Introduction
Breast cancer is a leading cause of
cancer-associated mortality in females worldwide and in excess of
one million new cases of breast cancer are diagnosed annually
(1). Breast cancer cell
progression is a coordinated process that involves cell cycle
dysregulation and a specific gene expression program to determine
tissue identity. Cell proliferation, differentiation, senescence
and apoptosis are cell cycle-dependent, and the basic regulatory
mechanisms of cell cycle progression rely on a multicomponent
system. At different phases, progression through the cell cycle is
regulated by sequential activation and subsequent inactivation of a
series of cyclin-dependent kinases (CDKs), whose activity depends
on interactions with cyclins and cyclin-dependent kinase inhibitors
(CDKIs) (2–4).
δ-crystallin enhancer factor 1 (δEF1), a member of
the zinc finger-homeodomain transcription factor family (5), regulates gene expression to modulate
cell differentiation and tissue-specific functions (6). Evidence has suggested that δEF1 is
important in breast cancer tumor growth and metastasis (7). To control breast cancer cell
proliferation, δEF1 downregulates p21 and concurrently upregulates
the expression of CDK2 and CDK4 (8). However, the direct molecular
mechanisms underlying the regulation of CDK4 expression by δEF1
have not yet been elucidated.
To address this issue, a series of different length
and E2-box-mutated CDK4 promoter luciferase reporter genes were
constructed in the present study. Luciferase assays were used to
assess the effect of δEF1 overexpression and knockdown on the
activity of the human CDK4 promoter. In addition, the effect of
human CDK4 promoter E2-box (CACGTG) deletion on the activation of
CDK4 transcription by δEF1 was investigated. The aim of the study
was to evaluate the role of the E2-box on the CDK4 promoter in the
promotion of CDK4 expression by δEF1.
Materials and methods
Cell culture
MDA-MB-231 cells (American Type Culture Collection,
Manassas, VA, USA) were maintained in Dulbecco's modified Eagle
medium (DMEM)-high glucose medium (Gibco-BRL, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo
Fisher Scientific, Inc., Waltham, MA, USA), penicillin (50
units/ml) and streptomycin (50 mg/ml). The MDA-MB-231 cells were
plated at a density of 5×104 cells/well in 24-well
plates for use in luciferase assays. This study was approved by The
Ethics Committee of Hebei United University (Tangshan, China).
Construction of plasmids
The generation of full-length δEF1 expression
vectors (δEF1-pcDNA6B) was performed as described previously
(6). The generation of
δEF1-specific small interfering RNA (siRNA) expression plasmids
(si-δEF1) was also performed as described previously (8).
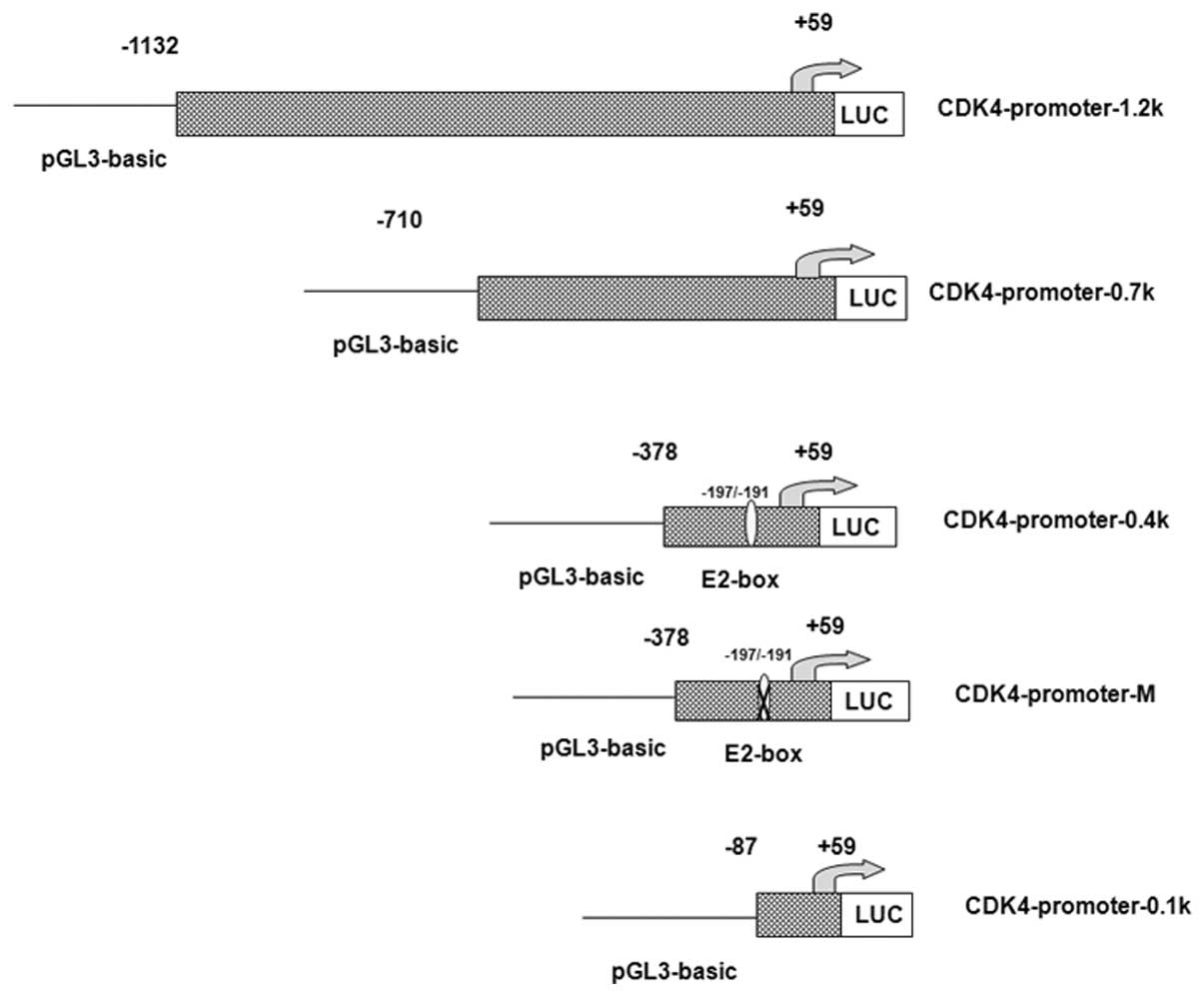
The human CDK4 promoter sequence was obtained by
polymerase chain reaction (PCR) from human blood genomic DNA and
cloned into pGL3-basic vectors (Promega Corp., Madison, WI, USA)
using the following primers: CDK4-promoter-1.2k (−1132),
5′-TTCGAGCTCGTGTTCTGG ACAGTGCTAAGTGC-3′ (forward);
CDK4-promoter-0.7k (−710),
5′-TTGGAGCTCGTCACTGAGCCTGTTGGATT-3′ (forward);
CDK4-promoter-0.4k (−378), 5′-TTGGAGCTCGCA
GACAGGCTGAAAGAC-3′ (forward); CDK4-promoter-0.1k (−87),
5′-TTGGAGCTCTCCCAGTCGAAGCACCTCC-3′ (forward); and
CDK4-promoter (+59), 5′-TGCAAGCTT TCACCCCCACCCTCACCAT-3′ (reverse;
bold text indicates SacI restriction enzyme sites).
Mutagenesis of the E2-box in the human CDK4 promoter was performed
using a QuikChange Site-Directed Mutagenesis kit (Stratagene Corp.,
La Jolla, CA, USA) with the following primers: 5′-GGG
TTGTGGCAGCCAGTCAAATGCCCGCGGC-3′ (forward) and
5′-GCCGCGGGCATTTGACTGGCTGCCACAACCC-3′ (reverse).
RNA extraction and semi-quantitative
PCR
MDA-MB-231 cells were transiently transfected with
δEF1-pcDNA6B or δEF1-specific siRNA expression plasmids in 24-well
plates using Lipofectamine 2000 (Invitrogen Life Technologies,
Carlsbad, CA, USA). At 24 h subsequent to transfection, the total
RNA was extracted using TRIzol reagent (Invitrogen Life
Technologies). A total of 0.5 μg total RNA from each sample was
used for first-strand cDNA synthesis using Moloney murine leukemia
virus (MMLV) reverse transcriptase (Promega Corp.). A specific
transcript of δEF1 was amplified by semi-quantitative PCR using the
following primers: 5′-GGCCCCAGGTGTAAGCGCAG-3′ (forward) and
5′-CGGGCAGGTGAGCAACTGGG-3′ (reverse). Verification of the
expression level of δEF1 was performed using semi-quantitative PCR.
GAPDH was used as an internal control.
Luciferase assay
MDA-MB-231 cells were cotransfected with
CDK4-promoter-1.2k, CDK4-promoter-0.7k, CDK4-promoter-0.4k,
CDK4-promoter-0.1k or the mutant human CDK4 promoter construct
CDK4-promoter-M in 24-well plates using Lipofectamine 2000
(Invitrogen Life Technologies). At 24 h subsequent to transfection,
lysates were prepared and the luciferase activity was measured
using a Dual-Luciferase Reporter Assay System (Promega Corp.)
according to the manufacturer's instructions. Luciferase activity
was normalized using the Renilla luciferase activity. The
luciferase activity of the extracts was assessed 24 h subsequent to
transfection using a Betascope analyzer (Betagen, Waltham, MA,
USA).
Results
Overexpression of δEF1 increases human
CDK4 promoter activity
In the present study, the CDK4 promoter region was
amplified from human blood genomic DNA. Four different regions of
the regulatory sequences in the CDK4 promoter, including ~1.2 kb of
the upstream region, were cloned into the dual luciferase
expression vector, pGL3-basic (Fig.
1). The overexpression of δEF1 mRNA was confirmed using
semi-quantitative PCR (Fig. 2A). A
comparison of the activity of these fragments in the dual
luciferase reporter assays revealed that δEF1 overexpression
significantly increased human CDK4 promoter activity of the
CDK4-promoter-1.2k, CDK4-promoter-0.7k and CDK4-promoter-0.4k
reporter genes. The increase was ~50% relative to the control (no
δEF1 transfection) (Fig. 2B). This
indicated that δEF1 was involved in the positive regulation of CDK4
transcription.
δEF1 promotes the transcription of CDK4
through the E2-box on the CDK4 promoter
δEF1 has been reported to function as a
transcriptional repressor by directly binding to the E2-box
[CA(C/G)(C/G)TG] in the promoter region of target genes. δEF1 binds
using its zinc finger clusters, which are located close to the N
and C termini of the molecule (9,10).
In the present study, a search using the transcription factor
databases Transcription Element Search System (TESS; http://www.cbil.upenn.edu/cgi-bin/tess/tess. Accessed
February 25, 2013) and TRANScription FACtor database (TRANSFAC;
http://www.cbrc.jp/research/db/TFSEARCH.html. Accessed
February 25, 2013) identified an E2-box (CACGTG) that is located at
position -197/-191 of the human CDK4 promoter. In order to
investigate whether δEF1 regulates the transcriptional activity of
the human CDK4 promoter through this putative response element, a
truncated CDK4 promoter reporter (CDK4-promoter-0.1k) was
constructed (Fig. 1). The results
showed that, by E2-box depletion, CDK4-promoter-0.1k exhibited no
increased luciferase activity relative to the CDK4-promoter-0.4k
(Fig. 2B). Furthermore,
δEF1-induced transactivation of CDK4-promoter-0.1k was almost
non-existent (Fig. 2B), indicating
that deletion of the E2-box on the human CDK4 promoter
significantly attenuated the activation of CDK4 transcription by
δEF1.
The E2-box on the human CDK4 promoter was mutated to
generate the CDK4-promoter-M construct as shown in Fig. 1. A luciferase assay demonstrated
that the activation of CDK4-promoter-M promoter activity by δEF1
was markedly decreased compared with the CDK4-promoter-0.4k
promoter (Fig. 2C). These data
indicated that δEF1 promoted the transcription of CDK4 through the
E2-box on the CDK4 promoter.
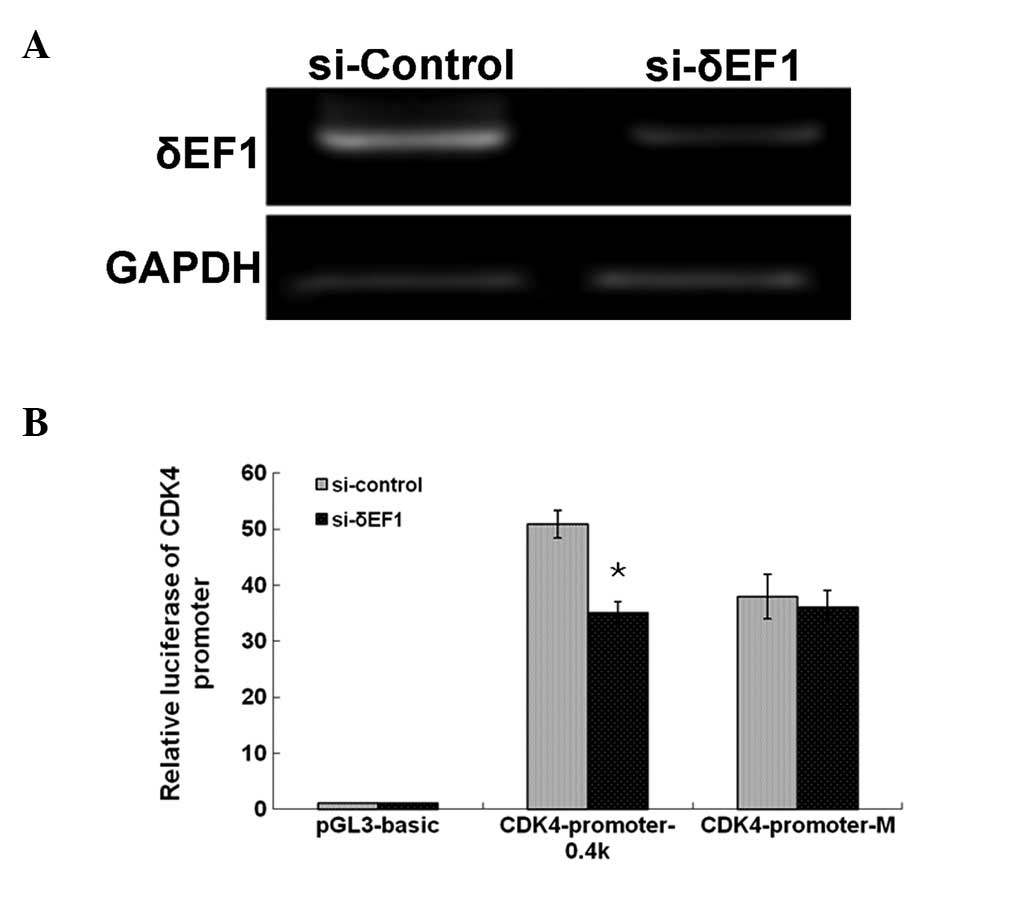
Knockdown of δEF1 inhibits the promoter
activity of CDK4
It was hypothesized that the knockdown of δEF1 using
RNA interference was likely to result in inhibition of the CDK4
promoter. To test this, an siRNA expression plasmid targeting δEF1
or a scrambled control siRNA plasmid was cotransfected with the
CDK4-promoter-0.4k reporter into the MDA-MB-231 cells. The
knockdown of δEF1 mRNA expression was confirmed by
semi-quantitative PCR (Fig. 3A).
The results showed that δEF1 depletion resulted in significant
inhibition of the promoter activity of CDK4 compared with the cells
transfected with the control (Fig.
3B). Therefore, the downregulation of endogenous δEF1 in breast
cancer cells is sufficient to allow inhibition of CDK4
expression.
Discussion
A previous study focusing on the mechanism by which
δEF1 promotes cell proliferation at the protein level have
indicated a possible signal transduction pathway involved in this
process (11). The aim of the
present study was to further elucidate this mechanism at the
transcriptional level. Investigations into the activity of the CDK4
promoter showed that δEF1 upregulated the activity of CDK4 promoter
fragments (−1132 to +59 bp), (−710 to +59 bp) and (−378 to +59 bp),
but not the fragment (−87 to +59 bp). These results indicated that
the core promoter region of δEF1-enhanced CDK4 expression may be
located within the (−378 to −87 bp) region of the CDK4 promoter.
Knockdown of δEF1 using RNA interference resulted in inhibition of
the CDK4 promoter. Furthermore, site-directed mutagenesis of the
E2-box (CACGTG, −197 to −191 bp) on the human CDK4 promoter was
performed and a luciferase assay demonstrated that δEF1 promoted
the transcription of CDK4 by engaging the E2-box on the CDK4
promoter. This data indicate that the E2-box on the CDK4 promoter
is important in the promotion of CDK4 expression by δEF1.
Traditionally, δEF1 has been identified as a widely
expressed transcriptional repressor in a number of cellular
processes, acting via interactions with corepressors or in
competition with activators for DNA binding sites (6,12,13).
However, a number of studies have indicated that δEF1 may also
function as a transcriptional activator in the regulation of
specific genes, including matrix metalloproteinase-1 (MMP-1) and
ovalbumin (7,14). These findings were consistent with
the results of our previous study, which showed that δEF1 activated
MMP-1 transcription during breast cancer epithelial-mesenchymal
transition (7) and induced micro
RNA 21 (miR-21) promoter activity by binding to the E2-box on the
miR-21 promoter (15). In
addition, the results of the present study demonstrated that CDK4
expression was upregulated by the action of δEF1 on the E2-box of
the CDK4 promoter.
In conclusion, the results of this study indicate
that the E2-box on the CDK4 promoter is the core region in which
δEF1 promotes CDK4 expression. These findings provide further
insight into the mechanism of δEF1 gene-promoted breast cancer
proliferation.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 81072093, 81302323 and
30671092) and the Natural Science Foundation of Hebei Province
(grant nos. C2009001260 and C2013209024).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Draetta G: Cell cycle control in
eukaryotes: molecular mechanisms of cdc2 activation. Trends Biochem
Sci. 15:378–383. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sherr CJ: G1 phase progression: cycling on
cue. Cell. 79:551–555. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caldon CE, Daly RJ, Sutherland RL and
Musgrove EA: Cell cycle control in breast cancer cells. J Cell
Biochem. 97:261–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Funahashi J, Kamachi Y, Goto K and Kondoh
H: Identification of nuclear factor delta EF1 and its binding site
essential for lens-specific activity of the delta 1-crystallin
enhancer. Nucleic Acids Res. 19:3543–3547. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang S, Zhao L, Yang J, Chai D, Zhang M,
Zhang J, Ji X and Zhu T: deltaEF1 represses BMP-2-induced
differentiation of C2C12 myoblasts into the osteoblast lineage. J
Biomed Sci. 14:663–679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu F, Wang C, Guo S, Sun W, Mi D, Gao Y,
Zhang J, Zhu T and Yang S: δEF1 promotes osteolytic metastasis of
MDA-MB-231 breast cancer cells by regulating MMP-1 expression.
Biochim Biophys Acta. 1809:200–210. 2011.
|
|
8
|
Hu F, Wang C, Du J, Sun W, Yan J, Mi D,
Zhang J, Qiao Y, Zhu T and Yang S: DeltaEF1 promotes breast cancer
cell proliferation through down-regulating p21 expression. Biochim
Biophys Acta. 1802:301–312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sekido R, Murai K, Kamachi Y and Kondoh H:
Two mechanisms in the action of repressor deltaEF1: binding site
competition with an activator and active repression. Genes Cells.
2:771–783. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sekido R, Murai K, Funahashi J, Kamachi Y,
Fujisawa-Sehara A, Nabeshima Y and Kondoh H: The delta-crystallin
enhancer-binding protein delta EF1 is a repressor of
E2-box-mediated gene activation. Mol Cell Biol. 14:5692–5700. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu F, Wang C, Du J, Sun W, Yan J, Mi D,
Zhang J, Qiao Y, Zhu T and Yang S: DeltaEF1 promotes breast cancer
cell proliferation through down-regulating p21 expression. Biochim
Biophys Acta. 1802:301–312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang S, Du J, Wang Z, Yuan W, Qiao Y,
Zhang M, Zhang J, Gao S, Yin J, Sun B and Zhu T: BMP-6 promotes
E-cadherin expression through repressing deltaEF1 in breast cancer
cells. BMC Cancer. 7:2112007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murray D, Precht P, Balakir R and Horton
WE Jr: The transcription factor deltaEF1 is inversely expressed
with type II collagen mRNA and can repress Col2a1 promoter activity
in transfected chondrocytes. J Biol Chem. 275:3610–3618. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dillner NB and Sanders MM: Transcriptional
activation by the zinc-finger homeodomain protein delta EF1 in
estrogen signaling cascades. DNA Cell Biol. 23:25–34. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du J, Yang S, An D, Hu F, Yuan W, Zhai C
and Zhu T: BMP-6 inhibits microRNA-21 expression in breast cancer
through repressing deltaEF1 and AP-1. Cell Res. 19:487–496. 2009.
View Article : Google Scholar : PubMed/NCBI
|