Introduction
Stress is a non-specific systemic adaptive response
of the body stimulated by a variety of internal and external
environmental, social and psychological factors, also known as the
stress response. Sympathetic and parasympathetic nervous system
balance and hypothalamic-pituitary-adrenal (HPA) axis function
alter under stress stimulation (1). Furthermore, external stimuli signals
are delivered through multiple pathways or channels into the cells
and trigger a series of cell reactions, including protein
synthesis, degradation and cytokine secretion (2,3).
These cause cardiomyocyte proliferation and hypertrophy or
apoptosis, resulting in myocardial injury.
microRNA (miRNA) is small non-coding RNA of 18–26
bp. miRNA causes the degradation of, or inhibits the translation
of, target mRNA by pairing with specific bases of the target mRNA
and is thus involved in the post-transcriptional regulation of gene
expression (4,5). Studies have observed that miRNA
expression has temporal and tissue specificity (1–6).
miRNA is important in various developmental stages of the
cardiovascular system, regulating cardiomyocyte proliferation,
differentiation and apoptosis under physiological and pathological
conditions (7–19). Dozens of miRNAs have been
identified in myocardial cells, including miR-133a, miR-133b,
miR-1d, miR-296, miR-21, miR-208 and miR-195 (4–18).
Studies have demonstrated that miRNA has an important role in the
regulation of heart development, cardiac hypertrophy, cardiac
electrophysiology and angiogenesis. The use of myocardial genomic
research in the study of stress-induced myocardial injury may
improve the understanding of the mechanisms of stress-induced
myocardial injury. However, there are currently no reports on the
role of miRNA underlying stress-induced myocardial injury.
In the present study, rat models of myocardial
injury induced by psychological stress were established and miRNA
microarrays were used to analyze the changes in the miRNA
expression profiles of injured myocardial tissues. Furthermore, the
role of miRNA in stress-induced myocardial injury was studied and
an attempt was made to locate the target miRNAs which regulate
stress-induced myocardial injury, which may provide the basis for
the development of new drugs for the prevention and treatment of
stress-induced myocardial injury.
Materials and methods
Animals
A total of 24 eight-week-old male Wistar rats with
body weights of 220±10 g were supplied by the Experimental Animal
Center of Jilin University Norman Bethune Medical Division
(Changchun, China). The laboratory was disinfected, quiet and
well-ventilated. The animals were provided with free access to food
and water and were housed at a temperature of 22±2°C, with 50%
humidity and a 12 h natural light-dark cycle. The rats were
randomly divided into the normal control group (NC; n=8), the
chronic stress group (CS; n=8) and the acute stress group (AS;
n=8). The Jilin University Animal Ethics Committee approved the
protocol for this study (no. 2012036).
Modeling
The method for modeling stress in a rat has been
described previously (21). For CS
modeling, the limbs of the rat were bandaged to a board, with the
tail kept free; subsequently the rats were hung upside down and
denied access to food and water randomly for 2 h each day for 14
consecutive days. For AS modeling, the rats fasted for 12–24 h with
access to water, and were subsequently hung upside down with their
limbs bound to a board for 6 h. In all groups, blood was collected
and the concentration of adrenocorticotropic hormone (ACTH) was
determined using an ELISA kit (R&D, Minneapolis, MN, USA) in
order to assess the stress state effect of the model.
Statistical processing for this section was
performed as follows: the SPSS 16.0 software package (IBM Corp.,
Chicago, IL, USA) was used for statistical analysis and measurement
data were stated as the mean ± standard deviation. Comparisons were
performed using the mean adopted t-test, and P<0.01 was
considered to indicate a statistically significant difference.
Extraction and qualification of total
RNA
The rats underwent thoracotomies under anesthesia in
order to remove their hearts. The hearts were washed using 0.2
mol/l phosphate buffer saline (pH 7.0) and stored in liquid
nitrogen. Next, ~100 mg myocardial tissues were frozen and crushed
using a BioPulverizer™ (Aoran Technology LTD, Shanghai, China),
mixed with 1 ml TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA), and homogenized. Total RNA was extracted using
the TRIzol reagent method. Extracted RNA was dissolved in
RNAse-free water and incubated at 60°C for 10 min.
Absorbance at 260, 280 and 230 nm was measured using
an ultraviolet spectrophotometer (ND-1000; NanoDrop, Wilmington,
DE, USA). A260/A280 and
A260/A230 ratios were calculated to determine
the purity of RNA. The RNA concentration was calculated according
to the formula: A260 × 40 ng/μl. Denaturing agarose gel
electrophoresis was performed. Briefly, 3 μg RNA were samples
stored in RNAse-free water mixed with formaldehyde-containing
loading buffer (containing 10 μg/ml ethidium bromide) at a volume
ratio of 1:3 and immediately incubated at 70°C for 15 min in order
to denature the samples. Electrophoresis was performed at a voltage
of 6 V/cm for 10–15 min, and gel images were captured using a gel
image processing system (UVP EC3 600 Imaging System, UVP LLC,
Upland, CA, USA).
miRNA labeling
miRNA was labeled using the miRCURY™ Array Power
Labeling kit (cat no. 208032-A; Exiqon, Inc., Woburn, MA, USA) in
accordance with the manufacturer’s instructions. Briefly, 4 μl calf
intestine phosphatase (CIP) reaction solution (1 μl total RNA, 0.5
μl CIP buffer, 0.5 μl CIP and 2 μl ddH2O) was incubated
at 37°C for 30 min and then at 95°C for 1 min to terminate the
reaction, and immediately placed in an ice bath for 10 min.
Following mild centrifugation (Sigma 4K 15CR; 200 × g), 3 μl
labeling buffer, 1.5 μl fluorescent labels (Hy3™ for the stress
group or Hy5™ for the control), 2 μl DMSO and 2 μl labeling enzyme
were sequentially added in an ice bath. The system was incubated at
16°C for 1 h and subsequently at 65°C for 15 min in order to
terminate the labeling reaction, and stored at 4°C following mild
centrifugation.
miRNA microarray hybridization and
scanning
The labeled RNAs and miRCURY Array chip (Shanghai
Kangcheng BioEngineering Co., Ltd., Shanghai, China) were
hybridized in accordance with the manufacturer’s instructions.
Briefly, 180 μl reaction mixture (25 μl labeled miRNA, 90 μl 2X
hybridization buffer and 65 μl nuclease-free buffer) was incubated
at 95°C in the dark for 2 min and subsequently placed in an ice
bath for 10 min. At the same time, the miRCURY Array chip was
assembled according to the manufacturer’s instructions (Shanghai
Kangcheng BioEngineering Co., Ltd.). The reaction mixture was
loaded through the loading port and the 1X hybridization buffer was
used to fill the hybridization chamber. The microarray chip was
packaged in a protective bag and placed vertically into water
(95°C) for 12 h, followed by drying in an oven at 56°C over night.
The chip was washed using the miRCURY Array Wash Buffer kit (cat
no. 208021, Exiqon, Inc., Woburn, MA, USA) according to the
manufacturer’s instructions, followed by centrifugation at 200 × g
for 5 min to dry the chip. The scanning procedure was performed
using an Axon GenePix 4000B microarray scanner (Axon Instruments,
Inc., Foster City, CA, USA) to obtain the scanning profiles,
followed by data analysis and analysis of the significant
difference using GenePix Pro6.0 software (Axon Instruments,
Inc.).
Data processing and analysis
An experienced operator utilized the GenePix Pro6.0
software for data processing and analysis. The signal points
included in the subsequent analysis should conform to two
prerequisites: i) The signal intensity of the red and green
channels is >0; and ii) the signal to noise ratio (SNR) of the
two channels is >1, or either SNR is >2. The weak signal
points that did not conform to the two prerequisites were excluded
from the subsequent analysis. miRNA was also excluded provided it
contained 3 or 4 weak signal points in a total of four repeat
points.
Lowess standardization (intra-chip standardization)
was performed to remove dye intensity-dependent deviation. Scale
standardization (inter-chip standardization) was used to reduce the
experimental or sample errors between the different chips. Four
repeat points in the chip were merged by calculating the median of
the Hy5/Hy3 ratio of each point. Subsequently the differentially
expressed miRNAs were determined using the statistical Student
t-test. P<0.05 was considered to indicate a statistically
significant difference. Fold change (stress/control) ≥1.5 indicated
upregulation and <0.67 indicated downregulation.
Results
Modeling
Rats in the AS group appeared to be in a stressed
state immediately following the surgery, but in the CS group,
stress appeared after five days. The HPA system excites the stress
stimulus and may be involved in abnormal secretion of
corticotropin-releasing hormone (CRH), ACTH and three
glucocorticoid hormones. The paraventricular nucleus of the
hypothalamus secretes CRH under stress. The adenohypophysis CRH
receptors perceive the change in ACTH and cause the adrenal cortex
to secrete ACTH. ACTH is a key hormone of the HPA axis. Therefore,
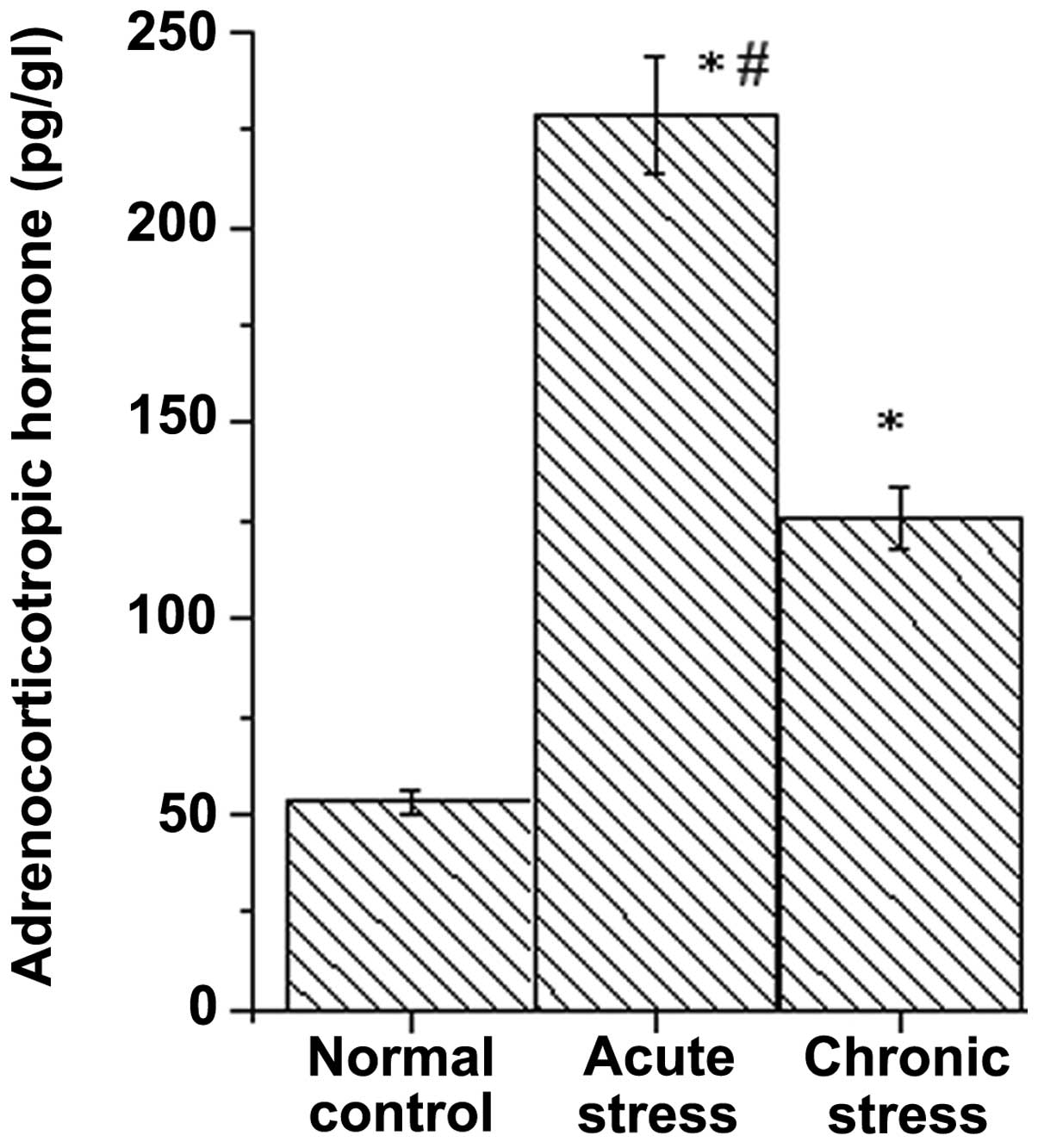
ACTH was selected to evaluate the stress state. As shown in
Fig. 1, the ACTH levels of the
stress model group increased significantly compared with the
control group (P<0.01), indicating that the model was in a
stressed state. ACTH levels of the AS model increased significantly
compared with the CS model (P<0.05).
Extraction and qualification of total
RNA
The A260/A280 ratio of RNA
solution is a method for detecting RNA purity and values close to
2.0 are considered to represent pure RNA. A ratio <1.8 indicates
sample contamination. A ratio >2.0 indicates RNA hydrolysis. The
ratio range between 1.8 and 2.1 is acceptable. In addition, the
A260/A230 ratio should be >1.8 for pure
RNA. As demonstrated in Table I,
the extracted RNAs conformed to the quality standards discussed and
thus qualified for the subsequent miRNA experiments. On the
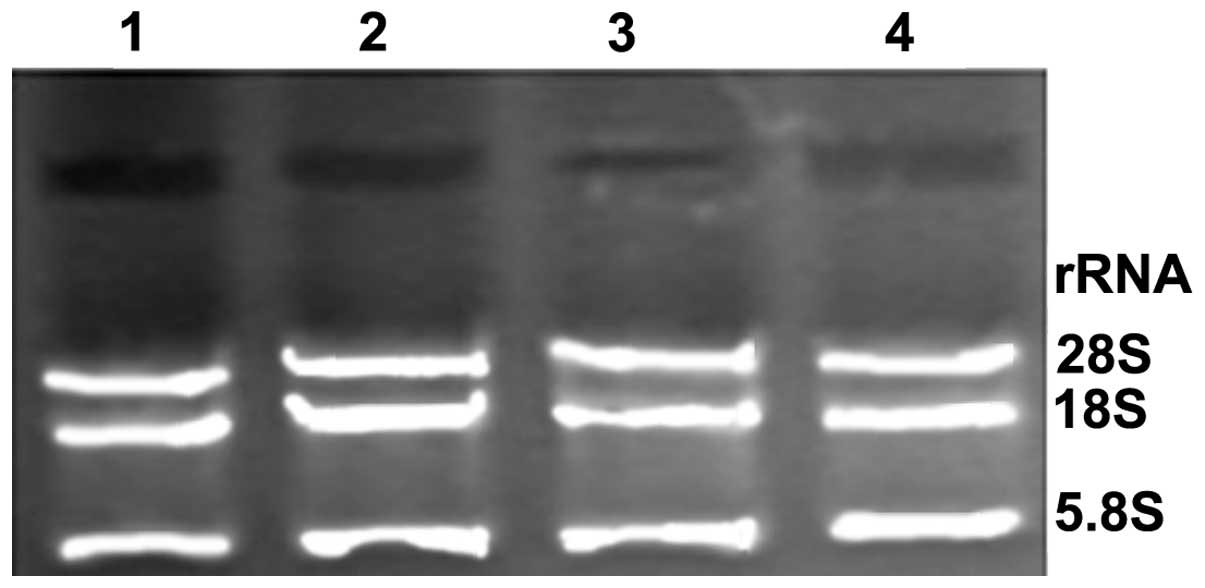
denaturing gel, the 28S, 18S and 5.8S ribosomal RNA (rRNA) bands
were bright (Fig. 2), and the rRNA
bands demonstrated no signs of impurity. This indicated that the
extracted total RNA was complete, RNA degradation and contamination
were low, and the extracted total RNA exhibited high levels of
purity and were of sufficient quality to qualify for use in
subsequent miRNA experiments.
 | Table IPurity of RNA in NC, AS and CS
models. |
Table I
Purity of RNA in NC, AS and CS
models.
| Sample ID | OD260/280
(ratio) | OD260/230
(ratio) | Concentration,
ng/μl | Volume, μl | Quantity, ng | QC result
(pass/fail) |
|---|
| NC1 | 2.03 | 2.24 | 1,063.73 | 15 | 15,955.95 | Pass |
| NC2 | 2.07 | 2.22 | 1,589.88 | 25 | 39,747.00 | Pass |
| AS | 2.06 | 2.22 | 1,533.42 | 50 | 76,671.00 | Pass |
| CS | 2.09 | 2.14 | 1,376.56 | 15 | 20,648.40 | Pass |
Differential expression of miRNAs
According to data processing and analysis, specific
miRNAs were identified to be differentially expressed in the stress
model by comparing them with the normal control group (Tables II and III). There were 68 differentially
expressed miRNAs in AS model, of these, 32 were upregulated and 36
were downregulated; there were 55 differentially expressed miRNAs
in the CS model, of these, 20 were upregulated and 35 were
downregulated. Of the 123 miRNAs, 15 were differentially expressed
in the AS and CS model groups, of these, four were significantly
upregulated (rno-miR-296, rno-miR-141, rno-miR-382 and
rno-miR-219-5p; Table IV) and 11
were downregulated (significantly downregulated, rno-miR-135a and
rno-miR-466b; Table V).
 | Table IIUpregulated miRNA in the AS and CS
models. |
Table II
Upregulated miRNA in the AS and CS
models.
| A, Upregulated miRNA
(AS). |
|---|
|
|---|
| Name | ID | Ratio scale slide 1
(AS/NC1) |
|---|
| rno-miR-21 | 5740 | 1.584 |
| rno-miR-132 | 10937 | 4.004 |
| rno-miR-141 | 10946 | 1.732 |
| rno-miR-19a | 10997 | 1.627 |
| rno-miR-221 | 11022 | 1.600 |
| rno-miR-223 | 11024 | 2.198 |
| rno-miR-31 | 11052 | 1.504 |
| rno-miR-32 | 11053 | 1.716 |
| rno-miR-379 | 11093 | 1.669 |
| rno-miR-129 | 11200 | 1.844 |
| rno-miR-322 | 11225 | 1.524 |
| rno-miR-336 | 11266 | 1.612 |
| rno-miR-201 | 13176 | 1.679 |
| rno-miR-376b-3p | 14304 | 1.725 |
| rno-miR-382 | 14307 | 2.007 |
| rno-miR-147 | 17411 | 1.732 |
| rno-miR-21 | 17896 | 1.697 |
| rno-miR-139-5p | 27542 | 1.618 |
| rno-miR-34b | 29153 | 1.693 |
| rno-miR-674-3p | 31053 | 1.503 |
| rno-miR-34c | 32772 | 1.882 |
| rno-miR-219-5p | 42509 | 1.701 |
| rno-miR-291a-3p | 42595 | 2.026 |
| rno-miR-20b-5p | 42640 | 1.730 |
| rno-miR-296 | 42713 | 1.997 |
| rno-miR-324-3p | 42719 | 1.602 |
| rno-miR-347 | 42763 | 1.907 |
| rno-miR-451 | 42866 | 1.548 |
| rno-miR-20a | 42876 | 2.142 |
| rno-miR-107 | 46629 | 1.566 |
| rno-miR-20a | 46793 | 1.639 |
| rno-miR-375 | 46918 | 1.536 |
|
| B, Upregulated miRNA
(CS). |
|
| Name | ID | Ratio scale slide 2
(CS/NC2) |
|
| rno-miR-141 | 10946 | 2.128 |
| rno-miR-182 | 10975 | 1.528 |
| rno-miR-194 | 10988 | 1.838 |
| rno-miR-377 | 11091 | 1.579 |
| rno-miR-448 | 11113 | 2.067 |
| rno-miR-351 | 11235 | 1.636 |
| rno-miR-344-3p | 11268 | 2.615 |
| rno-miR-381 | 14306 | 1.686 |
| rno-miR-382 | 14307 | 1.676 |
| rno-miR-200c | 17427 | 1.726 |
| rno-miR-877 | 30033 | 2.082 |
| rno-miR-25 | 42481 | 2.027 |
| rno-miR-219-5p | 42509 | 1.519 |
| rno-miR-671 | 42525 | 2.098 |
| rno-miR-330 | 42606 | 2.212 |
| rno-miR-615 | 42690 | 2.269 |
| rno-miR-296 | 42713 | 2.231 |
| rno-miR-218 | 42815 | 1.923 |
|
rno-miR-219-2-3p | 42834 | 2.271 |
| rno-miR-471 | 42916 | 2.519 |
 | Table IIIDownregulated miRNA AS and CS
models. |
Table III
Downregulated miRNA AS and CS
models.
| Downregulated
miRNA(AS) |
|---|
|
|---|
| Name | ID | Ratio scale slide 1
(AS/NC1) |
|---|
| rno-miR-146a | 10952 | 0.6450 |
| rno-miR-184 | 10978 | 0.6700 |
| rno-miR-300-3p | 11221 | 0.6230 |
| rno-miR-325-5p | 11226 | 0.6000 |
| rno-miR-329 | 11227 | 0.4570 |
| rno-miR-341 | 11229 | 0.6137 |
| rno-miR-297 | 11262 | 0.4580 |
| rno-miR-344-3p | 11268 | 0.6310 |
| rno-miR-338 | 17825 | 0.5270 |
| rno-miR-667 | 28944 | 0.6034 |
| rno-miR-708 | 29190 | 0.2780 |
| rno-miR-877 | 30033 | 0.5100 |
| rno-miR-761 | 32608 | 0.3230 |
| rno-miR-129 | 42467 | 0.5560 |
| rno-miR-204 | 42502 | 0.5240 |
| rno-miR-296 | 42528 | 0.6610 |
| rno-miR-196a | 42538 | 0.6040 |
| rno-miR-342-5p | 42576 | 0.4370 |
| rno-miR-466c | 42586 | 0.4200 |
| rno-miR-330 | 42606 | 0.6210 |
| rno-miR-30b-3p | 42626 | 0.4800 |
| rno-miR-878 | 42645 | 0.6120 |
| rno-miR-490 | 42703 | 0.4800 |
| rno-miR-325-3p | 42706 | 0.4530 |
| rno-miR-294 | 42707 | 0.6310 |
| rno-miR-34c | 42767 | 0.5980 |
| rno-miR-665 | 42770 | 0.3530 |
| rno-miR-150 | 42802 | 0.5930 |
| rno-miR-300-5p | 42826 | 0.5880 |
| rno-miR-135a | 42839 | 0.2680 |
| rno-miR-125b | 42845 | 0.6360 |
| rno-miR-743b | 42864 | 0.6190 |
| rno-miR-330 | 42875 | 0.4010 |
| rno-miR-551b | 42917 | 0.4400 |
| rno-miR-466b | 42933 | 0.3010 |
| rno-miR-742 | 42963 | 0.6440 |
|
| Downregulated miRNA
(CS) |
|
| Name | ID | Ratio Scale Slide 2
(CS/NC2) |
|
| rno-miR-9 | 4040 | 0.5790 |
| rno-miR-21 | 5740 | 0.5540 |
| rno-miR-130a | 10138 | 0.6440 |
| rno-miR-146b | 10306 | 0.6080 |
| rno-miR-136 | 10943 | 0.3230 |
| rno-miR-142-3p | 10947 | 0.4850 |
| rno-miR-146a | 10952 | 0.6090 |
| rno-miR-193 | 10986 | 0.5680 |
| rno-miR-204 | 11005 | 0.4770 |
| rno-miR-31 | 11052 | 0.3610 |
| rno-miR-33 | 11062 | 0.5510 |
| rno-miR-363 | 11077 | 0.4930 |
| rno-miR-341 | 11229 | 0.6460 |
| rno-miR-297 | 11262 | 0.5910 |
| rno-miR-10a-5p | 13485 | 0.4630 |
| rno-miR-488 | 17316 | 0.5520 |
| rno-miR-147 | 17411 | 0.5190 |
| rno-miR-425 | 17608 | 0.6640 |
| rno-miR-338 | 17825 | 0.6350 |
| rno-miR-142-5p | 19015 | 0.6220 |
| rno-miR-106b | 19582 | 0.6570 |
|
rno-miR-199a-5p | 19590 | 0.5520 |
| rno-miR-10a-3p | 28019 | 0.5170 |
| rno-miR-872 | 28250 | 0.6700 |
| rno-miR-144 | 29802 | 0.6640 |
| rno-miR-761 | 32608 | 0.5430 |
| rno-miR-129 | 42467 | 0.6670 |
| rno-miR-29b-1 | 42479 | 0.6100 |
| rno-miR-136 | 42512 | 0.4920 |
| rno-miR-338 | 42592 | 0.5560 |
| rno-miR-325-3p | 42706 | 0.6570 |
| rno-miR-150 | 42802 | 0.5740 |
| rno-miR-135a | 42839 | 0.1210 |
| rno-miR-330 | 42875 | 0.6280 |
| rno-miR-466b | 42933 | 0.3620 |
 | Table IVUpregulated miRNA in the AS and CS
models. |
Table IV
Upregulated miRNA in the AS and CS
models.
| Name | ID | Ratio scale slide
1(AS/NC1) | Ratio scale slide
1(CS/NC2) |
|---|
| rno-miR-141a | 10946 | 1.732 | 2.128 |
| rno-miR-382a | 14307 | 2.007 | 1.676 |
|
rno-miR-219-5pa | 42509 | 1.701 | 1.519 |
| rno-miR-296a | 42713 | 1.997 | 2.231 |
 | Table VDownregulated miRNA in the AS and CS
models. |
Table V
Downregulated miRNA in the AS and CS
models.
| Name | ID | Ratio scale slide 1
(AS/NC1) | Ratio scale slide 1
(CS/NC2) |
|---|
|
rno-miR-135aa | 42839 | 0.268 | 0.121 |
|
rno-miR-466ba | 42933 | 0.301 | 0.362 |
| rno-miR-146a | 10952 | 0.645 | 0.609 |
| rno-miR-341 | 11229 | 0.614 | 0.646 |
| rno-miR-338 | 17825 | 0.527 | 0.635 |
| rno-miR-761 | 32608 | 0.323 | 0.543 |
| rno-miR-129 | 42467 | 0.556 | 0.667 |
| rno-miR-325-3p | 42706 | 0.453 | 0.657 |
| rno-miR-150 | 42802 | 0.593 | 0.574 |
| rno-miR-330 | 42875 | 0.401 | 0.628 |
| rno-miR-297 | 11262 | 0.458 | 0.591 |
Discussion
Excessive psychological stress leads to myocardial
injury by changing the function of the sympathetic nervous system
and the HPA axis. Studies have demonstrated that miRNA expression
is tissue-specific and is involved in the formation and maintenance
of tissue specificity during biological development (4–18).
In myocardial cells numerous miRNAs have been identified to have an
important role in the various developmental stages of the
cardiovascular system by regulating cardiomyocyte proliferation,
differentiation and apoptosis under physiological and pathological
conditions (1–3).
The present study analyzed miRNA expression profiles
in the myocardial tissues of AS and CS rat models using miRNA
microarray technology and identified specific differentially
expressed miRNAs. In the differentially expressed miRNAs of the AS
and CS models, 15 miRNAs were differentially expressed in the AS
and CS models. Of these 15 miRNAs, rno-miR-296, rno-miR-141,
rno-miR-382 and rno-miR-219-5p were significantly upregulated,
particularly miR-296 (Table IV),
and 11 were downregulated (miR-135a and miR-466b were significantly
downregulated, particularly miR-135a; Table V). These results indicate that
miRNA changes caused by stress-induced myocardial injury are
different from the miRNA changes caused by other factors.
Therefore, the application of a reasonable stress-induced animal
model, to explore the changes in the miRNAs of myocardial tissues
under stress and identify specifically expressed miRNAs, is
conducive to the further use of RNA interference for the treatment
of stress-induced cardiovascular complications.
miR-125b, miR-146, miR-150, miR-199a, miR-21,
miR-129, miR-341 and miR-451 have been confirmed to play an
important role in the different developmental stages of the
cardiovascular system (4–18). They are also significantly
differentially expressed in the stress-induced model established in
this study, indicating that these miRNAs may be common targets for
myocardial injury caused by various factors. Stress can cause
myocardial injury, thus leading to changes in these miRNAs. This
finding also confirmed that stress itself is an important factor
for myocardial injury.
In two stress-induced models, rno-miR-296 was the
most significantly upregulated and miR-135a was the most
significantly downregulated. Würdinger et al (19) observed that miR-296 may reduce the
level of hepatocyte growth factor-regulated tyrosine kinase
substrate (HGS) and induce the decrease in expression level of
HGS-mediated vascular endothelial growth factor receptor-2 (VEGFR2)
and platelet-derived growth factor receptor β by interacting with
the substrate mRNA of target HGS. miR-296 promotes upregulation of
VEGFR and contributes to angiogenesis. In addition, the inhibition
of miR-296 has been found to reduce angiogenesis of xenograft
tumors (19). In the present
study, psychological stress-induced myocardial injury led to the
upregulation of miR-296.
NR3C2 is a ligand-dependent transcription factor
associated with steroid hormones. The transcription factor
regulates the balance of water and ions and affects blood pressure
by regulating water-sodium retention. Sõber et al (21) verified that NR3C2 may be the target
gene of miR-135a and is involved in the regulation of blood
pressure by inhibiting the in vitro translation of NR3C2 to
regulate the angiotensin-aldosterone system balance. In the present
study, miR-135a was significantly downregulated. We hypothesized
that miR-135a may interact with the target genes to inhibit
sympathetic nerve excitation and suppress the HPA axis and the
renin-vascular angiotensin system, resulting in the release of a
variety of stress hormones, including catecholamines, cortical
hormone, pancreatic glucagon and renin, thus protecting the
myocardium from injury.
In conclusion, rno-miR-296, rno-miR-141,
rno-miR-382, rno-miR-219-5p, miR-135a and miR-466b may be involved
in stress at the molecular level, thus causing myocardial injury.
The development of stress-induced myocardial injury is a complex
biological process and involves a variety of mechanisms. After
cells receive external stimuli, stimulatory signals are transferred
through multiple pathways or channels into the cells, causing a
series of reactions. Altering the conditions in these cells may
lead to the activation of the cell death pathway, particularly the
activation of the mitochondrial death mechanism, causing a death
cascade reaction, which includes cell necrosis and apoptosis. miRNA
may cause degradation of the target mRNA or inhibit its translation
by pairing with the specific base of target mRNA and thus play a
role in post-transcriptional regulation. Multiple miRNAs can
jointly regulate the same target gene, and multiple target genes
are capable of interacting with the same miRNA. By regulating the
level of mRNA transcription, miRNA controls the amount of protein
synthesis, thus regulating the occurrence and development of
cardiovascular diseases. Once the specific miRNA is screened out,
the target gene may be predicted. Furthermore, cardiomyocytes of
miRNA inhibition and overexpression, following gene transfer using
the miRNA mimic and miRNA inhibitors methods, may be cultured in
vitro to explore the relationship between the miRNA and target
genes in cardiomyocytes, which the authors believe should be
studied further. The specific miRNAs found in the present study are
the key to the further study of miRNA function.
Acknowledgements
This study was funded by a grant from the National
Natural Science Foundation of China (no. 30940041).
References
|
1
|
Mann DL: MicroRNAs and the failing heart.
N Engl J Med. 356:2644–2645. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thum T, Galuppo P, Wolf C, et al:
MicroRNAs in the human heart: a clue to fetal gene reprogramming in
heart failure. Circulation. 116:258–267. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carlson TR, Feng Y, Maisonpierre PC,
Mrksich M and Morla AO: Direct cell adhesion to the angiopoietins
mediated by integrins. J Biol Chem. 276:26516–26525. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen JF, Mandel EM, Thomson JM, et al: The
role of microRNA-1 and microRNA-133 in skeletal muscle
proliferation and differentiation. Nat Genet. 38:228–233. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Y, Ransom JF, Li A, et al:
Dysregulation of cardiogenesis, cardiac conduction, and cell cycle
in mice lacking miRNA-1-2. Cell. 129:303–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harfouche R, Gratton JP, Yancopoulos GD,
Noseda M, Karsan A and Hussain SN: Angiopoietin-1 activates both
anti- and proapoptotic mitogen-activated protein kinases. FASEB J.
17:1523–1525. 2003.PubMed/NCBI
|
|
7
|
Carè A, Catalucci D, Felicetti F, et al:
MicroRNA-133 controls cardiac hypertrophy. Nat Med. 13:613–618.
2007.
|
|
8
|
van Rooij E, Sutherland LB, Liu N, et al:
A signature pattern of stress-responsive microRNAs that can evoke
cardiac hyportrophy and heart failure. Proc Natl Acad Sci USA.
103:18255–18260. 2006.PubMed/NCBI
|
|
9
|
Thum T, Gross C, Fiedler J, et al:
MicroRNA-21 contributes to myocardial disease by stimulating MAP
kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tatsuguchi M, Seok HY and Callis TE:
Expression of microRNAs is dynamically regulated during
cardiomyocyte hypertrophy. J Mol Cell Cardiol. 42:1137–1141. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sayed D, Rane S, Lypowy J, et al:
MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen
H, Dean DB and Zhang C: MicroRNAs are aberrantly expressed in
hypertrophic heart: do they play a role in cardiac hypertrophy? Am
J Pathol. 170:183l–1840. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fiedler U, Reiss Y, Scharpfenecker M, et
al: Angiopoietin-2 sensitizes endothelial cells to TNF-alpha and
has a crucial role in the induction of inflammation. Nat Med.
12:235–239. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Rooij E, Sutherland LB, Qi X,
Richardson JA, Hill J and Olson EN: Control of stress-dependent
cardiac growth and gene expression by a microRNA. Science.
316:575–579. 2007.PubMed/NCBI
|
|
15
|
Duisters RF, Tijsen AJ, Schroen B, et al:
miR-133 and miR-30 regulate connective tissue growth factor:
implications for a role of microRNAs in myocardial matrix
remodeling. Circ Res. 104:170–178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin Z, Murtaza I, Wang K, Jiao J, Gao J
and Li PF: miR-23a functions downstream of NFATc3 to regulate
cardiac hypertrophy. Proc Natl Acad Sci USA. 106:12103–12108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harris TA, Yamakuchi M, Ferlito M, Mendell
JT and Lowenstein CJ: MicroRNA-126 regulates endothelial expression
of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA.
105:1516–1521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang S, Aurora AB, Johnson BA, et al: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Würdinger T, Tannous BA, Saydam O, et al:
miR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393.
2008.PubMed/NCBI
|
|
20
|
Chico TJ, Milo M and Crossman DC: The
genetics of cardiovascular disease: new insights from emerging
approaches. J Pathol. 220:186–197. 2010.PubMed/NCBI
|
|
21
|
Sõber S, Laan M and Annilo T: MicroRNAs
miR-124 and miR-135a are potential regulators of the
mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys
Res Commun. 391:727–732. 2010.PubMed/NCBI
|