e0138380)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hoffmann M, Hofmann-Winkler H, Smith JC,
Krüger N, Arora P, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler
M, Hempel T, et al: Camostat mesylate inhibits SARS-CoV-2
activation by TMPRSS2-related proteases and its metabolite GBPA
exerts antiviral activity. EbioMedicine. 65(103255)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Matsuyama S, Nagata N, Shirato K, Kawase
M, Takeda M and Taguchi F: Efficient activation of the severe acute
respiratory syndrome coronavirus spike protein by the transmembrane
protease TMPRSS2. J Virol. 84:12658–12664. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bilinska K, Jakubowska P, Von Bartheld CS
and Butowt R: Expression of the SARS-CoV-2 entry proteins, ACE2 and
TMPRSS2, in cells of the olfactory epithelium: Identification of
cell types and trends with age. ACS Chem Neurosci. 11:1555–1562.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ren HL, Wen GM, Zhao ZY, Liu DH and Xia P:
Can CD147 work as a therapeutic target for tumors through COVID-19
infection? Int J Med Sci. 19:2087–2092. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cheng Z, Zhou J, To KKW, Chu H, Li C, Wang
D, Yang D, Zheng S, Hao K, Bossé Y, et al: Identification of
TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1)
influenza and A(H7N9) influenza. J Infect Dis. 212:1214–1221.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cetinkaya EA: Coincidence of COVID-19
infection and smell-taste perception disorders. J Craniofac Surg.
31:e625–e626. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tolouian R, Tolouian AC and Ardalan M:
Blocking serine protease (TMPRSS2) by bromhexine; looking at
potential treatment to prevent COVID-19 infection. Marshall J Med.
6:11–14. 2020.
|
|
16
|
World Health Organization (WHO): Clinical
management of COVID-19: Living guideline, 18 August 2023. WHO,
Geneva, 2023.
|
|
17
|
Hannemann J, Schmidt-Hutten L, Hannemann
J, Kleinsang F and Böger R: Selection of reference genes for
normalization of gene expression after exposure of human
endothelial and epithelial cells to hypoxia. Int J Mol Sci.
26(1763)2025.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hampton TH, Koeppen K, Bashor L and
Stanton BA: Selection of reference genes for quantitative PCR:
Identifying reference genes for airway epithelial cells exposed to
Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol.
319:L256–L265. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Biji A, Khatun O, Swaraj S, Narayan R,
Rajmani RS, Sardar R, Satish D, Mehta S, Bindhu H, Jeevan M, et al:
Identification of COVID-19 prognostic markers and therapeutic
targets through meta-analysis and validation of Omics data from
nasopharyngeal samples. EBioMedicine. 70(103525)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bourgonje AR, Abdulle AE, Timens W,
Hillebrands JL, Navis GJ, Gordijn SJ, Bolling MC, Dijkstra G, Voors
AA, Osterhaus AD, et al: Angiotensin-converting enzyme 2 (ACE2),
SARS-CoV-2 and the pathophysiology of coronavirus disease 2019
(COVID-19). J Pathol. 251:228–248. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Saheb Sharif-Askari N, Saheb Sharif-Askari
F, Alabed M, Temsah MH, Al Heialy S, Hamid Q and Halwani R: Airways
expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 Is lower in
children than adults and increases with smoking and COPD. Mol Ther
Methods Clin Dev. 18:1–6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kimura H, Francisco D, Conway M, Martinez
FD, Vercelli D, Polverino F, Billheimer D and Kraft M: Type 2
inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells.
J Allergy Clin Immunol. 146:80–88.e8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Baratchian M, McManus JM, Berk MP,
Nakamura F, Mukhopadhyay S, Xu W, Erzurum S, Drazba J, Peterson J,
Klein EA, et al: Androgen regulation of pulmonary AR, TMPRSS2 and
ACE2 with implications for sex-discordant COVID-19 outcomes. Sci
Rep. 11(11130)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sarver D and Wong G: Obesity alters Ace2
and Tmprss2 expression in lung, trachea, and esophagus in a
sex-dependent manner: Implications for COVID-19. Biochem Biophys
Res Commun. 538:92–96. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Rossi ÁD, de Araújo JLF, de Almeida TB,
Ribeiro-Alves M, de Almeida Velozo C, Almeida JM, de Carvalho
Leitão I, Ferreira SN, da Silva Oliveira J, Alves HJ, et al:
Association between ACE2 and TMPRSS2 nasopharyngeal expression and
COVID-19 respiratory distress. Sci Rep. 11(9658)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Guo J, Huang Z, Lin L and Lv J:
Coronavirus disease 2019 (COVID-19) and cardiovascular disease: A
viewpoint on the potential influence of angiotensin-converting
enzyme inhibitors/ angiotensin receptor blockers on onset and
severity of severe acute respiratory syndrome coronavirus 2
infection. J Am Heart Assoc. 9(e016219)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Datta PK, Liu F, Fischer T, Rappaport J
and Qin X: SARS-CoV-2 pandemic and research gaps: Understanding
SARS-CoV-2 interaction with the ACE2 receptor and implications for
therapy. Theranostics. 10:7448–7464. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lu Y, Zhu Q, Fox DM, Gao C, Stanley SA and
Luo K: SARS-CoV-2 down-regulates ACE2 through lysosomal
degradation. Mol Biol Cell. 33(ar147)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Grace JA, Casey S, Burrell LM and Angus
PW: Proposed mechanism for increased COVID-19 mortality in patients
with decompensated cirrhosis. Hepatol Int. 14:884–885.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Triana S, Metz-Zumaran C, Ramirez C, Kee
C, Doldan P, Shahraz M, Schraivogel D, Gschwind AR, Sharma AK,
Steinmetz LM, et al: Single-cell analyses reveal SARS-CoV-2
interference with intrinsic immune response in the human gut. Mol
Syst Biol. 17(e10232)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kuba K, Yamaguchi T and Penninger JM:
Angiotensin-converting enzyme 2 (ACE2) in the pathogenesis of ARDS
in COVID-19. Front Immunol. 12(732690)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y,
Hou C, Wang H, Liu J, Yang D, et al: Role of angiotensin-converting
enzyme 2 (ACE2) in COVID-19. Crit Care. 24(422)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ji JY, Jo A, Won J, Gil CH, Shin H, Kim S,
Jeon YJ and Kim HJ: The nasal symbiont Staphylococcus species
restricts the transcription of SARS-CoV-2 entry factors in human
nasal epithelium. iScience. 24(103172)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Takabayashi T, Yoshida K, Imoto Y,
Schleimer RP and Fujieda S: Regulation of the expression of
SARS-CoV-2 receptor angiotensin-converting enzyme 2 in Nasal
Mucosa. Am J Rhinol Allergy. 36:115–122. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Fowler PC, Naluai ÅT, Oscarsson M,
Torkzadeh S, Bohman A, Bende M and Harandi AM: Differential
expression of angiotensin-converting enzyme 2 in nasal tissue of
patients with chronic rhinosinusitis with nasal polyps medRxiv,
2021.
|
|
37
|
Pavel AB, Glickman JW, Michels JR,
Kim-Schulze S, Miller RL and Guttman-Yassky E: Th2/Th1 cytokine
imbalance is associated with higher COVID-19 risk mortality. Front
Genet. 12(706902)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bestle D, Heindl MR, Limburg H, Van Lam
van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik
O, et al: TMPRSS2 and furin are both essential for proteolytic
activation of SARS-CoV-2 in human airway cells. Life Sci Alliance.
3(e202000786)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bunyavanich S, Do A and Vicencio A: Nasal
gene expression of angiotensin-converting enzyme 2 in children and
adults. JAMA. 323:2427–2429. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li MY, Li L, Zhang Y and Wang XS:
Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide
variety of human tissues. Infect Dis Poverty. 9(45)2020.PubMed/NCBI View Article : Google Scholar
|