Introduction
Lung cancer is one of the most malignant types of
tumor and presents a serious threat to the health of human beings.
In recent years, significant progress has been made in the
treatment of lung cancer; however, the therapies remain
unsatisfactory. The overall five-year survival rate for non-small
cell lung cancer (NSCLC; all stages combined) is ~15%, and even for
patients in the early stage of the disease, who have undergone
radical excision, the survival rate is only 50–70% (1). The reasons behind the failure of
therapy are recurrence and metastasis following surgery, which may
be associated with metastasis via the blood or the lymphatic system
in the early stage. Micrometastasis is an indication of a high
possibility of recurrence and metastasis, and is associated with
the pathological pattern, grade of differentiation and tumor node
metastasis (TNM) stage. The early discovery of micrometastasis has
an important clinical significance in recurrence and the prognostic
evaluation of lung cancer. In addition, it has particular value for
improving the prognosis of patients (2). At present, reverse
transcription-polymerase chain reaction (RT-PCR) is used for the
detection of micrometastases from solid tumors and to quantify the
expression of tumor markers and genes associated with tumors in the
peripheral blood. In the present study, the messenger RNA (mRNA)
expression levels of cytokeratin 19 (CK19), epidermal growth factor
receptor (EGFR) and lung-specific X protein (LUNX) were assessed
and the clinical significance of the mRNA levels was evaluated.
Materials and methods
Patients
A total of 42 patients (27 males and 15 females;
age, 23–81 years; average age, 58.1 years), who were diagnosed with
NSCLC by pathology from May 2008 to April 2011, were studied
retrospectively in Taizhou First People’s Hospital (Taizhou,
China). None of the patients presented with second primary tumors
and all the patients received initial treatment. There were 25
patients with squamous carcinomas and 17 patients with
adenocarcinomas. According to the TNM criteria, revised by the
International Union Against Cancer (UICC) in 1997, there were 4
cases in stage I, 12 cases in stage II, 17 cases in stage III and 9
cases in stage IV. Furthermore, there were 19 patients with
well-differentiated, 14 patients with moderately differentiated and
9 patients with poorly differentiated lung cancer. The positive
control group included four patients undergoing surgery for NSCLC.
The healthy control group included 40 individuals (27 males and 13
females; age, 25–79 years; average age, 52.35 years), while the
control group included 40 patients (23 males and 17 females; age,
27–75 years; average age, 55.6 years) with a benign disease. Of
these 40 control patients, 15 patients had pneumonia, 8 patients
had bronchiectasia, 4 patients had a lung abscess, 3 patients had a
tuberculoma and 10 patients had bullae of the lung. This study was
approved by the Ethics Committee of Taizhou First People’s Hospital
(Taizhou, China) and all participants gave written informed
consent.
Reagents and experimental apparatus
Lymphocyte separation medium was purchased from
Shanghai Hengxin Chemical Reagent Co, Ltd. (Shanghai, China) and
TRIzol™ reagent was obtained from Gibco (Carlsbad, CA, USA). Taq
DNA polymerase and a First Strand cDNA Synthesis kit were purchased
from Toyobo (Osaka, Japan). DNA marker was obtained from Beijing
Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China). The
LightCycler 480 instrument used for the RT-PCR assays was from
Roche (Mannheim, Germany). The Multiskan Spectrum was purchased
from Thermo Fisher (Waltham, MA, USA). The UV transmission
detection analyzer (ZF-3) was purchased from Shanghai Jihui
(Shanghai, China)
Blood samples
Prior to any treatment, a tube containing 5 ml
fasting peripheral venous blood was collected from each patient.
Following this, ethylenediaminetetraacetic acid (EDTA) was added
for anticoagulation. The mRNA expression levels of CK19, EGFR and
LUNX in the blood samples were subsequently assessed.
Extraction of total RNAs
Karyocytes were isolated from the blood samples
using lymphocyte separation medium and transferred into an
Eppendorf tube. Following centrifugation, the supernatant was
discarded, 1,500 μl TRIzol reagent was added and the sample was
mixed thoroughly and kept at room temperature for 10 min.
Chloroform (100 μl) was then added and the sample was centrifuged
for 10 min at 6,708 × g and 4°C. The supernatant was transferred to
a 1.5-ml EP tube and, following the addition of isopropanol, was
maintained at 0°C for 20 min. The volume of isopropanol was 3-fold
that of the supernatant. The supernatant and isopropanol were
subsequently centrifuged at 15,093 × g for 15 min, and then the
supernatant was discarded. The RNA precipitate was washed once with
200 μl 75% ethanol, centrifuged, dehumidified and dissolved by
adding 20 μl diethylpyrocarbonate (DEPC). Following surgery, the
NSCLC tissues were cryopreserved in liquid nitrogen. A total of 50
mg NSCLC tissues was used for grinding and the total RNAs were
extracted by the method described above. All the samples were
preserved at −70°C. The concentration and purity were measured
using the Multiskan Spectrum spectrophotometer. A260/A280 ratios in
the range of 1.8 to 2.0 were considered satisfactory for purity
standards in this study.
Synthesis of cDNA
The reverse transcription system (10 μl) included
2.0 μl 5X PrimeScript™ Buffer, 0.5 μl random 6 mers (100 μmol/l),
0.5 μl PrimeScript™ RT enzyme Mix I, 0.5 μl oligo(dT) primer (50
μmol/l), 500 ng total RNA and RNase-free dH2O. The
reaction conditions were 37°C for 15 min, followed by 85°C for 5
sec. cDNA samples were preserved at −20°C.
RT-PCR
The primers were as follows: for GAPDH: (sense:
5′-GGCTGGGACTGGCTGAGCCT-3′ and antisense:
5′-TGGCGACGCAAAAGAAGATG-3′); for CK19: (sense:
5′-GAAATCAGTACGCTGAGGGG-3′ and antisense:
5′-CCGGCTGGTGAACCAGGCTT-3′); for EGFR: (sense:
5′-AAATCCTGCATGGCGCCGTG-3′ and antisense:
5′-GGTGGTTCTGGAAGTCCATC-3′); for LUNX: (sense:
5′-AATGAGGTTCTCAGAGGCTT-3′ and antisense:
5′-TTAGACCTTGATGACAAACT-3′). The quantitative PCR system (20 μl)
included 10.0 μl 2X SYBR Premix Ex Taq, 0.4 μl forward primer (10
μmol/l), 0.5 μl reverse primer (10 μmol/l), 2.0 μl cDNA and 7.2 μl
ddH2O. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
was used as an internal control. The PCR conditions were as
follows: 40 cycles of predenaturation at 95°C for 1 min,
denaturation at 95°C for 5 sec and annealing and extension at 62°C
for 5 sec. Every sample required two duplicated tubes, and two
tubes of positive control and one tube of negative control (without
cDNA template) were included.
Gel electrophoresis
Following amplification, agarose gel electrophoresis
and ethidium bromide (EB) staining were performed. An ultraviolet
(UV) transmission instrument was used to record the results.
Standard curve
The cDNA samples of the NSCLC tissues were diluted
to the following gradients: 100, 101,
102 and 103. Following RT-PCR, amplification
curves were generated, respectively. The cDNA of all the samples in
the 20-μl PCR reaction solutions amounted to the cDNA obtained from
reverse transcription with different total RNA (100, 10, 1 and 0.1
ng respectively). The initial copy numbers of the mRNA in the
samples were set as 100, 10, 1 and 0.1, respectively, prior to the
standard curve being constructed by the LightCycle 480 instrument,
of formula: Y = KX + B, where X is the logarithm of the initial
copy number and Y is the value of the cycle threshold.
Repetitive experiments
Three tubes of positive control and one tube of
negative control were analyzed eight times, respectively, and the
value of the cycle threshold (Ct) was determined. The mRNA
expression levels of GAPDH, CK19, EGFR and LUNX mRNA were
determined.
Relative quantitative analysis
The positive control had an initial copy number of
100, and the relative expression of the target gene in a sample (F)
was defined as the ratio of the expression of the target gene in
the sample to that of the positive control. The expression of the
target gene in a sample was standardized using GAPDH as an internal
reference. From the standard curve, the initial copy number of the
target gene in a sample was obtained. The relative expression of
the target gene of a sample was then calculated using the formula:
10ΔYt/Bt - ΔYg/Bg (where ΔYt is the difference in the target gene
Ct value between the sample and the positive control, ΔYg is the
difference in the GAPDH Ct values between the sample and positive
control, Bt is the slope of the standard curve of the target gene
of the sample and Bg is the slope of the standard curve of
GAPDH).
Statistical analysis
SPSS 15.0 statistical software (SPSS, Inc., Chicago,
IL, USA) was used for data analysis. The quantitative data were
analyzed using one-way analysis of variance (ANOVA) and
χ2 tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
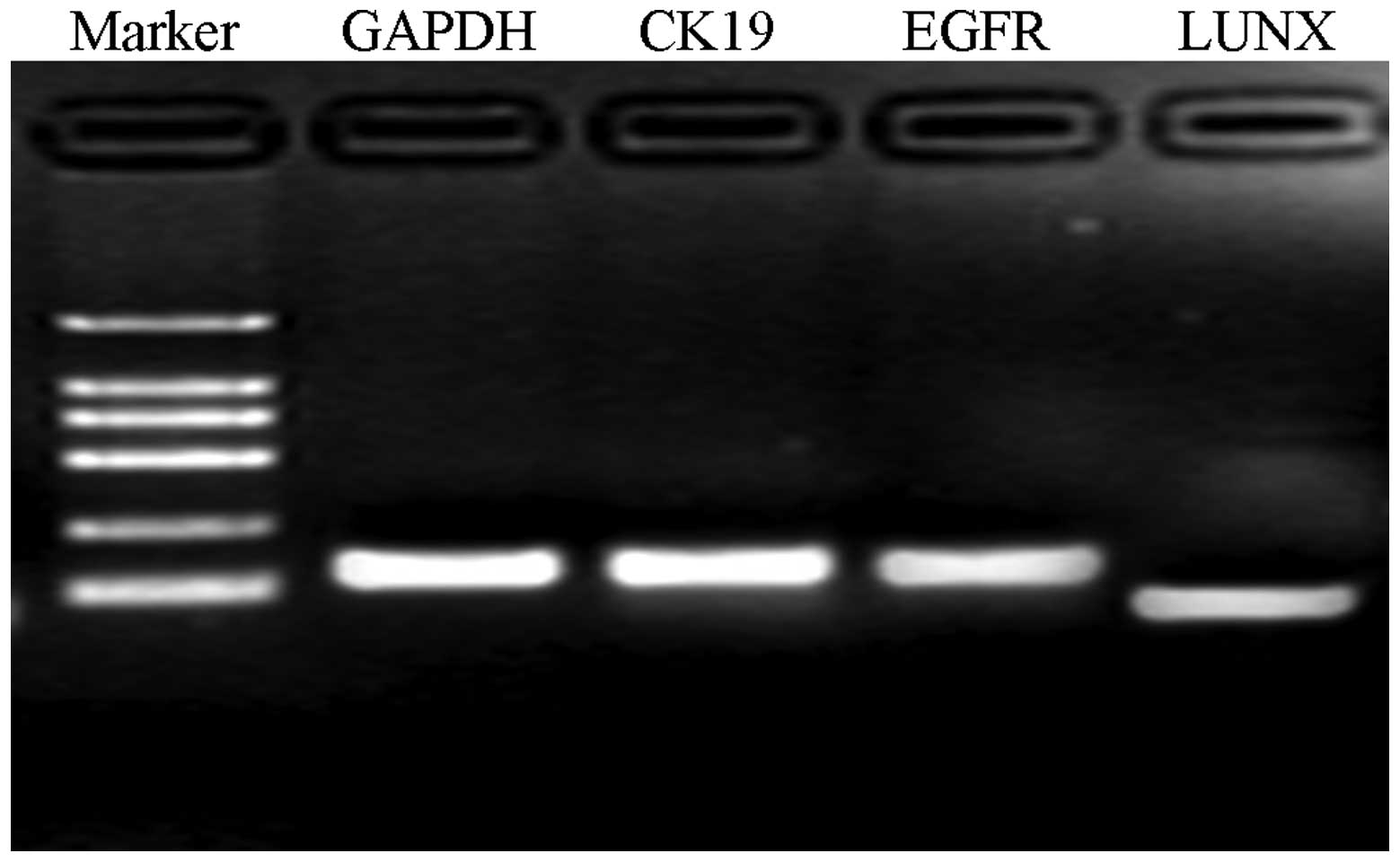
RT-PCR and gel electrophoresis
As shown in Fig. 1,
in the evaluation of the distribution of the mRNA, it was indicated
that the lengths of GAPDH, CK19, EGFR and LUNX mRNA were 150, 130,
126 and 90 bp, respectively. No evident dimer was observed
(Fig. 1).
Standard curve
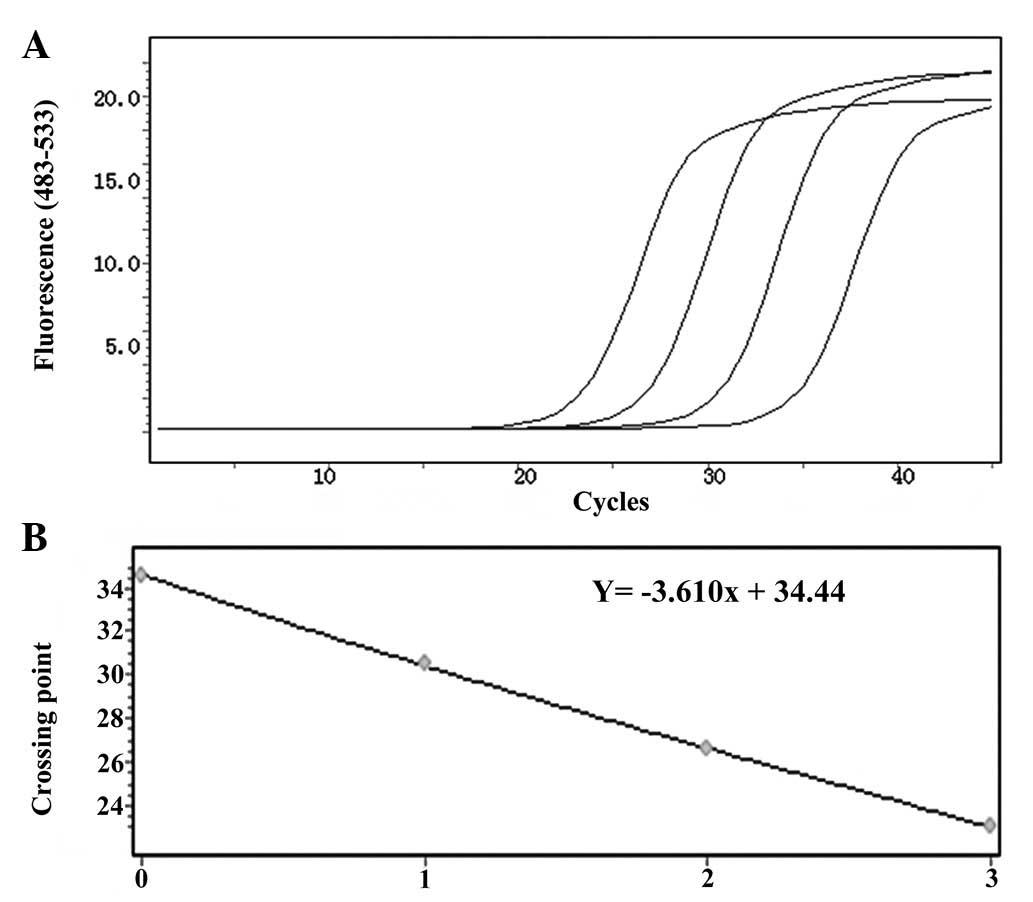
With regard to the amplification curves of the
positive samples, samples with different initial concentrations had
different Ct values, which was the beginning of the exponential
stage of PCR and the gradient (from left to right the initial copy
numbers were 100, 10, 1 and 0.1, respectively). If the difference
was approximated and the initial concentration was considerably
higher, the Ct value was likely to be much smaller. With regard to
the standard curve, there was a linear correlation between the Ct
value (Y) and the logarithm of the copy number of the sample (X).
The correlation coefficient was always 1. It was possible to obtain
the initial copy number of the sample from the standard curve from
the Ct value.
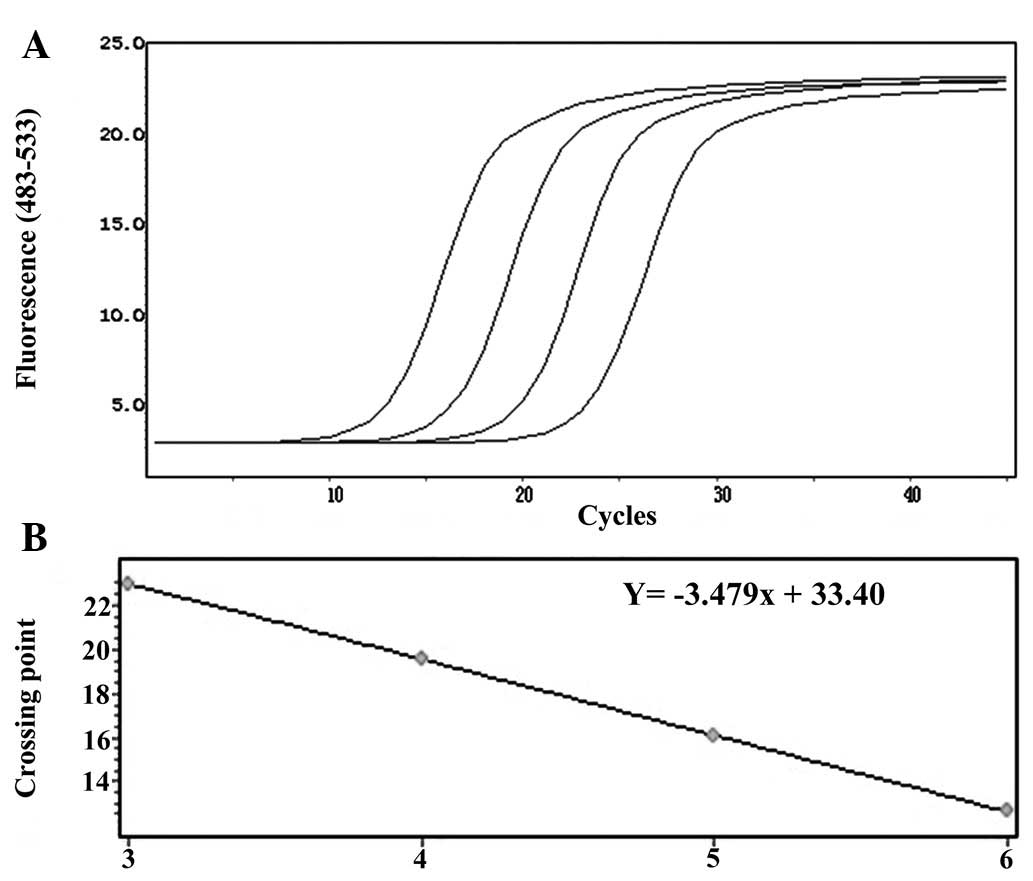
With regard to the amplification curve of GAPDH
(Fig. 2A), the standard curve had
good correlation (Fig. 2B) and the
regression equation was Y = −3.479× + 33.40. Furthermore, as shown
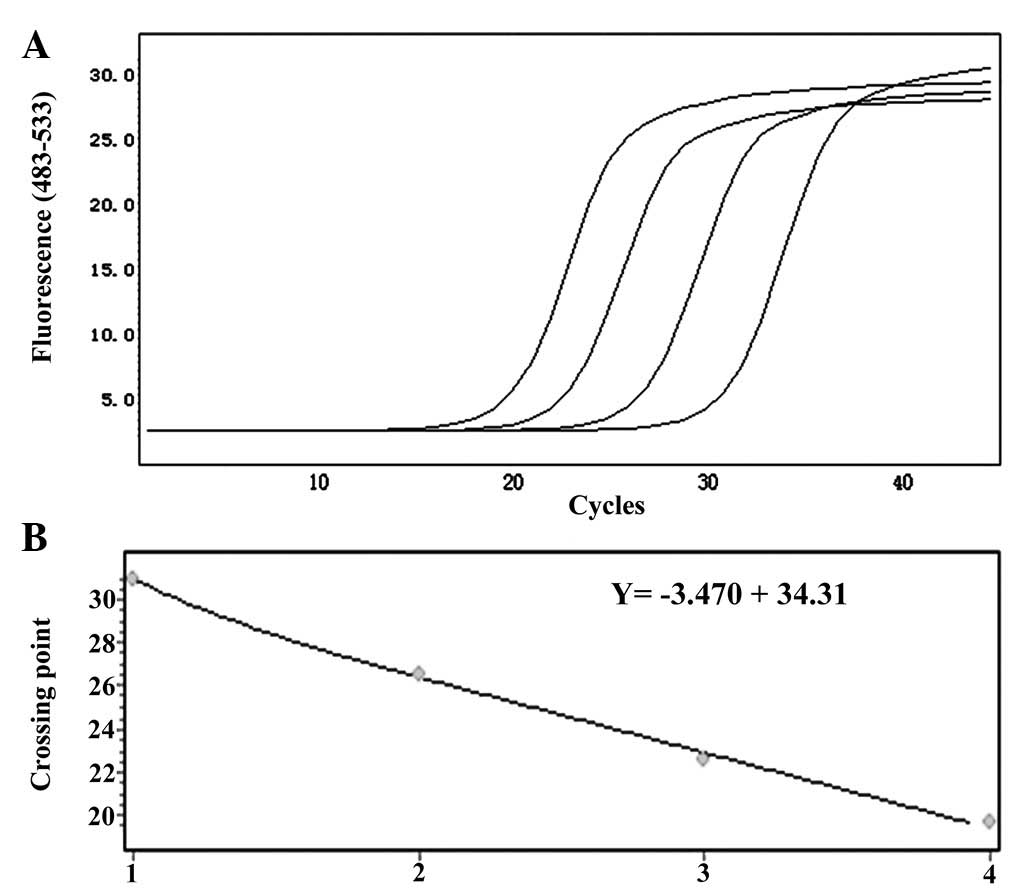
in Fig. 3A, the amplification
curve of CK19 mRNA and the standard curve showed good correlation
(Fig. 3B). The amplification and
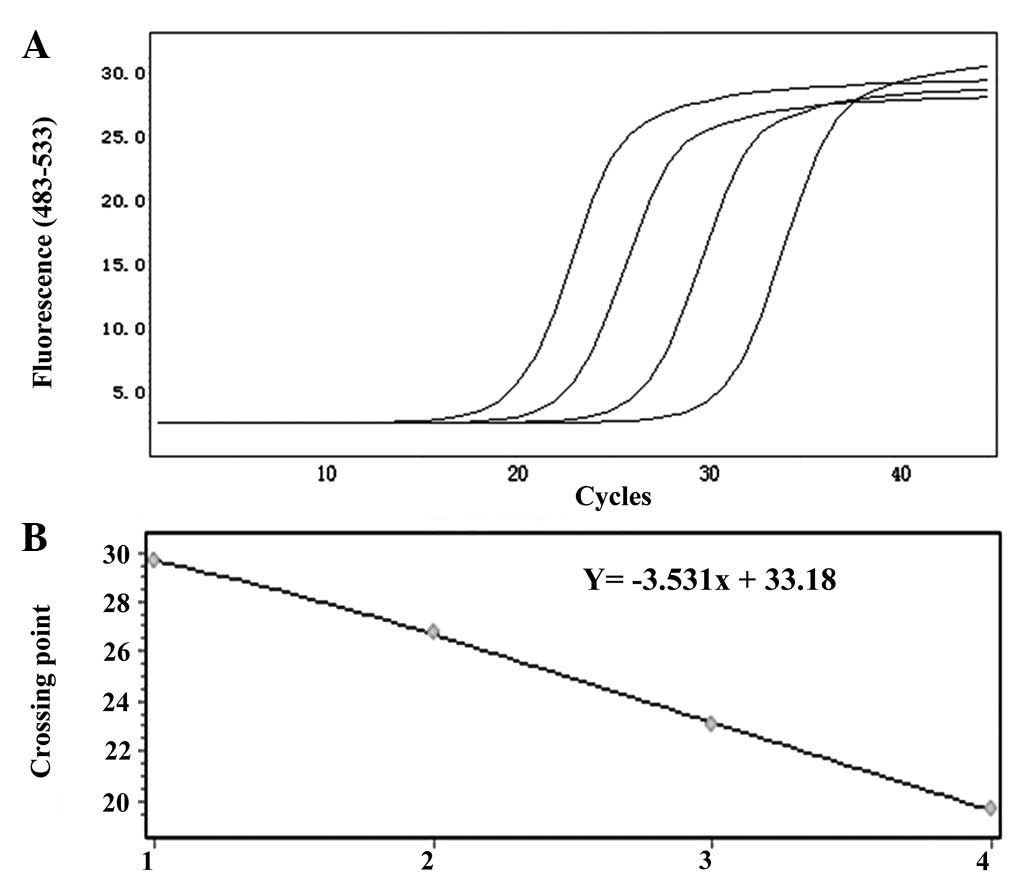
standard curves of EGFR and LUNX mRNA are shown in Figs. 4 and 5, respectively.
The results of the repeated experiments are shown in
Table I. These results indicated
that the Ct values of GAPDH, CK19 mRNA, EGFR mRNA and LUNX mRNA had
high repeatability and stability. The result of the healthy control
group was negative.
 | Table IResults of repetitive experiments. |
Table I
Results of repetitive experiments.
| Analyte | Mean of Ct value | Standard
deviation | Coefficient of
variation |
|---|
| GAPDH | 22.19 | 0.249 | 0.011 |
| CK19 | 27.11 | 0.162 | 0.006 |
| EGFR | 25.61 | 0.216 | 0.008 |
| LUNX | 28.22 | 0.045 | 0.002 |
Relative quantitative analysis
The positive rates of CK19 mRNA in the patient and
healthy control groups were 76.2 (32/42) and 15.0% (6/40)
respectively, which showed a significant difference (P<0.05).
The relative expression levels of CK19 mRNA were 1.72±0.41 and
0.27±0.13, respectively, in the two groups, which showed a
significant difference (P<0.05). The expression level of CK19
did not differ significantly according to the location, size,
clinical stage, differentiation of the primary tumor (all
P>0.05). The relative expression level of CK19 mRNA was higher
for central lung cancer, T3 + T4, stages III and IV and poorly
differentiated lung cancer, although with no significant
differences (all P>0.05). A significant difference was observed
between the expression of CK19 mRNA in squamous carcinoma and
adenocarcinoma (P<0.05), with a higher expression level in
squamous carcinoma. The positive rates of EGFR mRNA in the patient
and healthy control groups were 69.0 (29/42) and 12.5% (5/40)
respectively, which showed a significant difference (P<0.05).
The positive rates of LUNX mRNA in the patient and healthy control
groups were 40.5 (17/42) and 0% (0/40) respectively, which showed a
significant difference (P<0.05; data not shown). Expressions of
LUNX mRNA were observed in 3 cases of stage I and II NSCLC, and 6
cases of stage IV NSCLC.
Discussion
CK is a key component of intermediate fibers of the
cytoskeleton of epithelial cells and is expressed in normal
epithelial cells, epithelial tumors and metastatic cells (3). A false positive for CK expression may
be observed due to the contamination of epithelial cells, the
interference of a pseudogene and the low expression level of CK19
mRNA in the peripheral blood (4).
In order to avoid the contamination of epithelial cells caused by
blood collection in the present study, vacuum blood collection was
used in the clinic and a second tube of blood was used to analyze
the expression of CK19 mRNA. In the present study, the results
indicated that the expression of CK19 mRNA was associated with the
pathology of NSCLC. There was a significant difference in the level
of CK19 mRNA expression between squamous carcinoma and
adenocarcinoma. As a type of molecular marker for the
micrometastasis of NSCLC in the peripheral blood, CK19 mRNA was
shown to be important for the diagnosis of micrometastasis.
However, studies with large samples and follow-ups are required to
improve the specificity.
EGFR is the coreceptor of EGF and transforming
growth factor (TGF). The epidermal growth factor receptor is
commonly overexpressed in NSCLC. Since there are only small numbers
of tumor cells in micrometastases, it has not previously been
possible to use the routine examination of cell morphology to
detect tumor cells due to a low sensitivity (2). However, with the development of
molecular biology and immunology, tumor cells of micrometastases
have been able to be detected. The technologies of
immunohistochemistry, flow cytometry and RT-PCR have been applied
for the detection of the molecular markers of micrometastasis or
tumor cells in the lymph nodes, marrow and peripheral blood. The
positive rate of molecular markers has clinical value as a
predictive indicator of the prognosis (5).
LUNX is a novel human lung-specific gene that was
isolated by differential-display mRNA analysis in a study by Iwao
et al(6). The results of
that study indicated that NSCLC tumors and cancer-free lung tissues
were positive for LUNX mRNA. LUNX mRNA expression was enhanced in
NSCLC tumors. There was no expression of LUNX mRNA in the cells of
the peripheral blood. If LUNX mRNA was detected in the peripheral
blood, it indicated that there were tumor cells and micrometastasis
in the circulation (6). The study
by Cheng et al(7) provided
a detailed evaluation of the lung cancer tumor markers of LUNX,
CK19, carcinoembryonic antigen (CEA), vascular endothelial growth
factor (VEGF-C) and heterogeneous nuclear ribonucleoprotein (hnRNP)
A2/B1 mRNA and assessed the diagnostic utility of these markers in
patients with NSCLC. The results indicated that LUNX mRNA was the
most specific gene marker for lung cancer and had potential
diagnostic utility when measured in the peripheral blood and
pleural fluid of patients with NSCLC (7).
In the present study the results showed that the
positive rate of LUNX mRNA in NSCLC patients was 40.5% (17/42).
There were 3 cases showing the expression of LUNX mRNA out of 16
cases of stage I and II NSCLC, which indicated that there were
micrometastases in the peripheral blood in the early stages of
NSCLC. For the control group (with benign disease), there was no
expression of LUNX mRNA, which showed that the detection of LUNX
mRNA had high specificity. In the present study, there were 6 cases
that tested positive for LUNX mRNA expression out of 9 cases of
stage IV NSCLC, which indicated that the possibility of
micrometastasis in the peripheral blood increased in the advanced
stage. There have been a number of studies concerning the diagnosis
of lung cancer micrometastasis using the detection of LUNX mRNA
(6,8). The results of the study by Iwao et
al(6) showed that LUNX mRNA
was detected in 16/20 (80%) histologically positive lymph nodes and
21/84 (25%) histologically negative lymph nodes (6). The results of the study by Yang et
al(8) revealed that LUNX mRNA
was detected in the peripheral blood and regional lymph nodes, and
that there was no expression of LUNX mRNA in benign lung diseases
and the peripheral blood and lymph nodes of healthy people.
Therefore, the evaluation of mRNA expression of CK19, EGFR and LUNX
in the peripheral blood had important clinical value for the
diagnosis of micrometastasis and the prognosis of lung cancer.
Acknowledgements
This study was supported by the Science and
Technology Bureau of Taizhou Huangyan, China (grant no.
2009085).
References
|
1
|
Francis H and Solomon B: The current
status of targeted therapy for non-small cell lung cancer. Intern
Med J. 40:611–618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bi M and Wang Z: Advances on
micrometastasis of non-small cell lung cancer. Zhongguo Fei Ai Za
Zhi. 12:1041–1043. 2009.(In Chinese).
|
|
3
|
Niu Z, Zhou Q, Sun Z, Sun Z, Zhu W, Wang
Y, Che G, Qin J and Che X: Detection of mRNA expression of CK-19
and MUC1 gene for diagnosis of lymph node micrometastasis in NSCLC
patients by reverse transcriptase-polymerase chain reaction.
Zhongguo Fei Ai Za Zhi. 7:209–213. 2004.(In Chinese).
|
|
4
|
Marrakchi R, Ouerhani S, Benammar S,
Rouissi K, Bouhaha R, Bougatef K, Messai Y, Khadimallah I, Rahal K
and Ben Ammar-Elgaaied A: Detection of cytokeratin 19 mRNA and
CYFRA 21-1 (cytokeratin 19 fragments) in blood of Tunisian women
with breast cancer. Int J Biol Markers. 23:238–243. 2008.PubMed/NCBI
|
|
5
|
Zhong C, Jiang G and Tao G: Expression and
clinical significance of EGFR mRNA, SP-D mRNA, LUNX mRNA in
peripheral blood of lung cancer patients. Nantong University
Journal of Medical Sciences. 27:17–19. 2007.(In Chinese).
|
|
6
|
Iwao K, Watanabe T, Fujiwara Y, Takami K,
Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M and Tanigami
A: Isolation of a novel human lung-specific gene, LUNX, a potential
molecular marker for detection of micrometastasis in non-small-cell
lung cancer. Int J Cancer. 91:433–437. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng M, Chen Y, Yu X, Tian Z and Wei H:
Diagnostic utility of LunX mRNA in peripheral blood and pleural
fluid in patients with primary non-small cell lung cancer. BMC
Cancer. 8:1562008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang HX, Wu YL, Chen G, et al:
Transcriptase polymerase chain reaction assay designed for the
detection of Lunx-mRNA in peripheral blood to research the
micrometastasis in non-small cell lung cancer. Cancer Research On
Prevention and Treatment. 31:464–466. 2004.(In Chinese).
|