Introduction
Marrow stromal cells (MSCs) are the main source of
osteogenic cells in fracture healing and bone tissue reconstruction
(1). Under the action of related
growth factors, MSCs gradually differentiate into osteogenic cells
and synthesize extracellular matrix, including collagen, and
ultimately transform into mature bone tissue (2). Number and function abnormalities of
cytokines and growth factors directly cause weak differentiation of
MSCs to osteoblasts, leading to osteoporosis and fracture healing
disorders (3). A large amount of
growth factors, including bone morphogenetic protein (BMP),
fibroblast growth factor (FGF), vascular endothelial growth factor
(VEGF) and transforming growth factor-β (TGF-β) in the
extracellular matrix and on cell membrane surfaces are ingested and
controlled by the heparan sulfate (HS) lateral chains of heparan
sulfate proteoglycans (HSPGs) (4).
HSPGs are widely distributed in the cytolemma and extracellular
matrix, and the HS lateral chain has good affinity with growth
factors (5). In physiological and
pathological conditions, HSPGs upregulate the release and activity
of the above growth factors and participate in extracellular matrix
reconstruction, information transfer and signal transduction
(6). It has been identified that
HSPGs are involved in the regulation of endochondral ossification,
bone tissue reconstruction and fracture healing (7).
Heparanase (HPSE) is a unique endoglycosidase that
decomposes HS lateral chains of HSPGs and is closely related to the
function of HSPGs. It not only plays a significant role in tumor
metastasis and angiogenesis (8–10),
but also participates in fracture healing (11) and bone tissue formation (12). HPSE is expressed in normal human
osteoblasts (12). Whether HPSE is
expressed and plays a role in the osteogenic differentiation of
MSCs (precursor cells of osteoblasts) and whether the osteogenic
differentiation of MSCs is regulated by intervention of HPSE
expression, requires further investigation. In the current study,
the protein and mRNA expression levels of HPSE in the osteogenic
differentiation of rat MSCs were detected by western blot analysis
and reverse transcription-polymerase chain reaction (RT-PCR),
respectively. The aim of this study was to provide a foundation for
further study of HPSE.
Materials and methods
Reagents and animals
The HPSE primary antibody was purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). TRIzol was provided
by Invitrogen Life Technologies (Carlsbad, CA, USA). Alkaline
phosphatase (ALP) and an alizarin red staining kit were purchased
from Wuhan Boster Biological Technology, Ltd. (Wuhan, China). Three
male 2-month-old Sprague-Dawley (SD) rats and three male
10-month-old SD rats were provided by the Experimental Animal
Center of Nantong University.
MSC separation and induced osteogenic
differentiation
Single-cell suspensions of rat femur marrow were
prepared under aseptic conditions. The conventional adherent
culture method was used to separate and culture the MSCs. The cell
morphology was observed under an inverted microscope. The second
generation of MSCs was cultured in primary medium for 72 h,
followed by osteogenic induction (10−8 mol/l
dexamethasone, 10 mmol/l β-glycerophosphate sodium and 50
μg/ml vitamin C). After 1 week, the cell morphology was
observed under an inverted microscope. According to the
manufacturer’s instructions, the ALP activity in MSCs was
determined and alizarin red staining was conducted.
Determination of HPSE protein expression
by western blot analysis
Expression of the HPSE protein was determined using
western blotting on days 0, 1, 3, 7, 10, 14 and 21 of osteogenic
differentiation. Cell protein was extracted by the conventional
method, followed by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE; 15 μl sample in each well;
stacking gel, 80 V, 20 min; separating gel, 120 V, 60 min). The wet
electrical transfer method was used to transfer the protein on
separating gel to a polyvinylidene fluoride (PVDF) membrane
(constant voltage, 100 V for 90 min). Following Ponceau staining,
the clear red bands in the PVDF membrane indicated the successful
transfer. The PVDF membrane was immersed in 5% milk powder solution
at room temperature for 2 h of blocking. The primary antibody with
different dilution ratios was added, followed by incubation at 4°C
overnight. After washing with Tris-buffered saline with Tween-20
(TBST) containing 0.1% Tween-20 (3 times, 5 min each time), the
donkey anti-rabbit secondary antibody labeled with IRDye800
(1:5000; Rockland Immunochemicals, Boyertown, PA, USA) was added,
followed by incubation at 4°C overnight and washing with TBST. A
molecular Odyssey Infrared imaging system (Li-COR Biosciences,
Lincoln, NE, USA) was used to scan the PVDF membrane and analyze
the strips, using integrated optical density (IOD) as the relative
protein content.
Determination of HPSE mRNA expression by
quantitative real-time PCR
Expression of HPSE mRNA was determined by real-time
quantitative PCR on days 0, 1, 3, 7, 10, 14 and 21 of osteogenic
differentiation. Total RNA was extracted from the cell sample using
TRIzol according to the manufacturer’s instructions. The OD value
and concentration were determined using a spectrophotometer. Total
RNA (2 μg) was transcribed to cDNA using an Omniscript RT
kit. Primer 5 software was used to design the primers of the
internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
and the HPSE gene (Table I). The
quantitative real-time PCR was conducted in the following
conditions: 20 μl reaction system (containing 1 μl
cDNA); 1 μl upstream and downstream primers, respectively;
10 μl EvaGreen® qPCR Master Mix; and 7 μl
deionized water. The housekeeping gene GAPDH was used as the
internal control.
 | Table IPrimers and amplification fragment
length. |
Table I
Primers and amplification fragment
length.
| Gene | Upstream primer
(5′-3′) | Downstream primer
(5′-3′) | Amplified fragment
length (bp) |
|---|
| GAPDH |
GGCATCCTGGGCTACACT |
CCACCACCCTGTTGCTGT | 163 |
| HPSE |
CGGTTCTGACGGACTGCTT |
AAAACCCATAGGAAAAGGCG | 146 |
Statistical analysis
Statistical analysis was performed using SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). Analysis of
variance and t-test were performed to analyze the differences
between the two types of rat. P<0.05 was considered to indicate
a statistically significant difference.
Results
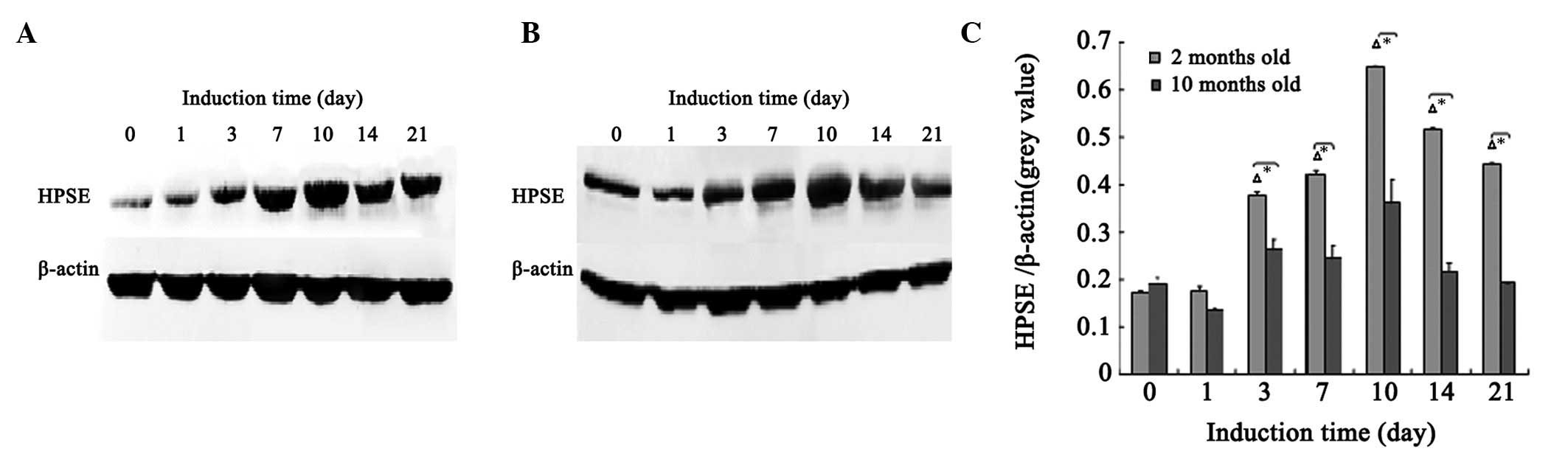
Expression of HPSE protein
The western blots in Fig. 1 show that, from day 3 of osteogenic
differentiation of MSCs, the protein expression levels of HPSE in
the 2-month-old rats were significantly increased compared with
basal levels (days 0 and 1; P<0.05). HPSE protein expression
reached a peak on day 10, followed by a gradual decline. On day 21,
the HPSE protein expression level continued to be significantly
higher than basal levels (days 0 and 1). A similar pattern was
presented in the 10-month-old rats; however, the differences from
basal levels were not significant (P>0.05). At each time point,
the protein levels of HPSE in the 2-month-old rats were
significantly higher compared with those in the 10-month-old rats
(P<0.05).
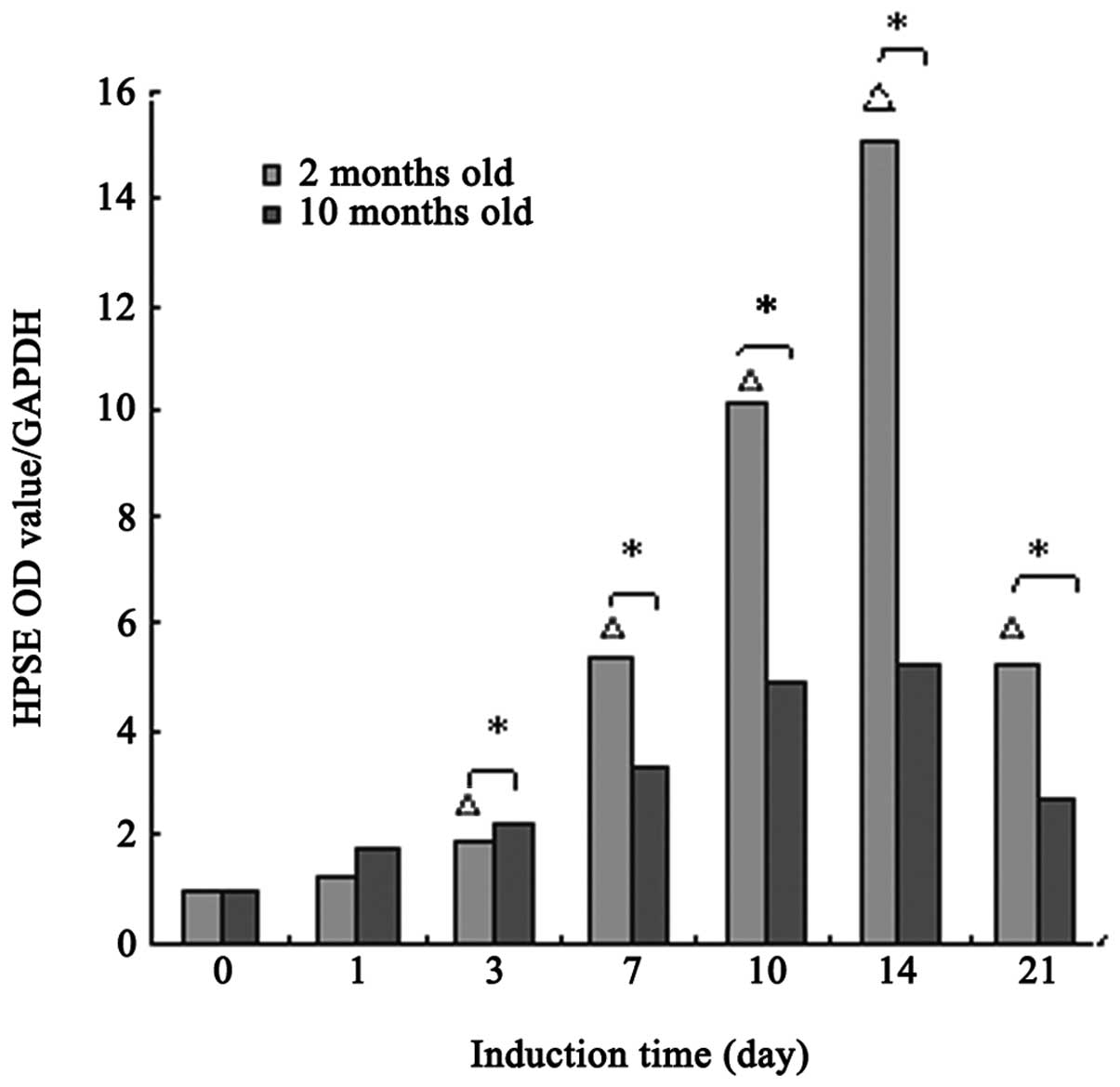
Expression of HPSE mRNA
As shown in Fig. 2,
the mRNA expression levels of HPSE in 2-month-old rats began to
increase on the third day, and was significantly different from
basal levels (days 0 and 1; P<0.05). The expression levels
remained significantly higher on day 21 (P<0.05). The same
pattern was observed in 10-month-old rats; however, there were no
significant differences from basal levels (P>0.05). As compared
with HPSE protein level, the mRNA level reached a peak on day 14.
This suggests that HPSE protein synthesis may be inhibited in the
post-transcriptional modification stage.
Discussion
Since Ogren and Lindahl first reported HPSE in mouse
mast cells and demonstrated its digestive function on
macromolecular heparin at specific sites (13), HPSE has been reported to be widely
expressed in cells of normal tissues and malignant tumors. The gene
sequences of the human HPSE gene were first determined in 1999, and
the molecular structure, synthesis and action mechanism of HPSE
were further studied (14,15). HPSE is the only known human
endoglycosidase that plays an irreplaceable role in physiological
and pathological processes. It has been a subject of intense
research in molecular biology (16).
HPSE digests HS at specific sites, adjusts the
release of HS-binding growth factors, including BMPs, FGFs and
VEGF, regulates cell differentiation, adhesion and proliferation,
and extracellular matrix reconstruction. In addition, HPSE has
independent activity unrelated to enzyme function and directly
activates corresponding receptors, increases AKT phosphorylation
and participates in malignant tumor metastasis (17). HPSE is involved in pathological
processes as follows: i) angiogenesis, metastasis and diffusion of
myeloma and malignancy of the gastrointestinal tract and mammary
gland (18); ii) tissue repair
processes, including liver tissue regeneration, skin wound healing
and hair regeneration (19,20);
and iii) molecular biological mechanisms of kidney diseases,
including diabetic nephropathy (21).
In addition, HPSE is involved in fracture healing
and normal bone tissue formation. Saijo et al(11) studied a mouse model of fracture and
detected HPSE mRNA in osteoclasts and precursor cells near the
fracture site on day 5 after fracture. In the callus formation
stage, a large amount of HPSE is synthesized in osteoclasts in
cartilage callus absorption and neovascularization areas; it
continues until the woven bone callus is transformed into a
cortical bone callus. This indicates that HPSE is synthesized in
osteoclasts in normal bone tissue and fracture sites. In the
osteochondral border area, the synthesized HPSE activates cartilage
absorption and bone formation and promotes the ossification of
cartilage. It is considered that HPSE may be one of the key
regulatory factors in bone tissue formation and regeneration. Smith
et al(12) identified that
the expression levels of HPSE mRNA in osteoporotic patients are
significantly reduced compared with those in healthy volunteers.
This indicates that HPSE mRNA expression is related to ALP
activity. Additionally, when human osteoblasts are exposed to
exogenous HPSE protein, the levels of phosphorylated histone H3 in
osteoblasts are increased, suggesting that HPSE may adjust bone
regeneration by regulating histone H3 phosphorylation. Kram et
al(7) successfully cultivated
HPSE-transgenic mice and identified that, compared with wild-type
mice, the trabecular bone volume, cortical bone thickness and bone
formation speed in HPSE-transgenic mice are significantly
increased, respectively. This indicates that HPSE is involved in
bone formation by regulating osteoblast activity.
In the present study, the expression of HPSE in the
osteogenic differentiation of MSCs was investigated. Results show
that from the third day of osteogenic differentiation, all HPSE
protein and mRNA expression levels in 2-month-old rats were
significantly increased compared with basal levels (days 0 and 1).
The HPSE protein levels peaked on day 10 while HPSE mRNA levels
peaked on day 14. This indicates that HPSE may be involved in the
osteoblastic differentiation of MSCs. There were no significant
differences of basal HPSE protein and mRNA expression levels
between the 2- and 10-month-old rats; however, the responses of
HPSE to osteogenic induction in the two ages of rat are different.
The patterns of expression for the 10-month-old rats were similar
to those of the 2-month-old rats; however, the differences compared
with basal levels were not statistically significant. This
indicates that the responses of HPSE to osteogenic induction in
aged rats are reduced. HPSE may play an important role in bone
tissue formation, which is consistent with results of the study by
Smith et al(12).
HPSE is involved in the osteogenic differentiation
of rat MSCs. The responses to osteogenic induction in aged rats are
weaker compared with those in young rats, which may be related to
the decline in osteogenic differentiation ability. The specific
mechanism of participation of HPSE in osteogenic differentiation is
worthy of further investigation. This is likely to contribute to an
in-depth understanding of fracture healing and osteoporosis
pathogenesis, as well as create conditions for exploring more
effective clinical treatment methods.
References
|
1.
|
Kagami H, Agata H and Tojo A: Bone marrow
stroma cells (bone marrow-derived multipotent mesenchymal stroma
cells) for bone tissue engineering: basic science to clinical
translation. Int Biochem Cell Biol. 43:286–289. 2011. View Article : Google Scholar
|
|
2.
|
Brown AJ, Alicknavitch M, D’Souza SS, et
al: Heparanase expression and activity influences chondrogenic and
osteogenic processes during endochondral bone formation. Bone.
43:689–699. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Nishimura R: Bone and calcium update; bone
research update. Regulatory mechanisms in osteoblast
differentiation. Clin Calcium. 21:103–112. 2011.(In Japanese).
|
|
4.
|
Moretti M, Sinnappah-Kang ND, Toller M,
Curcio F and Marchetti D: HPSE-1 expression and functionality in
differentiating neural cells. J Neurosci Res. 83:694–701. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Beauvais DM and Rapraeger AC: Syndecans in
tumor cell adhesion and signaling. Reprod Biol Endocrinol.
3:22004.
|
|
6.
|
Lamoureux F, Baud’huin M, Duplomb L,
Heymann D and Rédini F: Proteoglycans: key partners in bone cell
biology. Bioessays. 29:758–771. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kram V, Zcharia E, Yacoby-Zeevi O, et al:
Heparanase is expressed in osteoblastic cells and stimulates bone
formation and bone mass. J Cell Physiol. 207:784–792. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Kelly T, Miao HQ, Yang Y, et al: High
heparanase activity in multiple myeloma is associated with elevated
microvessel density. Cancer Res. 63:8749–8756. 2003.PubMed/NCBI
|
|
9.
|
Watanabe M, Aoki Y, Kase H and Tanaka K:
Heparanase expression and angiogenesis in endometrial cancer.
Gynecol Obstet Invest. 56:77–82. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Okawa T, Naomoto Y, Nobuhisa T, et al:
Heparanase is involved in angiogenesis in esophageal cancer through
induction of cyclooxygenase-2. Clin Cancer Res. 11:7995–8005. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Saijo M, Kitazawa R, Nakajima M, Kurosaka
M, Maeda S and Kitazawa S: Heparanase mRNA expression during
fracture repair in mice. Histochem Cell Biol. 120:493–503. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Smith PN, Freeman C, Yu D, et al:
Heparanase in primary human osteoblasts. J Othorp Res.
28:1315–1322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Ogren S and Lindahl U: Cleavage of
macromolecular heparin by an enzyme from mouse mastocytoma. J Biol
Chem. 250:2690–2697. 1975.PubMed/NCBI
|
|
14.
|
Vlodavsky I, Friedmann Y, Elkin M, et al:
Mammalian heparanase: gene cloning, expression and function in
tumor progression and metastasis. Nat Med. 5:793–802. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Toyoshima M and Nakajima M: Human
heparanase. Purification, characterization, cloning, and
expression. J Biol Chem. 274:24153–24160. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Vreys V and David G: Mammalian heparanase:
what is the message? J Cell Mol Med. 11:427–452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Gingis-Velitski S, Zester A, Flugelman MY,
Vlodavsky I and Ilan N: Heparanase induces endothelial cell
migration via protein kinase B/Akt activation. J Biol Chem.
279:23536–23541. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Nadir Y, Vlodavsky I and Brenner B:
Heparanase, tissue factor, and cancer. Semin Thromb Hemost.
34:187–194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Goldshmidt O, Yekilis R, Mawasi N, et al:
Heparanase expression during normal liver development and following
partial hepatectomy. J Pathol. 203:594–602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Zcharia E, Zilka R, Yaar A, et al:
Heparanase accelerates wound angiogenesis and wound healing in
mouse and rat models. FASEB J. 19:211–221. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kramer A, van den Hoven M, Rops A, et al:
Induction of glomerular heparanase expression in rats with
adriamycin nephropathy is regulated by reactive oxygen species and
the renin-angiotensin system. J Am Soc Nephrol. 17:2513–2520. 2006.
View Article : Google Scholar
|