Introduction
Lung cancer is the leading cause of cancer-related
death in the world (1). Despite
various advances in anticancer therapy, survival rates have not
improved during the last decade, and long-term survival remains
very poor (2–4). Local relapse may occur after surgical
removal of the primary tumor, and distant metastasis is not
unusual, arising from micrometastases that are undetectable when
the primary tumor is diagnosed. Thus, exploring factors that are
useful for predicting the progression and outcome of lung cancer is
critical.
LIM-domain only protein 7 (LMO7) is a fibrous
actin-binding protein that is widely expressed in adult tissues,
particularly at the apical surface of lung epithelial cells
(5). LMO7 is a member of a family
of nine proteins containing both PDZ and LIM domains that function
as protein-protein recognition modules (6,7). The
PDZ and LIM families are involved in forming the Z-band of muscles
through PDZ domains that bind to α-actinin or β-tropomyosin
(8). LMO7 is also involved in the
process of gene expression by acting as a nucleocytoplasmic shuttle
protein that regulates the transcription of emerin and
muscle-related genes (9).
Moreover, a yeast two-hybrid study demonstrated that LMO7 binds
afadin, the adaptor protein of nectins, at adherens junctions
through the LIM domain (8). These
observations suggest a role for LMO7 in the formation and
maintenance of epithelial architecture via remodeling of the actin
cytoskeleton.
On the other hand, a role of LMO7 in cancer
pathology has been well documented. The P100 LMO7 splice variant
(with a truncated C-terminal region) was originally identified by
subtraction and differential hybridization in Yoshida hepatoma
AH130W1 cells treated with transforming growth factor-β (TGF-β)
(10). TGF-β induced alternative
splicing of the LMO7 gene, as well as promoted the migration
of AH130W1 cells in an in vitro invasion assay (11,12).
In addition, increased expression of LMO7 (also known as pancreatic
cancer derived 1; PCD1) has been reported in cancer of the
colorectal region, breast, liver, lung, pancreas, stomach and
prostate, suggesting that PCD1 may play a role in cytoskeletal
reorganization during carcinogenesis (13–15).
Furthermore, the LMO7 gene is located on chromosome 13q22,
which is implicated in hereditary breast cancer (16,17),
although it remains controversial whether LMO7 is the only gene
responsible (16–18). Finally, LMO7-deficient mice
develop irregular and protruding epithelial lesions in the terminal
and respiratory bronchioles at a young age, and these mice tend to
develop lung adenocarcinoma at an older age, suggesting that LMO7
acts as a tumor-suppressor gene and that its deficiency confers a
genetic predisposition to lung cancer (5). Despite these findings, the role of
LMO7 in human lung carcinogenesis has yet to be studied.
We investigated the level of LMO7 protein expression
and its pattern of expression by immunohistochemistry in surgically
resected tissues obtained from 57 patients with primary lung
adenocarcinoma. We also analyzed how LMO7 expression in tumor
tissues influenced the progression of cancer and the prognosis of
these patients.
Materials and methods
Surgical specimens
A total of 57 formalin-fixed samples of primary lung
adenocarcinoma and adjacent normal lung tissues were obtained along
with clinicopathological data from patients who underwent surgery
at the Osaka Medical Center for Cancer and Cardiovascular Diseases
(Osaka, Japan) between April 2000 and July 2002. All patients
underwent potentially curative surgery without perioperative
adjuvant therapy. Further treatment was administered only when the
cancer recurred. The patients consisted of 31 men and 26 women,
42–79 years of age (mean 61.9).
Histologic classification of the tumor specimens was
based on the WHO criteria. Eight patients had tumors with an acinar
growth pattern, 42 had a papillary tumor pattern, 2 had
bronchioloalveolar tumors and 5 had solid tumors with mucin
formation (19). All tumors were
staged according to the pTNM pathological classification of the
International Union Against Cancer (20). There were 11 patients with p-stage
IA, 18 with p-stage IB, 1 with p-stage IIA, 9 with p-stage IIB, 10
with p-stage IIIA and 8 with p-stage IIIB disease. The median
postoperative follow-up period was 73 months (range 3–115).
Immunohistochemistry
To investigate LMO7 protein expression, thin
sections were cut from 10% formalin-fixed and paraffin-embedded
blocks of the surgical specimens, and were stained with a rabbit
anti-LMO7 antibody (clone #863) as previously described (5). This antibody was specific for a
recombinant protein containing amino acid residues of rat p100
#16/LMO7 (GeneBank accession no. AY609384), which was constructed
as a fusion protein with glutathione S transferase (GST) using the
pGEX plasmid vector (GE Healthcare UK Ltd., Buckinghamshire, UK)
and was then employed as an immunogen.
Sections (4 μm) of tissues were mounted on
poly-L-lysine-coated slides, air-dried and deparaffinized. Then,
endogenous peroxidase activity was blocked by incubation with 5%
hydrogen peroxide in 50% methanol for 20 min at room temperature.
Subsequently, antigen retrieval was performed by autoclaving at
120°C for 3 min in 10 mM citrate buffer. After blocking
non-specific binding by incubation with 5% skim milk in PBS for 60
min at room temperature, the sections were incubated with
polyclonal anti-LMO7 antibody overnight at 4°C. After rinsing with
PBS, the sections were incubated with biotinylated horse
anti-rabbit IgG (Vector, Burlingame, CA, USA) for 30 min at room
temperature, followed by washing with PBS. Immunoreactivity was
detected with an avidin-biotin system (NovaRED™; Vector). In every
control, sections were incubated with a 5-fold excess of GST-LMO7
fusion protein. Adjacent non-cancerous pulmonary tissues were also
examined as an internal positive control for LMO7 protein
expression.
Classification of immunohistochemical
findings
LMO7 expression was primarily detected in the
membranes of non-cancerous cells, such as pneumocytes, and some
bronchial epithelial cells, as well as in cancer cells. Certain
cancer cells also showed strong cytoplasmic immunostaining for
LMO7. Immunopositivity for LMO7 expression was classified on the
basis of the staining intensity. That is, cells with stronger
immunostaining were judged to be LMO7-positive, whereas cells with
weak or no immunostaining were classed as LMO7-negative.
When the percentage of LMO7-positive cancer cells in
a tumor specimen was ≥50%, the tumor was classified as LMO7
‘positive’, while tumors with <50% positive cells were
characterized as having low LMO7 expression. These semiquantitative
assessments were carried out by two independent investigators (H.N.
and K.H.) without knowledge of the clinicopathological data.
Statistical analysis
Associations between LMO7 immunostaining and
clinicopathological factors were assessed by the χ2
test, except age which was assessed by the Student’s t-test.
Univariate and multivariate analyses of the clinicopathological
factors associated with LMO7 expression were performed by the
logistic regression method. Survival curves were calculated from
the date of surgery to the time of death (or to final follow-up)
according to the Kaplan-Meier method (21), and differences in survival among
subgroups were analyzed by the log-rank test (22). Univariate and multivariate analyses
of the influence of variables on overall survival were performed
with the Cox proportional hazards regression model. Statistical
analyses were carried out with SAS software (Cary, NC, USA), and
p<0.05 was considered significant.
The surgical samples were obtained from patients
after providing informed consent. This study and the use of
clinical materials were approved by the relevant institutional
research ethics committees. For protection of privacy, identifying
information was removed from all samples before analysis in
accordance with the Ethical Guidelines for Human Genome/Gene
Research of the Japanese Government.
Results
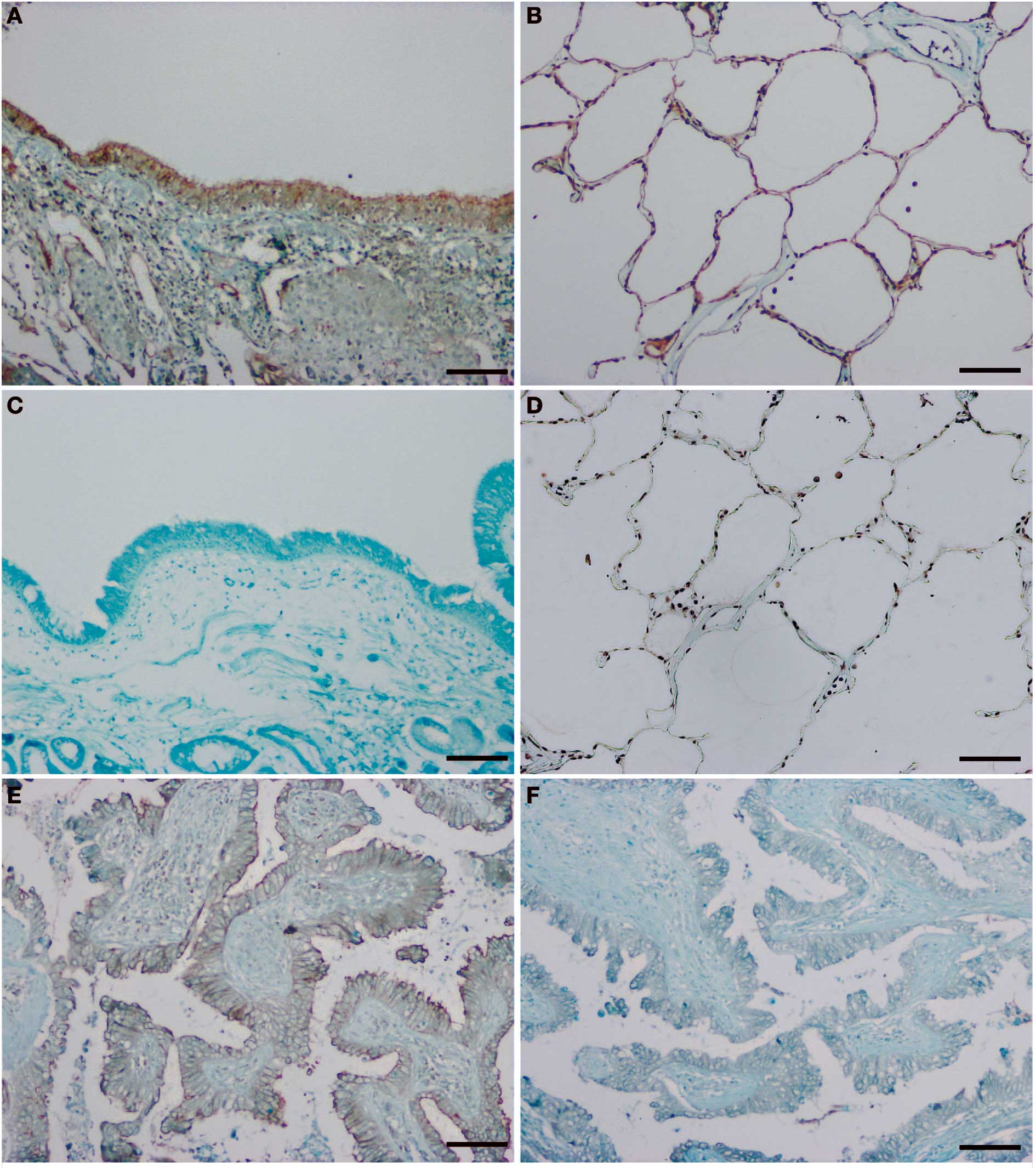
Immunohistochemistry for LMO7
We investigated the level and pattern of LMO7
protein positivity by immunohistochemistry of human lung
adenocarcinoma specimens because of our previous findings regarding
the expression and deficiency of LMO7 in mice (5). Normal bronchioalveolar epithelial
cells showed uniformly intense LMO7 positivity. LMO7 was usually
localized to the apical membranes of cells from the bronchial
epithelium (Fig. 1A), while normal
alveolar cells showed circumferential staining of the entire cell
membrane (Fig. 1B). The antibody
was confirmed to be specific for LMO7 protein because of the
complete lack of immunostaining in the adjacent sections after
pre-incubation with a 5-fold excess of GST-LMO7 fusion protein
(Fig. 1C and D). Certain stromal
cells, including the smooth muscle cells of blood vessels and
vascular endothelial cells, were also stained.
On the other hand, the adenocarcinoma cells of all
57 patients in this study showed circumferential LMO7 staining of
the plasma membrane (Fig. 1E).
Several patients also had tumor cells with LMO7 expression in the
cytoplasm or in part of the apical region. Immunostaining for LMO7
was also abolished by pre-incubation with a 5-fold excess of
GST-LMO7 fusion protein (Fig.
1F).
Correlation of decreased LMO7 expression
with a poor clinicopathological outcome
To investigate the biological and
clinicopathological significance of LMO7, we evaluated the relative
intensity of LMO7 staining of the tumor specimens, and classified
33 patients (58%) and 24 patients (42%) into a low LMO7 group and
an LMO7-positive group, respectively, as described in Materials and
methods. The relationship between LMO7 expression and various
clinicopathological factors is summarized in Table I. Patients in the low LMO7 group
had significantly more advanced tumors than those in the
LMO7-positive group with regard to T factor (p=0.011), nodal
involvement (p=0.026) and p-stage (p=0.010). There was no
significant relation between LMO7 expression and tumor histology
(p=0.9580; χ2 test).
 | Table IAssociation between LMO7 expression
and clinico-pathological factors. |
Table I
Association between LMO7 expression
and clinico-pathological factors.
| LMO7
expressiona
| p-value |
|---|
| Positive (n=24) | Low (n=33) |
|---|
| Age (mean ± SD;
years) | 62.2±10.2 | 61.7±11.3 | NS |
| Gender | | | |
| Male | 13 | 18 | |
| Female | 11 | 15 | 0.977 |
| T factor |
| T1 | 11 | 5 | |
| T2 | 11 | 19 | |
| T3 and T4 | 2 | 9 | 0.011c (T1 vs. T2-4) |
| Nodal
involvement | | | |
| N0 | 18 | 15 | |
| N1 | 4 | 5 | |
| N2 and N3 | 2 | 13 | 0.026b (N0 vs. N1-3) |
| Histological
differentiation | | | |
| Well, moderate | 21 | 27 | |
| Poor | 3 | 6 | 0.561 |
| p-stage | | | |
| IA and IB | 17 | 12 | |
| IIA and IIB | 4 | 6 | |
| IIIA and IIIB | 3 | 15 | 0.010c (I vs. II and III) |
Logistic regression analysis was performed with the
outcome variable being the LMO7 level (‘positive’ vs. ‘low’) and
the covariates being various clinicopathological factors. On
univariate analysis, the T factor and nodal involvement were
positively associated with the level of LMO7 expression. According
to multivariate analysis, LMO7 expression was independently
associated with the T factor at the time of surgery (p=0.041)
(Table II). The results revealed
that T1 status was linked to the LMO7-positive group, while T2-4
status was associated with lower expression of LMO7.
 | Table IIUnivariate and multivariate analyses
of prognostic indicators with the Cox proportional hazards
model. |
Table II
Univariate and multivariate analyses
of prognostic indicators with the Cox proportional hazards
model.
| Variables | Risk ratio (95%
CI) | p-value |
|---|
| Univariate model | | |
| T factor (T1 vs.
T2-4) | 2.893
(1.116–9.855) | 0.027a |
| Nodal involvement
(N0 vs. N1-3) | 2.276
(1.092–4.896) | 0.028a |
| Histological
differentiation (well, moderate vs. poor) | 3.226
(1.255–7.358) | 0.017b |
| LMO7 expression
(positive vs. low) | 2.348
(1.071–5.676) | 0.033a |
| Multivariate
model | | |
| T factor (T1 vs.
T2-4) | 2.996
(1.085–10.642) | 0.033a |
| Nodal involvement
(N0 vs. N1-3) | 1.509
(0.698–3.344) | 0.296 |
| Histological
differentiation (well, moderate vs. poor) | 3.889
(1.424–9.612) | 0.010b |
| LMO7 expression
(positive vs. low) | 1.853
(0.813–4.629) | 0.146 |
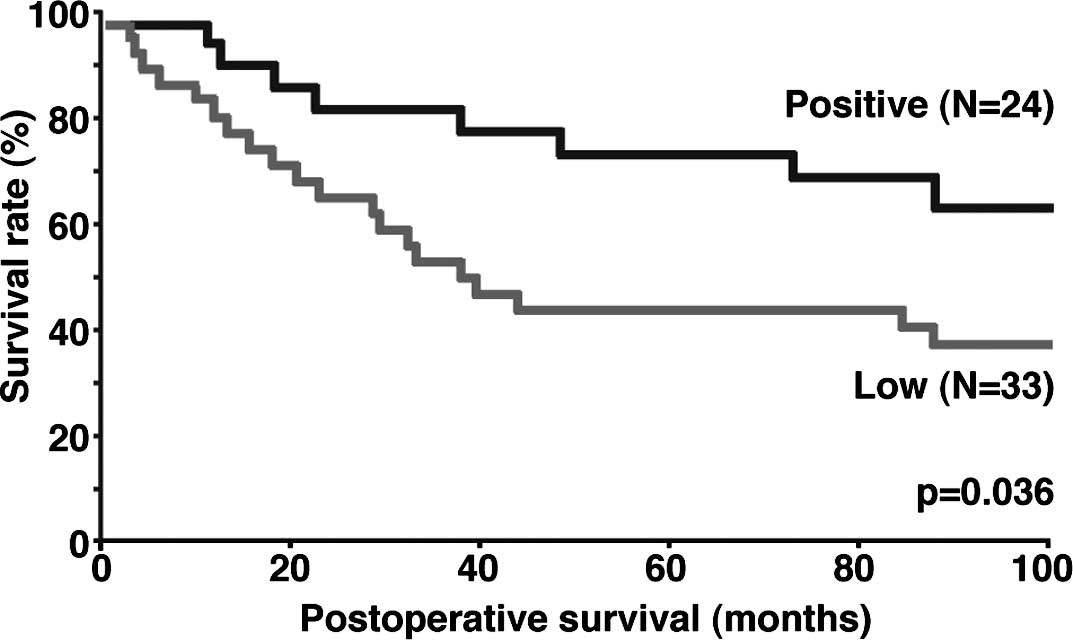
When postoperative overall survival curves were
drawn according to the level of LMO7 expression in the 57 patients
with lung adenocarcinoma, patients in the low expression group had
a significantly worse prognosis than those in the LMO7-positive
group (p=0.036) (Fig. 2).
When the Cox proportional hazards model was employed
for multivariate analysis, it showed that LMO7 expression was not
an independent prognostic factor, although LMO7 was significantly
associated with the prognosis according to univariate analysis
(Table II). However, the T factor
that was independently associated with LMO7 expression according to
logistic regression analysis (Table
III) was an independent prognostic indicator in the Cox
proportional hazards model (Table
II).
 | Table IIIResults of univariate and multivariate
logistic regression analyses. |
Table III
Results of univariate and multivariate
logistic regression analyses.
| Model | Odds ratio (95%
CI) | p-value |
|---|
| Univariate model | | |
| Age | 1.004
(0.956–1.056) | 0.867 |
| Gender (male vs.
female) | 0.985
(0.341–2.856) | 0.977 |
| T factor (T1 vs.
T2-4) | 0.211
(0.056–0.703) | 0.011b |
| Nodal involvement
(N0 vs. N1-3) | 0.278
(0.083–0.845) | 0.024a |
| Histological
differentiation(well, moderate vs. poor) | 0.643
(0.124–2.744) | 0.557 |
| Multivariate
model | | |
| T factor (T1 vs.
T2-4) | 0.262
(0.065–0.946) | 0.041a |
| Nodal involvement
(N0 vs. N1-3) | 0.391
(0.107–1.333) | 0.134 |
| Histological
differentiation (well, moderate vs. poor) | 0.734
(0.121–3.749) | 0.714 |
Discussion
To clarify the role of LMO7 in the pathogenesis of
human lung adenocarcinoma, we examined LMO7 protein expression with
an anti-LMO7 antibody that effectively detects LMO7 under a variety
of experimental conditions (5).
Our immunohistochemical analysis showed that LMO7 was localized
circumferentially in the plasma membrane of adenocarcinoma cells,
and that decreased expression of LMO7 was significantly correlated
with tumor progression and a poor prognosis of patients with lung
adenocarcinoma. LMO7 is mainly expressed by normal bronchiolar and
alveolar epithelial cells in the lungs of humans as well as in
mice, and a knockout study indicated that LMO7 acts as a tumor
suppressor for murine lung adenocarcinoma (5). Therefore, our findings are not only
consistent with earlier observations, but also demonstrate that
down-regulation of LMO7 expression is related to the
clinicopathological features of lung adenocarcinoma and to the
prognosis. Of course, this study did not exclude the possibility
that down-regulation of LMO7 expression could also be a useful
prognostic indicator for other types of cancers, particularly
hereditary human breast cancer (16–18).
The low LMO7 expression group was significantly
associated with more advanced tumors in the present study.
Multivariate logistic regression analysis showed that LMO7
expression was independently associated with the T factor.
Furthermore, Kaplan-Meier analysis of survival revealed a
significant difference between the LMO7-positive group and the low
expression group in the patients with lung adenocarcinoma. These
results support a potential influence of LMO7 on tumor progression
and the prognosis of human lung carcinoma. Although Cox
proportional hazards analysis failed to identify LMO7 expression as
an independent prognostic indicator, the T factor was an
independent variable.
Immunohistochemistry does not provide us with data
on the molecular mechanisms underlying changes in protein levels.
It is thus unclear whether LMO7 immunolabelling is correlated with
the level of native LMO7 protein, splice variants or mutant
protein. Although up-regulation of LMO7 has been observed by
immunohistochemistry in various types of human cancers (13–15),
the biological significance of these findings remains unknown. Our
studies have demonstrated that overexpression of P100 LMO7, a
splice variant induced by TGF-β, enhances the migration,
proliferation and invasion of MDCK cells (unpublished data). Based
on these oncogenic properties, up-regulation of LMO7 in various
types of cancers may reflect a role in tumor progression.
In conclusion, down-regulation of LMO7 is related to
the clinicopathological features of human lung adenocarcinoma and
to the prognosis, but it remains unclear whether LMO7 may be a
candidate for molecular-targeting therapy. Further studies are
required to elucidate the role of LMO7 in the pathogenesis of human
lung adenocarcinoma.
Abbreviations:
|
LMO7
|
LIM-domain only protein 7;
|
|
PCD1
|
pancreatic cancer derived 1;
|
|
TGF-β
|
transforming growth factor-β;
|
|
GST
|
glutathione S-transferase
|
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer Statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
2
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001.PubMed/NCBI
|
|
3
|
Naruke T, Tsuchiya R, Kondo H and Asamura
H: Prognosis and survival after resection for bronchogenic
carcinoma based on the 1997 TNM-staging classification: the
Japanese experience. Ann Thorac Surg. 71:1759–1764. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH: Comparison of
four chemotherapy regimens for advanced non-small cell lung cancer.
N Engl J Med. 346:92–98. 2002. View Article : Google Scholar
|
|
5
|
Tanaka-Okamoto M, Hori K, Ishizaki H,
Hosoi A, Itoh Y, Wei M, Wanibuchi H, Mizoguchi A, Nakamura H and
Miyoshi J: Increased susceptibility to spontaneous lung cancer in
mice lacking LIM-domain only 7. Cancer Sci. 100:608–616. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harris BZ and Lim WA: Mechanism and role
of PDZ domains in signaling complex assembly. J Cell Sci.
114:3219–3231. 2001.PubMed/NCBI
|
|
7
|
Kadrmas JL and Beckerle MC: The LIM
domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell
Biol. 5:920–931. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holaska JM, Rais-Bahrami S and Wilson KL:
Lmo7 is an emerin-binding protein that regulates the transcription
of emerin and many other muscle-relevant genes. Hum Mol Genet.
15:3459–3472. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ooshio T, Irie K, Morimoto K, Fukuhara A,
Imai T and Takai Y: Involvement of LMO7 in the association of two
cell-cell adhesion molecules, nectin and E-cadherin, through afadin
and α-actinin in epithelial cells. J Biol Chem. 279:365–373.
2004.
|
|
10
|
Nakamura H, Mukai M, Komatsu K,
Tanaka-Okamoto M, Itoh Y, Ishizaki H, Tatsuta M, Inoue M and
Miyoshi J: Transforming growth factor-β1 induces LMO7 while
enhancing the invasiveness of rat ascites hepatoma cells. Cancer
Lett. 220:95–99. 2005.
|
|
11
|
Akedo A, Shinkai K, Mukai M, Mori Y,
Tateishi R, Tanaka K, Yamamoto R and Morishita T: Interaction of
rat ascites hepatoma cells with cultured mesothelial cell layers: a
model for tumor invasion. Cancer Res. 46:2416–2422. 1986.PubMed/NCBI
|
|
12
|
Mukai M, Shinkai K, Komatsu K and Akedo H:
Potentiation of invasive capacity of rat ascites hepatoma cells by
transforming growth factor-β. Jpn J Cancer Res. 80:107–110.
1989.
|
|
13
|
Kang S, Xu H, Duan X, Liu J-J, He Z, Yu F,
Zhou S, Meng X-Q, Cao M and Kennedy G: PCD1, a novel gene
containing PDZ and LIM domains, is overexpressed in several human
cancers. Cancer Res. 60:5296–5302. 2000.PubMed/NCBI
|
|
14
|
Furuya M, Tsuji N, Endoh T, Moriai R,
Kobayashi D, Yagihashi A and Watanabe N: A novel gene containing
PDZ and LIM domains, PCD1, is overexpressed in human colorectal
cancer. Anticancer Res. 22:4183–4186. 2002.PubMed/NCBI
|
|
15
|
Sasaki M, Tsuji N, Furuya M, Kondoh K,
Kamagata C, Kobayashi D, Yagihashi A and Watanabe N: PCD1, a novel
gene containing PDZ and LIM domains, is overexpressed in human
breast cancer and linked to lymph node metastasis. Anticancer Res.
23:2717–2721. 2003.PubMed/NCBI
|
|
16
|
Kainu T, Juo SH, Desper R, et al: Somatic
deletions in hereditary breast cancers implicate 13q21 as a
putative novel breast cancer susceptibility locus. Proc Natl Acad
Sci USA. 97:9603–9608. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rozenblum E, Vahteristo P, Sandberg T, et
al: A genomic map of a 6-Mb region at 13q21–q22 implicated in
cancer development: identification and characterization of
candidate genes. Human Genet. 110:111–121. 2002.PubMed/NCBI
|
|
18
|
Thompson D, Szabo CI, Mangion J, et al:
Evaluation of linkage of breast cancer to the putative BRCA3 locus
on chromosome 13q21 in 128 multiple case families from the breast
cancer linkage consortium. Proc Natl Acad Sci USA. 99:827–831.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Travis WD, Colby TV, Corrin B, Shimosato Y
and Brambilla E: Histological typing of lung and pleural tumors.
World Health Organization International Histological Classification
of Tumors. 3rd edition. Springer; Berlin: 1999
|
|
20
|
Sobin L and Wittekind CH: TNM
Classification of Malignant Tumours. 6th edition. Wiley-Liss; New
York: 2002
|
|
21
|
Kaplan EL and Meier P: Nonparametric
estimation for incomplete observations. J Am Stat Assoc.
53:457–481. 1958. View Article : Google Scholar
|
|
22
|
Peto R, Pike MC, Armitage P, Breslow NE,
Cox DR, Howard SV, Mantel N, McPherson K, Peto J and Smith PG:
Design and analysis of randomized clinical trials requiring
prolonged observation of each patient. II. Analysis and examples.
Br J Cancer. 35:1–39. 1977. View Article : Google Scholar
|