Introduction
Hepatocellular carcinoma (HCC) is associated with a
dismal patient outcome, and its incidence rate continues to rise
(1). At present, only 10–20% of
patients with HCC are eligible for surgical therapy (2). The results of therapeutic treatment
for liver cancer remain unsatisfactory.
MicroRNAs (miRNAs) are single-stranded RNA
molecules, approximately 21–23 nucleotides in length, which are
able to regulate gene expression at either the transcriptional or
post-transcriptional level. miRNAs are encoded by genes that are
transcribed from DNA, but are not translated into protein. This
type of non-coding RNA was first described in 1993 by Lee et
al (3), which was later
referred to as microRNA (4).
Subsequent studies in recent years have demonstrated a significant
difference in miRNA expression profiles in human malignancies,
including chronic lymphocytic leukaemia, pediatric Burkitt’s
lymphoma, gastric cancer, lung cancer and large-cell lymphoma.
These findings suggest that miRNAs may play an important role in
carcinogenesis as a novel class of oncogenes or tumor-suppressor
genes (5,6). Thus, miRNAs have been considered as
novel targets for cancer therapy.
Coptidis rhizoma (CR) has been used in
Chinese medicine for thousands of years. Previous studies have
demonstrated that CR exhibits a variety of pharmacological actions,
including anti-neoplastic, anti-oxidative, anti-bacterial,
anti-viral, anti-inflammatory, anti-hyperglycemic,
anti-hypertensive and liver protective effects (7–9). The
anti-tumor action of CR has been extensively reported. CR induces
multiple signal transduction pathways to promote cancer cell
apoptosis, inhibiting tumor cell proliferation and neoplastic
angiogenesis. Yet, few studies have reported the effects of CR in
the treatment of liver cancer (10,11).
In the present study, we used a sensitive miRNA
on-chip array to examine the alteration in the miRNA expression
profile in HCC MHHCC-97 cells after treatment with Coptidis
rhizoma aqueous extract (CRAE). In the miRNA on-chip array, the
elevated expression of miR-21 and miR-23a was detected in the
MHCC97-L cells after treatment with 175 μg/ ml of CRAE. The
altered expression of these miRNAs was further validated by
quantitative real-time PCR (qRT-PCR) analysis. Using TargetScan and
PicTar databases, analysis indicated that several signal
transduction pathways may be altered by CRAE via miRNA expression.
Our results suggest the therapeutic potential of CRAE by targeting
specific miRNAs in liver cancer cells.
Materials and methods
Preparation of the Coptidis rhizoma
aqueous extract
To prepare the CRAE, raw material was cut into small
pieces, and 50 g of crude CR was boiled in distilled (10 times)
water (w/v) at 100°C for 1 h and then filtered. Next, the filtrate
was evaporated to dryness, and the residue was dissolved in water
containing 0.1% dimethyl sulfoxide (DMSO). The quality control of
CRAE was reported in our previous study (9).
Cell culture and drug intervention
Human HCC MHCC97-L cells were maintained in high
glucose Dulbecco’s modified Eagle’s medium (Invitrogen, USA)
supplemented with 10% fetal bovine serum and 1% antibiotics. Cells
were incubated in a humidified incubator containing 5%
CO2 at 37°C. For drug intervention, cells were seeded in
T25 culture flasks with ∼70% confluence. Cells were starved
overnight in serum-free medium and then were exposed to 175
μg/ml of CRAE for 48 h. Then, the cells were collected for
analysis using cell scrappers. Cells treated with vehicle (0.1%
DMSO) for 48 h were collected as the control.
Cell viability assay
The MTT assay was used to determine the cytotoxicity
of CRAE in the MHCC97-L cells. Cells were seeded in a 96-well plate
at a density of 10,000 cells per well, and a series of
concentrations (0, 1.75, 3.5, 7, 14, 28, 56, 112, 224 and 448
μg/ml) of CRAE were added to the cells after a 48-h
incubation with the control 0.1% DMSO. Fifteen microliters of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
5 mg/ml; Sigma, USA) was added to each well, and the cells were
further incubated at 37°C for 4 h. The culture medium was then
discarded, and 200 μl DMSO was added to dissolve the
crystals. The Multiskan MS microplate reader (Labsystems, Finland)
was used to measure the absorbance of formazan formed at 595
nm.
Preparation of the miRNA-enriched total
RNA sample
miRNA-enriched total RNAs of MHCC97-L cells
untreated and treated with 175 μg/ml CRAE were extracted and
purified by the miRNeasy Mini kit (Qiagen, Germany) according to
the manufacturer’s instructions.
On-chip microarray analysis
The NCode™ Rapid miRNA Labeling System (Invitrogen)
was used for Poly (A) tailing of total RNA and ligation for on-chip
microarray analysis. Briefly, 1 μl of NCode™ Multi-Species
miRNA microarray controls (2 fmol/μl) was spiked, and Poly
(A) tailing reactions were set up for each total RNA sample. The
Poly (A) tailed reactions were separately ligated to the labeled
DNA polymers using the 6X Alexa Fluor® 3 (A3) Rapid
Ligation mix or 6X Alexa Fluor® 5 (A5) Rapid Ligation
mix, according to the manufacturer’s instructions. For
hybridization of the labeled miRNAs with the miRNA microarray, the
two differentially labeled reactions were combined into one tube,
and the volume was reduced by half in a SpeedVac®
Concentrator. BSA (50 mg/ml) was then added to a total volume of
28.5 μl. The samples were incubated with 28.5 μl of
2X enhanced hybridization buffer at 65°C for 10 min and then loaded
onto NCode™ Human miRNA Microarrays V3. The arrays were mounted
with the Maui Mixer SL and hybridized overnight (16–20 h) at 52°C
with constant mixing. The arrays were then washed according to the
standard protocol recommended by the manufacturer. The arrays were
scanned using a GenePix® 4000B microarray scanner
(Molecular Devices, USA). Alexa Fluor® 3 (green channel)
and Alexa Fluor® 5 (red channel) excitation and emission
maxima were identical to Alexa Fluor® 546 and 647,
respectively. The scanner was programmed accordingly at 556 nm
(excitation) and 573 nm (emission) for Alexa Fluor® 3,
and 650 nm (excitation) and 665 nm (emission) for Alexa
Fluor® 5. The scanned images were saved as TIFF files
with the array barcode number included in the name of the file,
then annotated and analyzed using GenePix® software with
the GAL files containing the array list samples for the human
samples. The data output file was saved as a GPR file and exported
as a text file for easy access and analysis using Microsoft
Excel.
Quantitative real-time PCR for miRNA
analysis
The miRCURY LNA™ Universal RT microRNA PCR system is
designed for sensitive and accurate detection of miRNA by
quantitative real-time PCR using SYBR Green (Exiqon, Denmark). The
miRCURY LNA Universal RT miRNA PCR portfolio is comprised of three
types of reagent kits, including the Universal cDNA synthesis kit
and the SYBR Green master mix kit. Single-stranded cDNA was
generated from the miRNA-enriched total RNA sample by reverse
transcription using the Universal cDNA synthesis kit according to
the manufacturer’s instructions. The expression of the miRNAs was
detected using the SYBR Green master mix kit in a 96-well optical
plate. Finally, relative miRNA expression levels were calculated by
comparison to the expression of the endogenous control of U6
snRNA.
Target prediction
To predict the function of the selected miRNAs
related to the anti-tumor action of CRAE, we indentified the
possible targets of miR-21 and miR-23a based on two miRNA
databases, TargetScan (http://www.targetscan.org/index.html) and PicTar
(http://pictar.mdc-berlin.de), according
to the manufacturer’s instructions. Several target genes which were
common to both TargetScan and PicTar were identified for further
study.
Validation of the predicted targets
To validate the predicted target genes from the two
miRNA databases, RT-PCR was used to detect the expression of the
predicted targets. Reverse-transcription reaction was performed
using the Roche Reverse Transcription kit (Roche, USA) to prepare
the cDNA samples. qRT-PCR was conducted using the SYBR Green PCR
kit (Roche) with 1 μM of primers for PLAG1 (left: 5′-GTCCAG
CCCGAAATATGAGA-3′, right: 5′-CAGCACCAAGAGGCA ACC-3′, Invitrogen),
or RP2 (left: 5′-AAGAGACGGAAG GCTGACAA-3′, right:
5′-GAACATGTAGTCTTTTGGATCA ACC-3′, Invitrogen), or STAT3 (left:
5′-CCCTTGGATTGA GAGTCAAGA-3′, right: 5′-AAGCGGCTATACTGCTGGTC-3′,
Invitrogen), or SATB1 (left: 5′-GGGTACGCGATGAACTG AA-3′, right:
5′-TTCTGAAAGCAAGCCCTGA-3′, Invitrogen), or NTF3 (left:
5′-AAAAACGGTTGCAGGGGTAT-3′, right: 5′-GGTTTGGGATGTTTTGCACT-3′,
Invitrogen), or POU4F2 (left: 5′-CCCTTTGAACCCCACCTC-3′, right:
5′-CTT CCTGCAAACAGCCATCT-3′, Invitrogen), or NEK6 (left:
5′-CTGGGCTGTCTGCTGTACG-3′, right: 5′-AGTCACACT GCTCGATCTTCTG-3′,
Invitrogen), or SLC6A14 (left: 5′-AGC AAAGAGGTGGATATTCTGG-3′,
right: 5′-CACCAATGA CCAGATAAATATTGC-3′, Invitrogen), or SET8 (left:
5′-CCA TCAAGGGCAAACAGG-3′, right: 5′-GACTGCAGCTCG GCTTTG-3′,
Invitrogen) on the LightCycler 480 real-time PCR system (Roche).
The expression of GAPDH was used as the endogenous control
(forward: 5′-GCTAGGGACGGC CTGAAG-3′; reverse:
5′-GCCCAATACGACCAAATCC-3′; Invitrogen) for the normalization of
gene expression of the above primers.
Statistical analysis
Statistical differences among groups were determined
by the Student’s t-test. Differences were considered to be
significant at p<0.05.
Results
Cytotoxicity of CRAE on HCC MHCC97-L
cells
In order to determine the cytotoxicity of CRAE on
MHCC97-L cells, the MTT assay was conducted to detect the cancer
cell viability after CRAE intervention. A dose-dependent
suppression in the viability of MHCC97-L cells was observed after
treatment with various concentrations of CRAE for 48 h. Since the
major compound in CRAE is berberine, which was found to exhibit a
potent anti-cancer effect in our previous study (11), the concentration of CRAE was
normalized by the content of berberine, and the IC50
value was calculated accordingly. Our result showed that ∼175
μg/ml of CRAE was able to induce 50% MHCC97-L cell death
after a 48-h treatment, indicating that CRAE exhibits a potent
anti-cancer action at a low dose (Fig.
1).
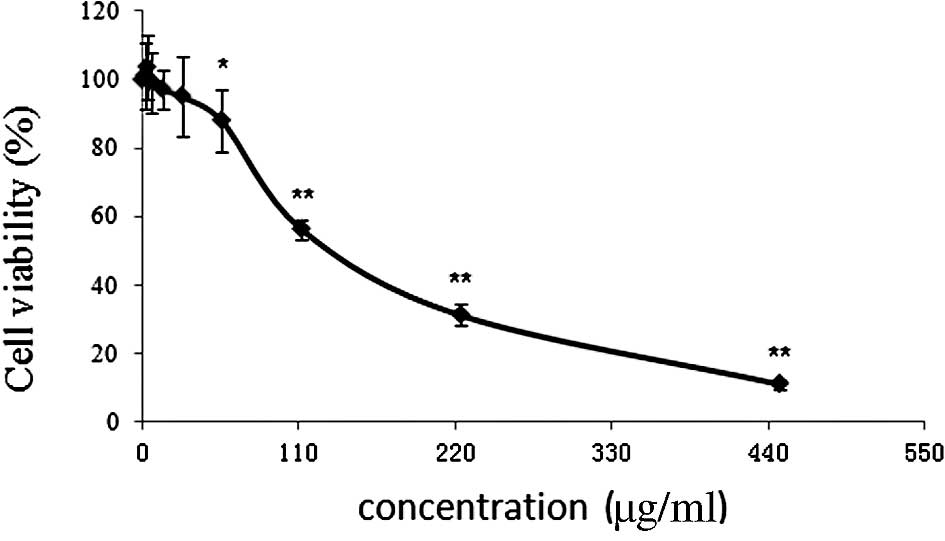 | Figure 1.Cytotoxicity of CRAE in hepatocellular
carcinoma MHCC97-L cells. Cells were seeded in 96-well plates at a
density of 10,000 cells per well and treated with different
concentrations of CRAE (0, 1.75, 3.5, 7, 14, 28, 56, 112, 224 and
448 μg/ml) for 48 h. MTT assay was introduced to determine
the cell viability after CRAE intervention (*p<0.05,
**p<0.01 compared to the control group). |
Expression of miRNA profiling in MHCC-97
cells with or without treatment of CRAE
In order to delineate the responses of miRNAs in the
MHCC97-L cells to CRAE treatment, miRNA-enriched total RNAs were
extracted from untreated MHCC97-L cells and cells treated with 175
μg/ml CRAE for 48 h. The values for each individual miRNA
probe sequence were printed in triplicates on the array. Cluster
analysis was conducted on the expression profiles of the control
and experimental groups. The NCode™ Human miRNA Microarray V3 chip
was used. Those miRNAs which exhibited significant changes
(p<0.05) are shown in Fig. 2.
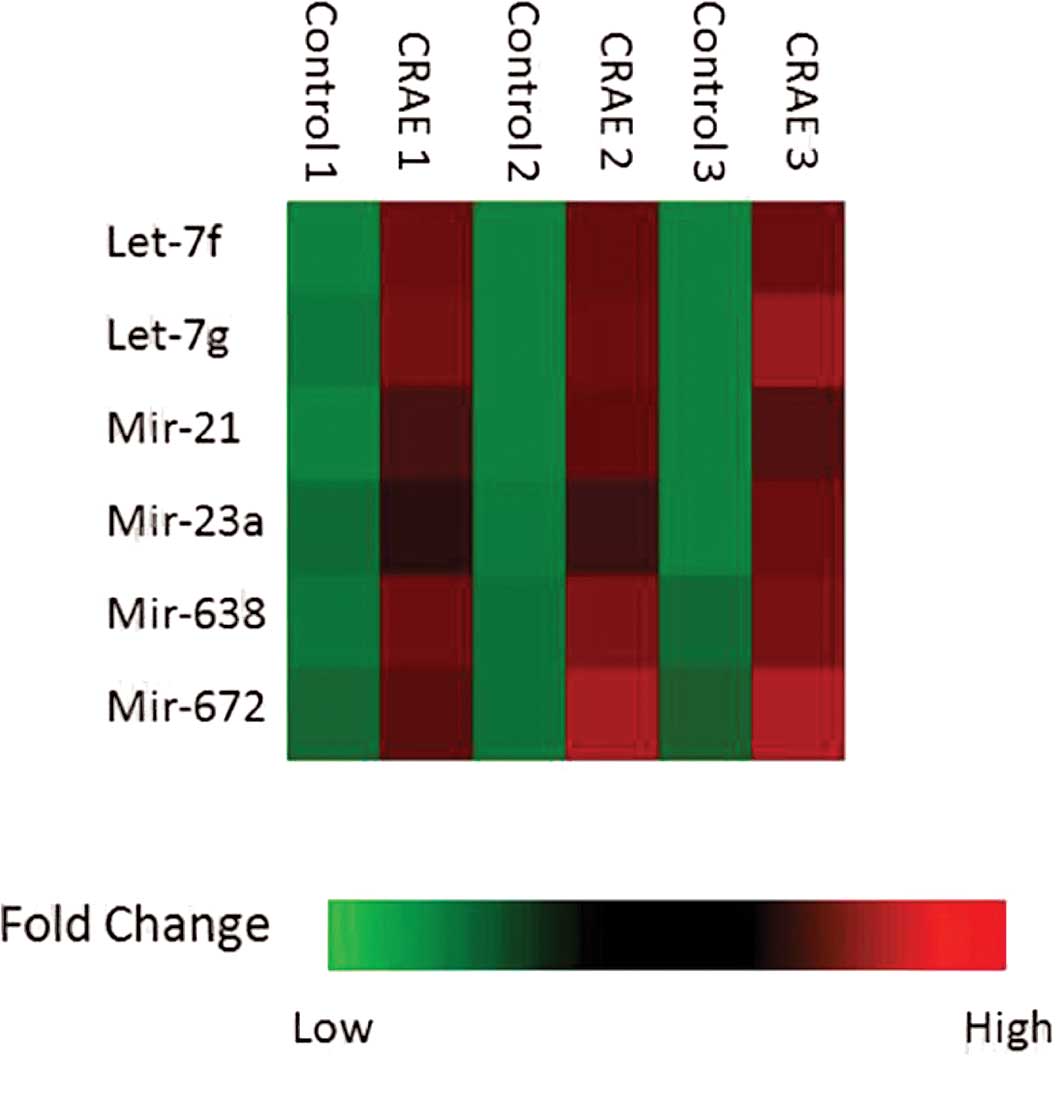
Six miRNAs, including hsa-let-7f, hsa-let-7g, hsa-mir-21,
hsa-mir-23a, hsa-mir-638 and has-mir-672, were strikingly
up-regulated after 175 μg/ml CRAE treatment for 48 h as
shown in Fig. 3.
Validation of the differential expression
of miRNAs by qRT-PCR
The expression levels of the six miRNAs were
determined by qRT-PCR using SYBR® Green 1 with U6 RNA as
an endogenous control. The same miRNA-enriched RNAs were reversely
transcribed and amplified in a 96-well plate for SYBR®
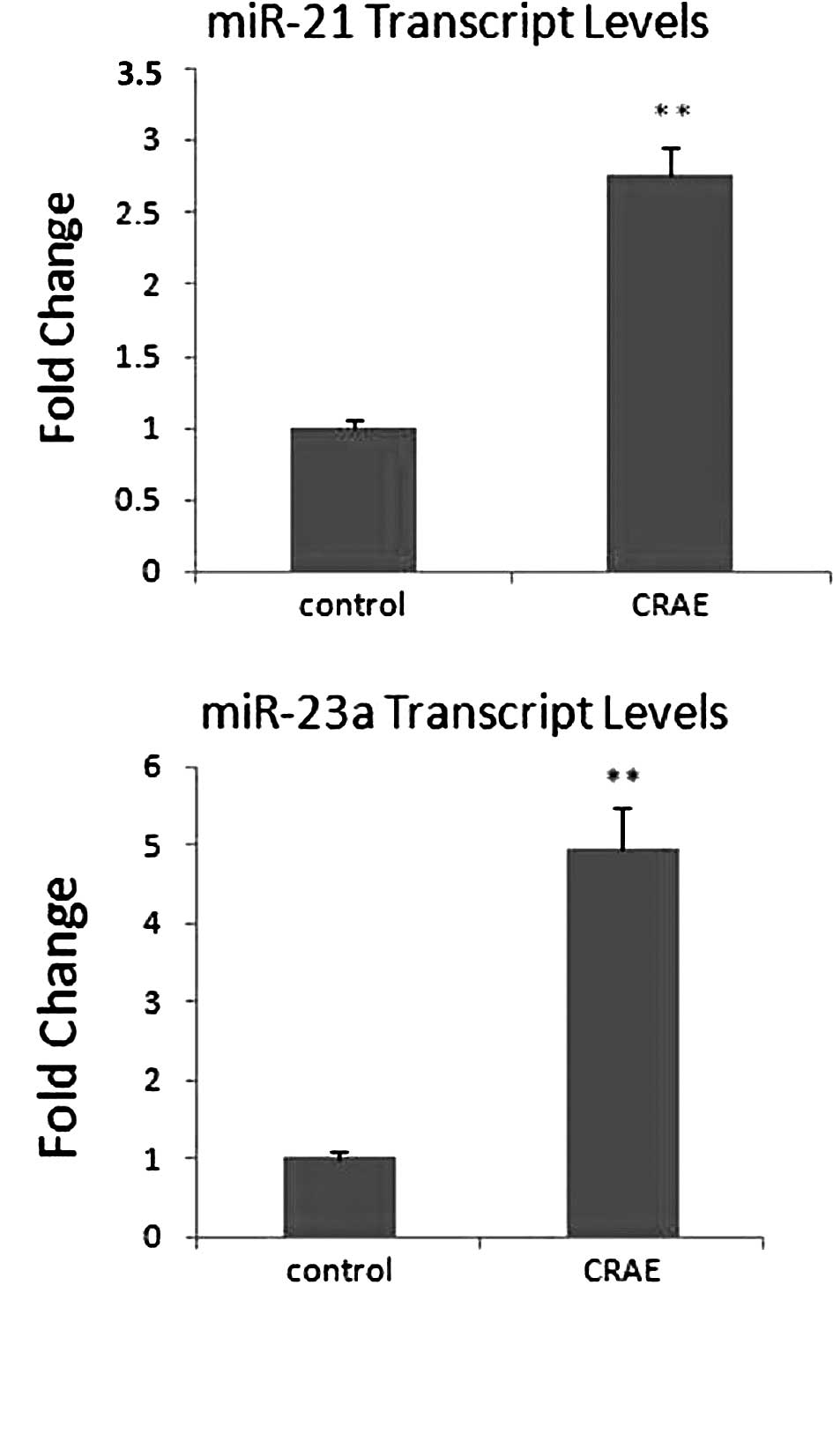
Green 1 real-time PCR analysis. Among these six miRNAs, elevated
expression levels of miR-21 and miR-23a after CRAE treatment were
confirmed. A 3-fold up-regulation of miR-21 expression was detected
in the MHCC97-L cells treated with 175 μg/ml CRAE in
comparison to the control group (Fig.
4A). Strikingly, the transcription level of miR-23a was
increased 5-fold in the MHCC97-L cells treated with CRAE (Fig. 4B). However, alteration in the
expression levels of hsa-let-7f, hsa-let-7g, hsa-mir-638 and
hsa-mir-672 was not confirmed in the qRT-PCR analysis. These
results revealed that miR-21 and miR-23a expression was
significantly up-regulated by 175 μg/ml CRAE treatment.
Prediction of potential target genes of
miR-21 and miR-23a
To predict the function of the selected miRNAs
related to the anti-tumor action of CRAE, identification of the
target genes were attempted using related miRNA databases. Two
miRNA databases, TargetScan (http://www.targetscan.org/index.html) and PicTar
(http://www.pictar.mdc-berlin.de), were
searched according to the manufacturer’s instructions, and several
target genes which were common to both TargetScan and PicTar were
identified. The most promising five candidate targets for miR-21
and miR-23a are listed in Tables I
and II, respectively.
 | Table I.Prediction of miR-21 target
genes. |
Table I.
Prediction of miR-21 target
genes.
| Target gene | Accession no. | Target gene
name |
|---|
| PLAG1 | NM_002655 | Homo sapiens
pleiomorphic adenoma gene 1 |
| RP2 | NM_006915 | Homo sapiens
retinitis pigmentosa 2 (X-linked recessive) |
| STAT3 | NM_139276 | Homo sapiens signal
transducer and activator of transcription 3 (acute-phase response
factor) |
| SATB1 | NM_002971 | Homo sapiens
special AT-rich sequence binding protein 1 (binds to nuclear
matrix/scaffold-associating DNA’s) |
| NTF3 | NM_002527 | Homo sapiens
neurotrophin 3 |
 | Table II.Prediction of miR-23a target
genes. |
Table II.
Prediction of miR-23a target
genes.
| Target gene | Accession no. | Target gene
name |
|---|
| SEMA6D | NM_153618 | Homo sapiens sema
domain, transmembrane domain (TM), and cytoplasmic domain,
(semaphorin) 6D |
| POU4F2 | NM_004575 | Homo sapiens POU
domain, class 4, transcription factor 2 |
| NEK6 | NM_014397 | Homo sapiens NIMA
(never in mitosis gene a)-related kinase 6 |
| SLC6A14 | NM_007231 | Homo sapiens solute
carrier family 6 (neurotransmitter transporter), member 14 |
| SET8 | NM_020382 | Homo sapiens PR/SET
domain containing protein 8 |
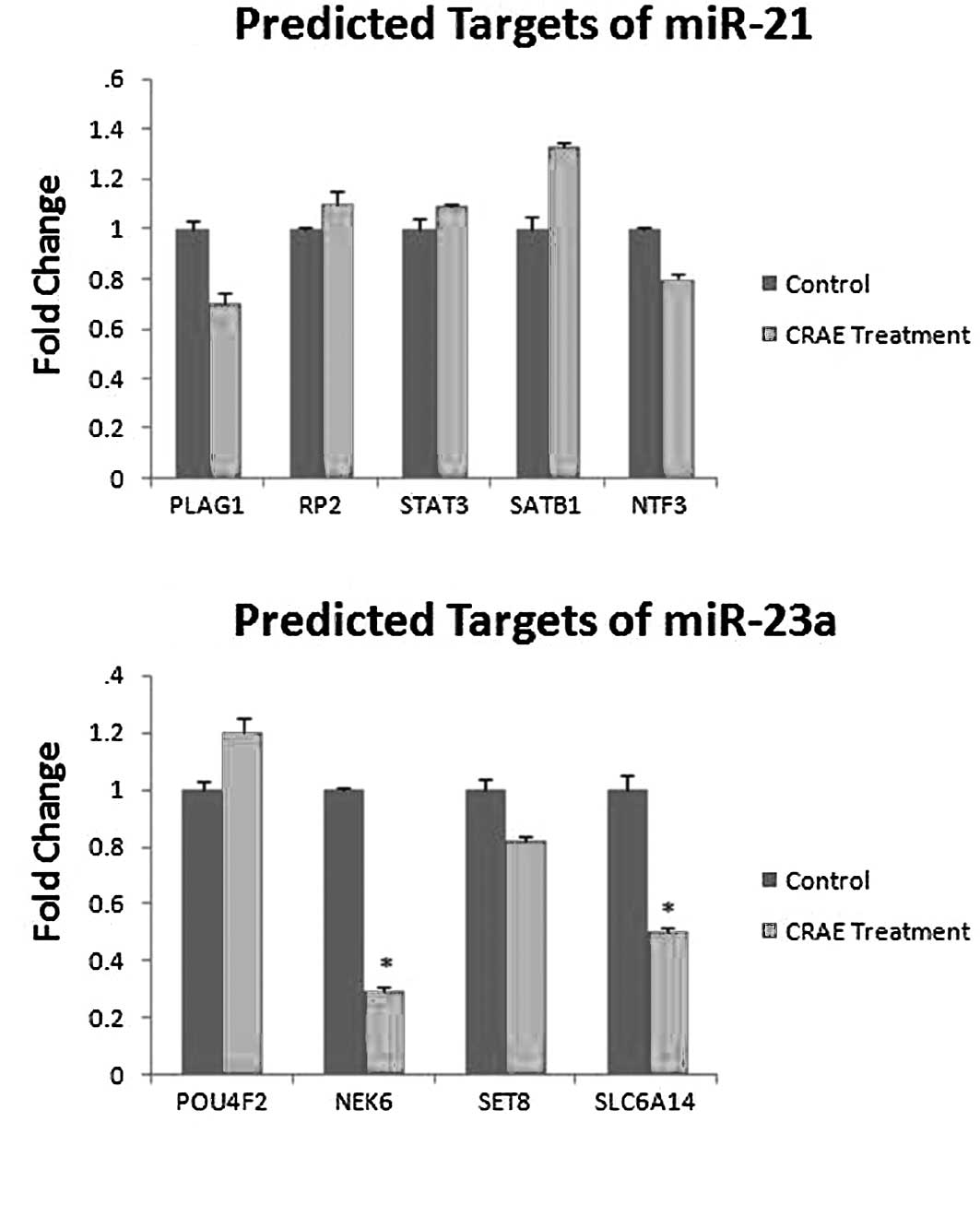
Validation of the predicted targets
The expression of PLAG1, RP2, STAT3, SATB1 and NTF3,
predicted target genes of miR-21, and POU4F2, NEK6, SLC6A14 and
SET8, predicted target genes of miR-23a, was determined by qRT-PCR
using SYBR® Green 1 with GAPDH as an endogenous control.
We found that only the expression of NEK6 and SLC6A14 exhibited a
significant change after treatment with 175 μg/ml CRAE,
while the predicted targets of miR-21 exhibited no change (Fig. 5A and B).
Discussion
Discovered in 1993 for the first time (4), miRNA has been shown to play a crucial
role in the regulation of physiological processes in cell biology
(12,13). Moreover, evidence supports the
involvement of miRNA in the initiation and progression of human
cancers. For example, miR-155 is overexpressed in breast cancer and
has the potential to be a target of breast cancer therapy (14). miR-126, together with epidermal
growth factor-like domain 7, inhibit proliferation in lung cancer
(15). miR-15a and miR-16 are
underexpressed in ovarian cancer by down-regulating the expression
of bim-1 (16). Bhattacharyya
et al (17) compared the
levels of miRNA in tumor cells and adjacent non-tumor cells from
liver cancer patients with a history of hepatitis. They reported
that patients with poor disease-free survival had low overall
levels of 19 particular miRNAs compared to those showing better
survival after 16 years of follow-up. A unique pattern of miRNA
expression is found in different hematopoietic cell lineages and
plays a role in their proliferation and differentiation (17). All the currently available data
confirm the involvement of miRNAs in cancer progression and
demonstrate the potential of miRNAs as markers for diagnosis,
prognosis and perspective treatment targets for cancer therapy.
Medicinal plants have had an extensive history of
use in cancer therapy, and plant-derived compounds have played an
important role in the development of various clinically useful
anti-cancer agents (18,19). Coptidis rhizoma (CR) is a
species of goldthread native to China (Botanical Dermatology
Database; http://bodd.cf.ac.uk) and one of the 50
fundamental herbs used in traditional Chinese medicine (20). It has been shown that CR is a
promising Chinese herb used in cancer therapy; however,
comprehensive studies on the molecular mechanism of the anti-tumor
action of CRAE are limited (10,21).
We previously reported that CR had the strongest cytotoxicity among
a selection of 16 anti-cancer Chinese herbs in rat leukemia L-1210
cells, and a much higher inhibitory activity for growth of tumor
cells was present in the water extract of CR, compared to extracts
using other solvents (22). Our
previous studies revealed that CRAE and its active compound,
berberine, are promising anti-cancer drugs due to their low
toxicity and liver protective properties (9–11,18,21).
In the present study, we confirmed that regulation of certain
specific miRNAs may also contribute to an anti-cancer effect
(23,24). The microarray results revealed a
differential expression profile for various miRNAs between cells
treated with and without CRAE. The six potential miRNAs were
further evaluated by qRT-PCR, indicating that CRAE treatment
up-regulates the expression of miR-21 and miR-23a. Previous reports
have revealed that miRNA expression profiles are influenced by the
intervention of natural products isolated from medicinal plants,
such as curcumin (25), isoflavone
(26), indole-3-carbinol,
3,3′-diindolylmethane (27),
(-)-epigallocatechin-3-gallate (28) and resveratrol (29). Our results indicate that herbal
extract CRAE has unique effects on the miRNA expression profile in
HCC cells.
Prediction of the potential target genes of the
selected miR-21 and miR-23a was conducted through a search of the
TargetScan and PicTar databases. These genes may contribute to the
development of anti-cancer drugs and cancer-targeted therapy. We
found that the expression of NEK6 and SLC6A14 in MHCC97-L cells
decreased after treatment with 175 μg/ ml CRAE. NEK6 encodes
a serine or threonine kinase which mediates the initiation of
mitosis. SLC6A14 is a member of the Na(+) and Cl(−)-dependent
neurotransmitter transporter family. Certain related target genes
may play a critical role in the development of CRAE as an
anti-cancer agent; this warrants further study. The tumor
suppressor tropomyosin 1 (TPM1) was previously identified as a
potential miR-21 target. TPM1 is an isoform of tropomyosin which
belongs to a family of actin filament-binding proteins. A binding
site for miR-21 was identified in the 3-UTR of the TPM2 transcript
and this was shown to be necessary for miR-21-mediated
translational repression (30).
Another target of miR-21 that has been recently identified in
breast cancer cell lines is the tumor-suppressor gene, programmed
cell death-4 (PDCD4) (31). miR-21
appears to down-regulate PDCD4 at both the mRNA and protein levels,
and this is at least partly responsible for some of the effects of
miR-21 on cellular proliferation. In addition, miR-21 may interfere
with the expression of p63 and may function as a tumor-suppressor
gene in some types of cancers (32).
In conclusion, the miRNA expression profiling in the
human HCC MHCC97-L cells after treatment with CRAE was determined
by miRNA on-chip array and qRT-PCR. Strikingly, overexpression of
miR-21 and miR-23a in the MHCC97-L cells was detected after
exposure to 175 μg/ml of CRAE for 48 h. Prediction of the
target genes of these selected miRNAs indicated that several
tumor-suppressor genes may be affected by CRAE through the altered
expression of miRNAs. To the best of our knowledge, up-regulation
of miRNAs, miR-21 and miR-23a, in human liver cancer cells treated
with CRAE has not been previously reported in the literature. Our
present findings may shed light on the novel target of CRAE in
cancer therapy; however, further studies on the function of miR-21
and miR-23a, as well as their potential target genes, are
warranted.
Acknowledgements
The present study was financially
supported by grants from the research council of the University of
Hong Kong (project code, 200907176140), the Research Grants Council
(RGC) of Hong Kong SAR, China (project code, 764708M), the Pong
Ding Yueng Endowment Fund for Education and Research in
Chinese-Western Medicine (project code, 20005274) and the Hong Kong
Government-Matching Grant Scheme (4th Phase, project code,
20740314). The cell line MHCC97-L was a kind gift from the Liver
Cancer Institute Fudan University, Shanghai, China. The authors
would like to express thanks to Dr Ka-Yu Siu, Ms. Cindy Lee, Mr.
Keith Wong and Mr. Freddy Tsang for the technical support.
References
|
1.
|
Cahill BA and Braccia D: Current treatment
for hepatocellular carcinoma. Clin J Oncol Nurs. 8:393–399. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
3.
|
Lee RC, Feinbaum RL and Ambros V: The
C. Elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
4.
|
Ruvkun G: Molecular biology: glimpses of a
tiny RNA world. Science. 294:797–799. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Chang HM and Paul But PH: Pharmacology and
Applications of Chinese Materia Medica. World Scientific
Publishing; Singapore: pp. 1061–1077. 2001
|
|
8.
|
Kim HY, Shin HS, Park H, Kim YC, Yun YG,
Park S, Shin HJ and Kim K: In vitro inhibition of coronavirus
replications by the traditionally used medicinal herbal extracts,
Cimicifuga rhizoma, Meliae cortex, Coptidis rhizoma and
Phellodendron cortex. J Clin Virol. 41:122–128. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Ye X, Feng Y, Tong Y, Ng KM, Tsao S, Lau
GK, Sze C, Zhang Y, Tang J, Shen J and Kobayashi S:
Hepatoprotective effects of Coptidis rhizoma aqueous extract
on carbon tetrachloride-induced acute liver hepatotoxicity in rats.
J Ethnopharmacol. 124:130–136. 2009.
|
|
10.
|
Tang J, Feng Y, Tsao S, Wang N, Curtain R
and Wang Y: Berberine and Coptidis rhizoma as novel
antineoplastic agents: a review of traditional use and biomedical
investigations. J Ethnopharmacol. 126:5–17. 2009.
|
|
11.
|
Tsang CM, Lau EP, Di K, Cheung PY, Hau PM,
Ching YP, Wong YC, Cheung AL, Wan TS, Tong Y, Tsao SW and Feng Y:
Berberine inhibits Rho GTPases and cell migration at low doses but
induces G2 arrest and apoptosis at high doses in human cancer
cells. Int J Mol Med. 24:131–138. 2009.PubMed/NCBI
|
|
12.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Li Y, Zhu X, Gu J, Dong D, Yao J, Lin C,
Huang K and Fei J: Anti-miR-21 oligonucleotide sensitizes leukemic
K562 cells to arsenic trioxide by inducing apoptosis. Cancer Sci.
101:948–954. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu
H, Liu MF and Wang ED: MicroRNA-155 functions as an oncomiR in
breast cancer by targeting the suppressor of cytokine signaling 1
gene. Cancer Res. 70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Sun Y, Bai Y, Zhang F, Wang Y, Guo Y and
Guo L: miR-126 inhibits non-small cell lung cancer cell
proliferation by targeting EGFL7. Biochem Biophys Res Commun.
391:1483–1489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bhattacharya R, Nicoloso M, Arvizo R, Wang
E, Cortez A, Rossi S, Calin GA and Mukherjee P: MiR-15a and MiR-16
control Bmi-1 expression in ovarian cancer. Cancer Res.
69:9090–9095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Bhattacharyya SN, Habermacher R, Martine
U, Closs EI and Filipowicz W: Relief of microRNA-mediated
translational repression in human cells subjected to stress. Cell.
125:1111–1124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Feng Y, Cheung KF, Wang N, Liu P,
Nagamatsu T and Tong Y: Chinese medicines as a resource for liver
fibrosis treatment. Chin Med. 4:162009.PubMed/NCBI
|
|
19.
|
Cragg GM and Newman DJ: Plants as a source
of anti-cancer agents. J Ethnopharmacol. 100:72–79. 2005.PubMed/NCBI
|
|
20.
|
Yang LQ, Singh M, Yap EH, Ng GC, Xu HX and
Sim KY: In vitro response of Blastocystis hominis against
traditional Chinese medicine. J Ethnopharmacol. 55:35–42. 1996.
|
|
21.
|
Feng Y, Luo WQ and Zhu SQ: Explore new
clinical application of Huanglian and corresponding compound
prescriptions from their traditional use. Zhongguo Zhong Yao Za
Zhi. 33:1221–1225. 2008.PubMed/NCBI
|
|
22.
|
Luo WQ, Hui SC, Chan TY and Feng Y:
Inhibitory effect of water extract from golden thread (Huanglian)
on leukemia L-1210 cells cultured in vitro. Pharmacologist.
44:1262002.
|
|
23.
|
Gao P, Tchernyshyov I, Chang TC, Lee YS,
Kita K, Ochi T, Zeller KI, De Marzo AM, van Eyk JE, Mendell JT and
Dang CV: c-Myc suppression of miR-23a/b enhances mitochondrial
glutaminase expression and glutamine metabolism. Nature.
458:762–765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Sun M, Estrov Z, Ji Y, Coombes KR, Harris
DH and Kurzrock R: Curcumin (diferuloylmethane) alters the
expression profiles of microRNAs in human pancreatic cancer cells.
Mol Cancer Ther. 7:464–473. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Li Y and Sarkar FH: Down-regulation of
invasion and angiogenesis-related genes identified by cDNA
microarray analysis of PC3 prostate cancer cells treated with
genistein. Cancer Lett. 186:157–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Garikapaty VP, Ashok BT, Tadi K, Mittelman
A and Tiwari RK: 3,3′-Diindolylmethane downregulates pro-survival
pathway in hormone independent prostate cancer. Biochem Biophys Res
Commun. 340:718–725. 2006.
|
|
28.
|
Katiyar SK, Afaq F, Perez A and Mukhtar H:
Green tea polyphenol (-)-epigallocatechin-3-gallate treatment of
human skin inhibits ultraviolet radiation-induced oxidative stress.
Carcinogenesis. 22:287–294. 2001. View Article : Google Scholar
|
|
29.
|
Kundu JK and Surh YJ: Cancer
chemopreventive and therapeutic potential of resveratrol:
mechanistic perspectives. Cancer Lett. 269:243–261. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Papagiannakopoulos T, Shapiro A and Kosik
KS: MicroRNA-21 targets a network of key tumor-suppressive pathways
in glioblastoma cells. Cancer Res. 68:8164–8172. 2008. View Article : Google Scholar : PubMed/NCBI
|