Introduction
Menin, encoded by the multiple endocrine
neoplasia type I (MEN1) gene, is a nuclear protein which is mutated
in patients with a dominantly inherited disorder characterized by
the appearance of combinations of tumors in a number of endocrine
tissues, including the pituitary and parathyroid glands and the
pancreas (1,2), and occasionally in non-endocrine
glands (3,4). The human MEN1 gene has been
identified by positional cloning and is localized to chromosome
11q13. The gene consists of 10 exons and codes for an mRNA 2.8 kb
in length (5). The protein product
of the MEN1 gene, menin, is highly conserved in organisms
from Drosophila to humans. As menin does not show a
clear homology to any known protein motifs, it has been challenging
to elucidate the mechanism by which menin acts as a tumor
suppressor (6).
Numerous studies have revealed that menin is
mainly located in the nucleus and possesses nuclear localization
signals (NLSs) in its carboxy-terminal region (7). It has also been reported that
menin is found in membrane and cytoplasmic fractions
(8), although at lower levels than
in nuclear fractions, indicating that menin may also play a
role outside of the nucleus. Multiple lines of evidence suggest
that menin, as a scaffold protein, interacts with
cytoskeletal proteins, including glial fibrillary acid protein
(GFAP), vimentin (8) and IQGAP1
(9). However, less is known
concerning the expression and biochemical function of menin
expression in non-endocrine cells. To elucide the mechanism by
which menin functions as a tumor suppressor, in the present
study, we report the detection of endogenously expressed
menin in 13 human cancer cell lines. In particular, we
further investigated the subcellular localization of menin
in cancer cell lines.
Materials and methods
Cell lines and cell culture
The following cell lines were obtained from the
Institute of Cell Biology of the Chinese Academy of Sciences
(Shanghai, China): AGS, BGC-823 and SGC-7901 (human gastric
cancer), Huh7 and Hep3B2.1–7 (liver cancer), MCF-7 and MDA-MB-231
(breast cancer), SW480 (colon cancer), SGH44 (brain glioma), SKOV-3
(ovarian cancer) and HeLa (cervical cancer). The transformed human
gastric epithelial cell line GES-1 was a gift from Dr Yong-Chang
Chen (Jiangsu University, China). The MNK-28 cell line was a gift
from Dr Xiao-Ying Li (Rui-Jin Hospital, Shanghai Jiao-Tong
University School of Medicine, Shanghai, China). The cells were
cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand
Island, NY, USA) supplemented with 10% newborn calf serum (NBCS;
Minhai Bio-engineering Co., Lanzhou, China) and maintained at 37°C
in a humidified atmosphere of 5% CO2. The medium was
changed every two days and the cells were subcultured until
reaching confluency.
RNA extraction and reverse-transcription
polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagents
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Total RNA (1 μg) was reverse transcribed into cDNA
using dNTPs (1 mM), 5X reverse transcription buffer (500 mM
Tris-HCL pH 8.3, 250 mM KCl, 50 mM MgCl2 and 50 mM DTT),
16 units RNasin and 2.5 units of AMV reverse transcriptase (Gibco
BRL, Life Technologies, Carlsbad, CA, USA). PCR was carried out in
a total volume of 20 μl, containing 2 μl 10X buffer, 0.4 μl dNTP,
0.5 μl primers, 1 μl cDNA, 0.2 μl Taq enzyme and 15.4 μl
H2O (Shenergy Biocolor Biological Science and Technology
Co., Shanghai, China). The specific primers were: menin
gene, forward 5′-GCCTGGGTAGTGTTTGGGC-3′ and reverse
5′-CACAGCGCATGTATGATCCTT-3′, product of 452 bp; β-actin, forward
5′-TGCGTGACATTAAGGAGAAG-3′ and reverse 5′-GCTCGTAGCTCTTCTCCA-3′,
product of 247 bp. Following an initial denaturation step of 5 min
at 95°C, 30 cycles of amplification for the primer pairs were
carried out. Each cycle included a denaturation step of 30 sec at
95°C, annealing for 1 min at 55°C and an elongation step of 1 min
at 72°C, with a final extension for 5 min at 72°C. The products
were separated on 1.5% agarose gel and the relative gene expression
was measured using a digital image (Perkin-Elmer, Wellesley, MA,
USA). The experiments were performed in triplicate and the mean
value was calculated.
Immunofluorescence microscopy
The cells grown on cover-slips were fixed with
freshly prepared paraformaldehyde (40 g/l in PBS) for 1 h prior to
being penetrated with 0.3% Triton X-100 and blocked with 3% bovine
serum albumin (BSA) in PBS. The cells were then incubated with the
primary antibody at 4°C overnight and then with the Cy3-conjugated
secondary antibody for 1 h at room temperature (RT), with three
washes following each incubation. The distribution of the target
protein in the cells was analyzed by fluorescence microscopy.
Preparation of nuclear and cytoplasmic
samples
The cells were extracted by Dounce homogenization in
HEM buffer (10 mmol/l HEPES pH 7.5, 2 mmol/l EDTA, 1 mmol/l
MgCl2) as described previously (10). The homogenate was centrifuged at
500 x g at 4°C for 5 min to obtain the nuclear proteins and the
supernatant was centrifuged at 37,000 x g at 4°C for 30 min. The
supernatant and the pellet from the second centrifugation are
referred as cytosol and membrane preparations, respectively. The
membrane preparation was washed twice with HEM buffer to remove
contaminating cytosol. The protein concentrations were determined
and equal amounts of protein from each preparation (30 μg) were
subjected to SDS-PAGE.
Western blotting
Sample proteins were run on 10% SDS-polyacrylamide
gels and transferred to a polyvinylidene difluoride membrane (PVDF,
Amersham, Piscataway, NJ, USA) by electronic transfer. The
membranes were blocked with 5% non-fat dried milk and then
incubated with an antibody against menin (dilution 1:200)
and GAPDH (dilution 1:5,000) overnight at 4°C and with the
secondary antibody for 1 h at RT, with three washes following each
incubation. Electrochemiluminescence reagents were used to show the
positive bands on the membrane. The bands on film were analyzed
with GeneSnap/Gene Tool software from Syngene (Cambridge, UK).
Statistical analysis
All of the numerical data are expressed as the mean
± SD. Statistical analysis was performed using SPSS 16.0 edition
program (SPSS Inc., Chicago, IL, USA) for ANOVA with the Scheffé
multiple comparison test. P<0.05 was considered to indicate a
statistically significant result.
Results
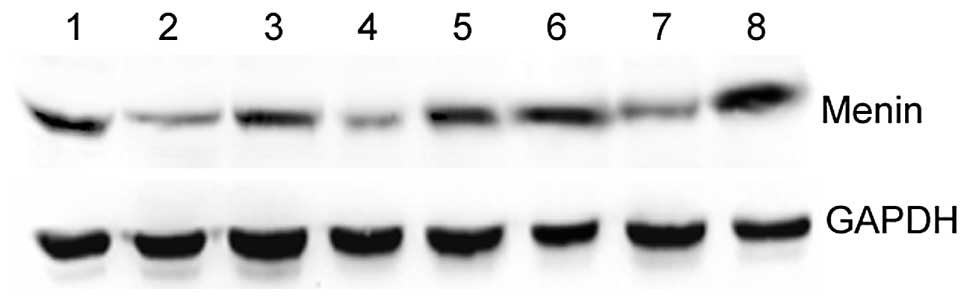
Expression of menin mRNA in human cancer
cells
As shown in Fig. 1,
menin mRNA was examined in 13 human cancer cell lines.
RT-PCR results revealed that menin was positively expressed
in all the cell lines examined in this experiment. In the gastric
cancer cell lines, the expression of menin mRNA was
gradually increased from AGS, BGC-823, SGC-7901 and MNK-28 to
GES-1. It was significantly higher in GES-1 than the other four
gastric cancer cell lines (P<0.05). The expression of
menin may be correlated with the malignancy of the
cells.
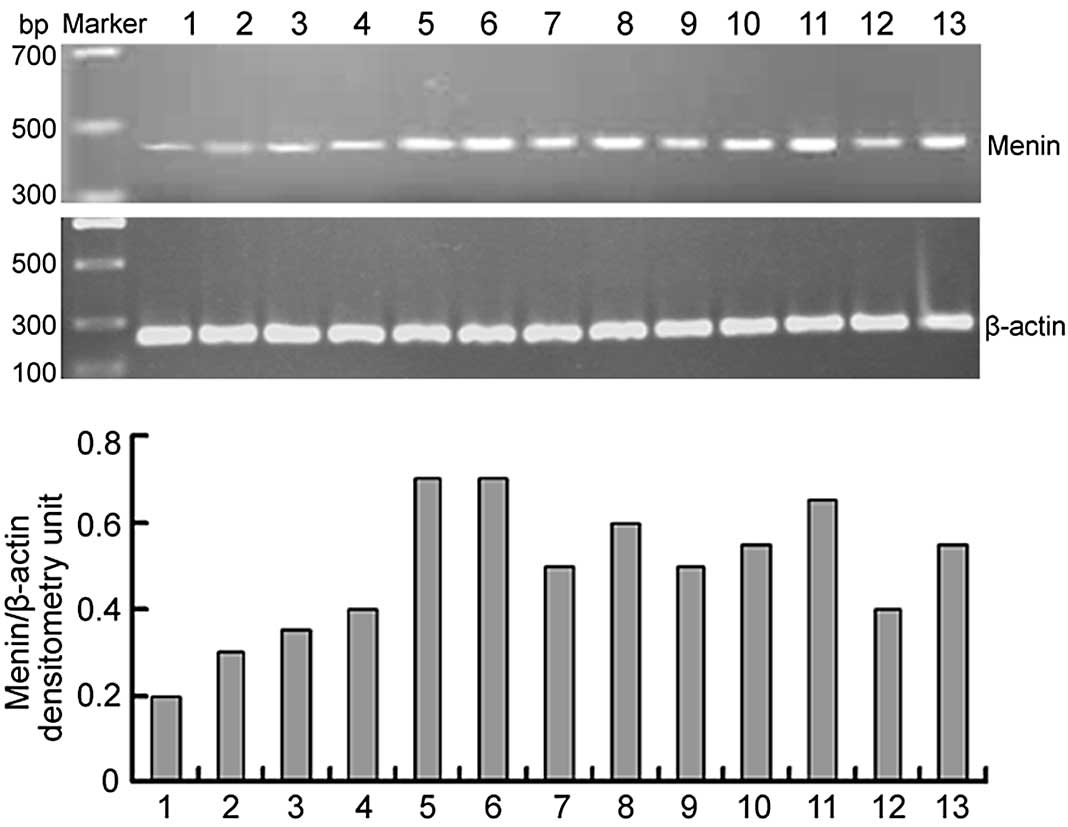 | Figure 1.Expression of menin mRNA in 17
human cancer cell lines was detected by RT-PCR. The 452-bp human
menin-specific sequence and a 247-bp β-actin sequence were
amplified from the cDNA of cancer cell lines, separated by agarose
gel electrophoresis and visualized by ethidium bromide staining.
Densitometry of menin transcripts was standardized to
β-actin. Lane 1, AGS; lane 2, BGC-823; lane 3, SGC-7901; lane 4,
MKN-28; lane 5, GES-1; lane 6, Huh7; lane 7, Hep3B2.1–7; lane 8,
MCF-7; lane 9, MDA-MB-231; lane 10, SW480; lane 11, SGH44; lane 12,
SKOV-3; lane 13, HeLa. RT-PCR, reverse transcription-polymerase
chain reaction. |
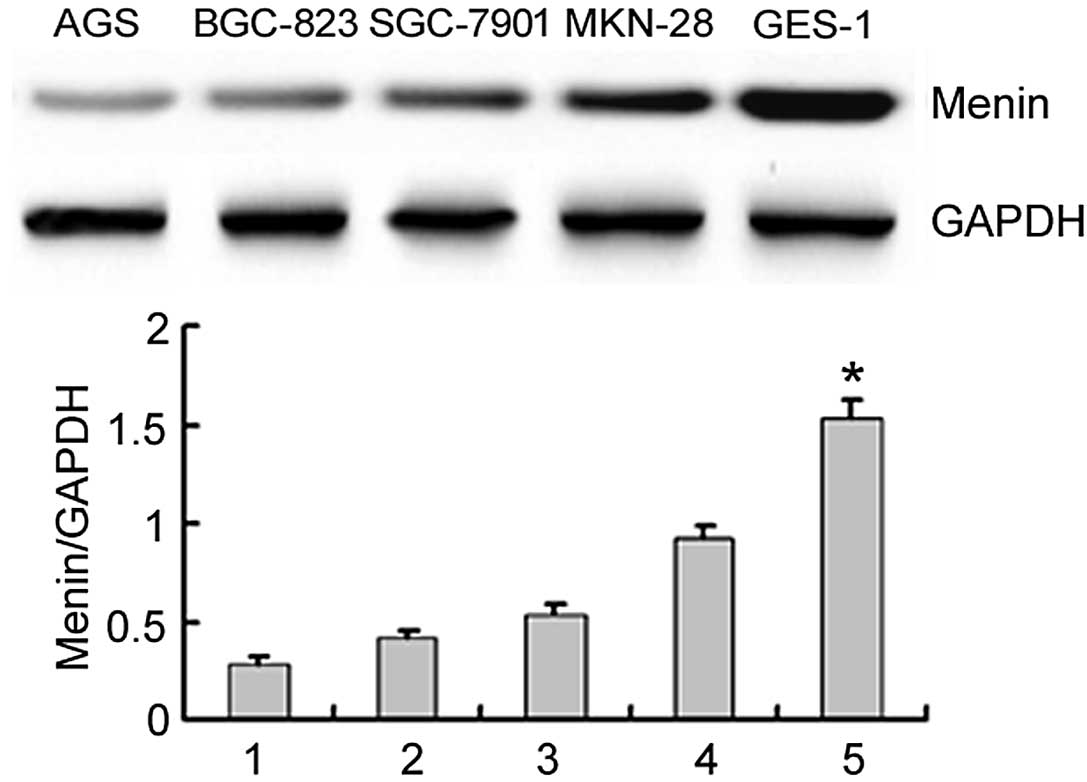
Menin is generally distributed in cancer
cell lines
The expression of the menin protein in cancer
cell lines was further confirmed by immunoblotting, which revealed
a 74-kDa protein band. The results also revealed that the
menin protein was expressed in different cancer cell lines
(Figs. 2 and 3). Moreover, the menin protein was
expressed not only in the nucleus, but also in the cytosol and
membrane in the GES-1, MCF-7, SGH44 and HeLa cell lines (data not
shown). The expression of the menin protein was
significantly higher in GES-1 cells than in the other gastric
cancer cell lines (P<0.05; Fig.
2).
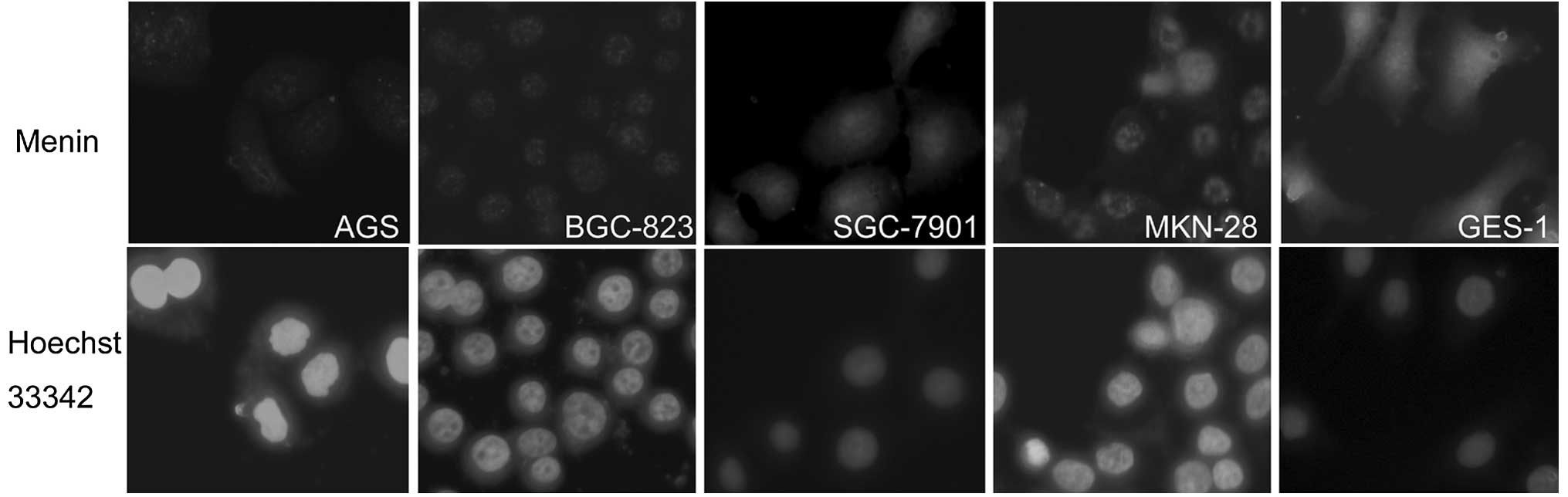
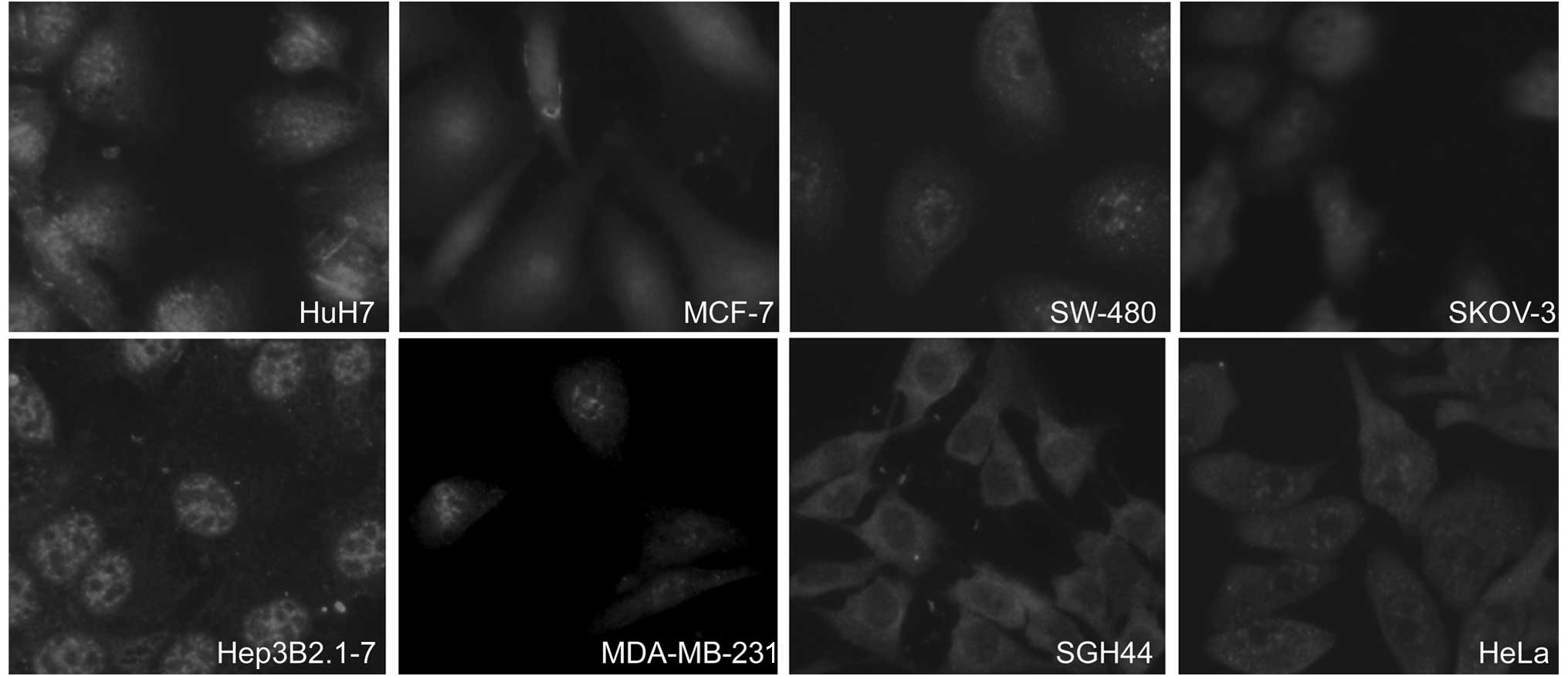
The subcellular localization of menin in
different cancer cell lines
Immunofluorescence microscopy revealed that
menin was located primarily in the nucleus. However, in the
four human cancer cell lines (GES-1, MCF-7, SGH44 and HeLa),
menin was localized not only in the nucleus, but also in the
cytosol and membrane. Moreover, in the SGH44 cells more
menin was located in the cytosol than that in the nucleus
(Figs. 4 and 5).
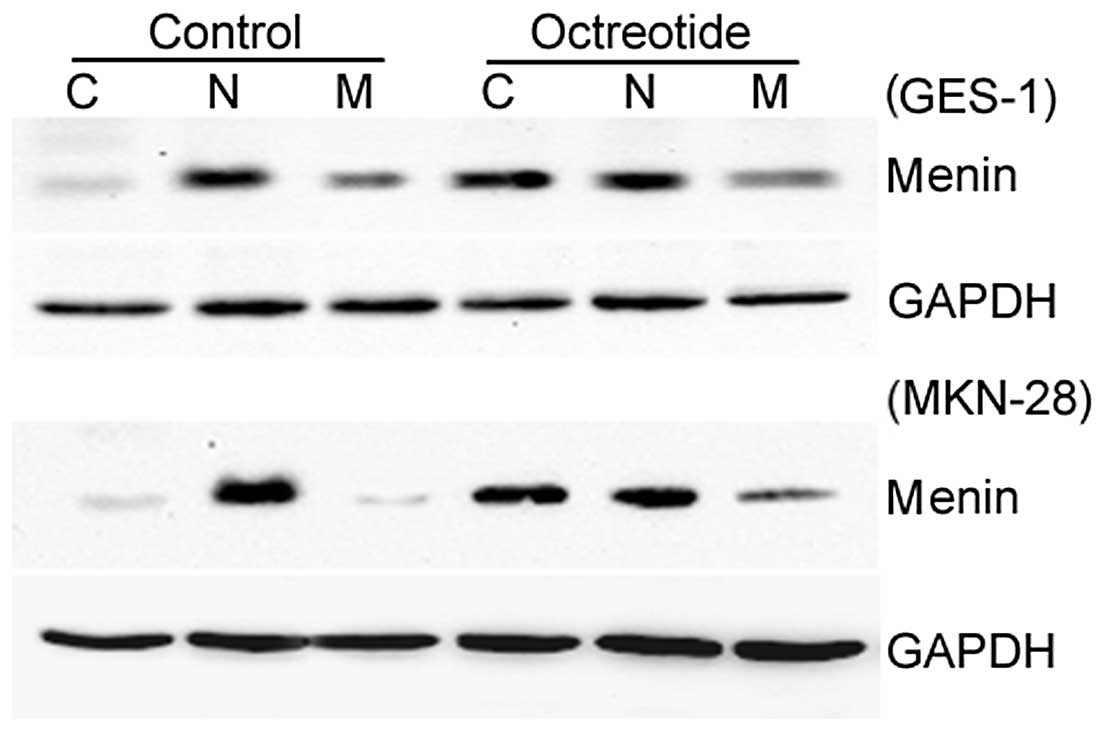
More menin protein is distributed in the
cytosol with octreotide treatment
The relative protein expression levels of
menin in the GES-1 and MKN-28 cell lines were significantly
increased in the octreotide group compared with the control group.
Moreover, more menin protein was located in the cytosol with
octreotide treatment (Fig. 6).
Discussion
The present study detected the expression and
distribution of menin in 13 cancer cell lines, including
AGS, BGC-823, SGC-7901 and MNK-28 (gastric cancer), GES-1
(transformed gastric epithelium), Huh7 and Hep3B2.1–7 (liver
cancer), MDA-MB-231 and MCF-7 (breast cancer), SW480 (colon
cancer), SKOV-3 (ovarian cancer), HeLa (cervical cancer) and SGH44
(brain glioma) cells. The results of RT-PCR, western blotting and
immunofluorescence microscopy revealed that menin was
positively expressed in all of the cell lines examined in this
study. The nuclear localization of the menin protein was
extensive and general, with a discrepancy between the gastric
cancer and transformed gastric epithelial cell lines and the cancer
and SGH44 (brain glioma) cell lines. Furthermore, we found that the
somatostatin analog octreotide increased the expression of
menin, particularly in the cytosol. Consistent with the
findings of Mensah-Osman et al (11), octreotide induces menin
expression by the suppression of PKA activation.
The MEN1 gene encodes the menin protein,
which is thought to be involved in a number of mechanisms that are
dysregulated in cancer cells, including genome stability and the
regulation of gene transcription, cell proliferation and apoptosis
(12–15). These diverse menin functions
were largely attributed to the crucial role of menin in
endocrine cells and tissues. The results of the present study
support the hypothesis that the nucleus-cytoplasm-membrane treble
distribution of menin was a general phenomenon and that more
menin protein was localized to the nucleus of non-endocrine
cancer cells. Although the primary function of menin as a
regulator of gene transcription, cell proliferation, apoptosis and
genome stability has been determined (16,17),
the role of menin protein in the cytoplasm-membrane
localization remains unknown. These observations provide novel
insights into how menin suppresses tumorigenesis.
Menin is predominantly located in the nucleus
and contains two classic NLSs, NLS1 and NLS2 (18). Classic NLSs comprise positively
charged amino acid residues which bind to a soluble transport
receptor complex made up of importins α and β, causing the protein
to be translocated to the nucleus (19). The nuclear localization of
menin is thought to be necessary for its ability to regulate
gene transcription, as the protein regulates gene expression by
associating with chromatin and the nuclear matrix and binding
double-stranded DNA (20).
Whereas, the simultaneous deletion of NLS1 and NLS2 in menin
attenuates menin translocation into the nucleus, on the
contrary, accumulating quantities of menin protein are noted
in the cytosol. Our results showed that menin was also
localized to the cytosol and membrane in GES-1, MCF-7, SGH44 and
HeLa cells. In particular, the level of expression of menin
was higher in the cytosol than in the nucleus in SGH44 cells. These
results suggest that menin NLS deletion mutants and the
deletion of a stretch of amino acid residues may affect the
expression and general structure of menin. On the other
hand, menin has several NLSs, suggesting that its
transcriptional activity may be regulated by its ability to move in
and out of the nucleus (18).
Mensah-Osman et al (11) found that octreotide-suppressed PKA
activation may markedly increase the numbers of
menin-expressing cells and levels of menin mRNA and
menin protein expression. This study also revealed that when
the cells were treated with octreotide, the expression of the
menin protein was increased and the protein was rapidly
transported out of the nucleus and congregated in the cytosol and
on the membrane. Generally, stimuli that promote the expression of
a protein are also able to promote its intracellular movement (most
are cytoplasm-to-nucleus translocations) (21,22).
Octreotide may promote the reverse translocation of the
menin protein from the nucleus to the cytoplasm. Consistent
with our results, Yan et al (9) observed the co-localization of IQGAP1
with a non-nuclear pool of menin in β cell lines. The
co-localization of menin at the plasma membrane was observed to be
extensive, particularly at cell-cell junctions. The authors also
reported that menin interacts with IQGAP1, a scaffold
protein, reducing its interaction with GTP-Rac1 and increasing its
interaction with E-cadherin/β-catenin. This suggests the existence
of a menin-IQGAP1 pathway which influences cell migration
and cell-cell adhesion in endocrine tissue. According to its new
localization, our results provide new insight into some unknown
functions of menin protein in non-endocrine cells.
Together, these results demonstrated that
menin was positively expressed in all of the cell lines
examined in this study. The nuclear localization of the
menin protein was extensive and general and its expression
in the cytoplasm may play a more significant role in coordinating
the activation and repression of gene transcription than merely
targeting menin to the nucleus. The precise roles of
menin in the cytoplasm and the membrane are not yet fully
understood. Further detailed analysis must be carried out to
establish the functions of menin and its role in the
cytoplasm.
Acknowledgements
This study was supported by grants
from the Natural Science Foundation of Jiangsu Province (no.
BK2008115) and the Medical Science Foundation of Wuxi, Jiangsu
Province (no. YGM1111).
References
|
1.
|
Wu T and Hua X: Menin represses
tumorigenesis via repressing cell proliferation. Am J Cancer Res.
1:726–739. 2011.PubMed/NCBI
|
|
2.
|
Pannett AA and Thakker RV: Multiple
endocrine neoplasia type 1. Endocr Relat Cancer. 6:449–473. 1999.
View Article : Google Scholar
|
|
3.
|
Tsukada T, Nagamura Y and Ohkura N: MEN1
gene and its mutations: basic and clinical implications. Cancer
Sci. 100:209–215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X and
Jin GH: Lung cancer cell migration is regulated via repressing
growth factor PTN/ RPTP β/ζ signaling by menin. Oncogene.
29:5416–5426. 2010.PubMed/NCBI
|
|
5.
|
Hory B and Drüeke TB: Menin and MEN 1
gene: a model of tumour suppressor system. Nephrol Dial Transplant.
13:2176–2179. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Yang Y and Hua X: In search of tumor
suppressing functions of menin. Mol Cell Endocrinol. 265–266:34–41.
2007.PubMed/NCBI
|
|
7.
|
Guru SC, Goldsmith PK, Burns AL, et al:
Menin, the product of the MEN1 gene, is a nuclear protein. Proc
Natl Acad Sci USA. 95:1630–1634. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Lopez-Egido J, Cunningham J, Berg M, Oberg
K, Bongcam-Rudloff E and Gobl A: Menin’s interaction with glial
fibrillary acidic protein and vimentin suggests a role for the
intermediate filament network in regulating menin activity. Exp
Cell Res. 278:175–183. 2002.
|
|
9.
|
Yan J, Yang Y, Zhang H, et al: Menin
interacts with IQGAP1 to enhance intercellular adhesion of
beta-cells. Oncogene. 28:973–982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Chen YC, Ren F, Sang JR, Tao Y and Xu WR:
Type II cGMP-dependent protein kinase inhibits proliferation of the
gastric cancer cell line BGC-823. Mol Med Report. 3:361–366.
2010.PubMed/NCBI
|
|
11.
|
Mensah-Osman E, Zavros Y and Merchant JL:
Somatostatin stimulates menin gene expression by inhibiting protein
kinase A. Am J Physiol Gastrointest Liver Physiol. 295:G843–G854.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Sowa H, Kaji H, Hendy GN, et al: Menin is
required for bone morphogenetic protein 2- and transforming growth
factor beta-regulated osteoblastic differentiation through
interaction with Smads and Runx2. J Biol Chem. 279:40267–40275.
2004. View Article : Google Scholar
|
|
13.
|
Kim YS, Burns AL, Goldsmith PK, et al:
Stable overexpression of MEN1 suppresses tumorigenicity of RAS.
Oncogene. 18:5936–5942. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
La P, Yang Y, Karnik SK, et al:
Menin-mediated caspase 8 expression in suppressing multiple
endocrine neoplasia type 1. J Biol Chem. 282:31332–31340. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Guru SC, Crabtree JS, Brown KD, et al:
Isolation, genomic organization, and expression analysis of Men1,
the murine homolog of the MEN1 gene. Mamm Genome. 10:592–596. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Zhang H, Li W, Wang Q, et al:
Glucose-mediated repression of menin promotes pancreatic β-cell
proliferation. Endocrinology. 153:602–611. 2012.PubMed/NCBI
|
|
17.
|
Francis J, Lin W, Rozenblatt-Rosen O and
Meyerson M: The menin tumor suppressor protein is phosphorylated in
response to DNA damage. PLoS One. 6:e161192011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
La P, Desmond A, Hou Z, Silva AC, Schnepp
RW and Hua X: Tumor suppressor menin: the essential role of nuclear
localization signal domains in coordinating gene expression.
Oncogene. 25:3537–3546. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Komeili A and O’Shea EK: New perspectives
on nuclear transport. Annu Rev Genet. 35:341–364. 2001. View Article : Google Scholar
|
|
20.
|
La P, Silva AC, Hou Z, et al: Direct
binding of DNA by tumor suppressor menin. J Biol Chem.
279:49045–49054. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Tell G, Damante G, Caldwell D and Kelley
MR: The intracellular localization of APE1/Ref-1: more than a
passive phenomenon? Antioxid Redox Signal. 7:367–384. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Li Y, Chen Y, Tao Y, Xu J and Chen M: RhoA
protein is generally distributed in the nuclei of cancer cells.
Oncol Rep. 24:1005–1009. 2010.PubMed/NCBI
|