Introduction
Gastric cancer is the fourth most common type of
cancer causing ~800,000 mortalities worldwide each year (1). In general, gastric cancer has a
5-year survival rate of ~15% and for patients with advanced gastric
cancer, the median overall survival is <1 year (2,3).
Considering these statistics, increased research into potential
preventative methods for gastric cancer is required, as well as
improved early detection and more effective treatments.
Human epidermal growth factor receptor 2 (ERBB2) is
an important biomarker not only for breast cancer, but for other
types of cancer prognosis and patient treatment decisions as well.
ERBB2 is a member of the epidermal growth factor receptor (EGFR)
family that is associated with increased proliferation of tumor
cells. As determined by various studies, between 6 and 35% of
patients with gastric and gastroesophageal junction cancers exhibit
ERBB2 gene amplification and protein overexpression (3–6).
MicroRNAs (miRNAs) are short non-coding RNA
molecules that suppress the expression of protein coding genes by
partial complementary binding, particularly to 3′ untranslated
regions (UTRs) of mRNAs. Alterations in miRNA expression are
involved in the initiation, progression and metastasis of human
cancer and it has been hypothesized that miRNAs function as tumor
suppressors and oncogenes in cancer development (7,8).
Based on 160 paired samples of non-tumor mucosa and cancer cells,
Ueda et al reported that 22 miRNAs were upregulated and 13
miRNAs were downregulated in gastric cancer, indicating that
specific miRNAs are associated with the progression and prognosis
of gastric cancer (9).
A number of studies have revealed that the
expression of miR-375 is reduced in several human cancers,
including head and neck squamous cell carcinoma, esophageal cancer
and hepatocellular carcinoma (10–12).
In addition, previous studies have indicated that miR-375 may be
one of the most important miRNAs involved in the progression of
gastric cancer (13,14). Therefore, in the present study, the
expression and mechanisms of miR-375 were investigated in gastric
cancer with the aim of providing a novel candidate for the
diagnosis and treatment of human gastric cancer.
Materials and methods
Tissue samples
For miR-375 detection, 30-paired gastric tissue
samples were collected (cancer lesions and adjacent non-tumor
mucosae) from patients that had undergone gastrectomy at Renji
Hospital (Shanghai, China) from March 2011 to January 2013. All the
samples were collected in the same manner and snap-frozen
immediately in liquid nitrogen. The samples were stored at −80°C
until required for RNA and protein extraction. Since
microdissection is difficult to perform in diffuse-type gastric
cancer, bulk tissue was used in all the cases for technical
uniformity. Approval for the study was provided by the Ethics
Committee of Renji Hospital and every patient provided written
informed consent. Diagnosis of gastric cancer was confirmed by at
least two pathologists and staging was based on pathological
observations according to the 7th American Joint Committee on
Cancer guidelines (15).
Gastric cancer cell lines
The BGC-823 human gastric adenoma cell line was
purchased from the Cell Bank of Shanghai (Shanghai, China). Cells
were routinely cultured in RPMI 1640 medium, supplemented with 10%
fetal bovine serum (Hyclone, Logan, UT, USA), at 37°C in a
humidified atmosphere with 5% CO2.
ERBB2 expression vector construction
The full length coding region of human ERBB2 was
amplified by reverse transcription polymerase chain reaction (PCR)
and cloned into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA,
USA), which was then designated as pcDNA3.1-ERBB2. This vector and
the control vector, pcDNA3.1, were transfected into cells using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA), according to the manufacturer’s instructions.
3′-UTR luciferase reporter assays
To generate the 3′-UTR luciferase reporter, the full
length of the 3′-UTR from ERBB2 was cloned into the downstream
region of the firefly luciferase gene using the pGL3-control vector
(Promega Corporation, Madison, WI, USA). Mutant miR-375 target
sites in the 3′-UTR of ERBB2 were used as corresponding controls.
An miR-375 mimic and inhibitor were synthesized by Shanghai
GenePharma Co., Ltd (Shanghai, China) and a pRL-TK plasmid
(Promega), containing Renilla luciferase, was cotransfected
for data normalization. For the luciferase reporter assays, BGC-823
cells were seeded in 48-well plates. Luciferase reporter vectors
were cotransfected with miR-375 mimic or miR-375 inhibitor using
Lipofectamine 2000. After two days, the cells were harvested and
assayed with the Dual-Luciferase Assay (Promega Corporation).
Experiments were performed in triplicate and the results are
expressed as relative luciferase activity (Firefly luciferase
activity/Renilla luciferase activity).
Western blot analysis
Protein extracts were boiled in
SDS/β-mercaptoethanol sample buffer, and 30-μg protein samples were
loaded into each lane of the 8% polyacrylamide gels. Proteins were
separated by electrophoresis and then blotted onto polyvinylidene
fluoride membranes (Amersham Pharmacia Biotech, Amersham, UK) by
electrophoretic transfer. The membranes were incubated with mouse
anti-ERBB2 (Abcam, Cambridge, MA, USA) or mouse anti-β-actin
monoclonal antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) for 1 h at 37°C. Specific protein-antibody complexes were
then detected using horseradish peroxidase-conjugated rabbit
anti-mouse secondary IgG. Detection was performed using an enhanced
chemiluminescence kit (Pierce Manufacturing, Inc., Appleton, WI,
USA) and the β-actin signal was used as a loading control.
RNA extraction and miR-375 expression
detection
Quantitative PCR analysis was used to determine the
relative expression levels of miR-375. Total RNA was extracted from
the tissue samples using TRIzol reagent (Invitrogen Lift
Technologies), according to the manufacturer’s instructions. The
expression level of miR-375 was detected using TaqMan miRNA
quantitative PCR. Single-stranded cDNA was synthesized using a
TaqMan microRNA reverse transcription kit (Applied
Biosystems, Inc., Foster City, CA, USA), which was then amplified
using TaqMan Universal PCR Master Mix (Applied Biosystems,
Inc.) with miRNA-specific TaqMan minor groove binder probes
(Applied Biosystems, Inc.). U6 spliceosomal RNA was used for
normalization. Each sample was measured in triplicate for the
detection of miR-375 expression.
Cell proliferation assays
BGC-823 cells were seeded in 96-well plates at a low
density of 5×103 cells/well in Dulbecco’s modified
Eagle’s medium (Hyclone) and allowed to attach overnight. The cells
were then transfected with miR-375 mimic or miR-375 mimic plus
pcDNA3.1-ERBB2, with nonsense short RNA and miR-375 plus pcDNA-3.1
used as controls. Next, 20 μl MTT (5 mg/ml; Sigma-Aldrich, St.
Louis, MO, USA) was added to each well 48 h following transfection
and the cells were incubated for a further 4 h. Absorbance was
recorded at 570 nm with a 96-well plate reader following the
addition of dimethyl sulfoxide.
Statistical analysis
Data were analyzed using SPSS software, version 16
(SPSS, Inc., Chicago, IL, USA). Results from the independent groups
were analyzed using the t-test. Tissue miR-375 expression levels
were analyzed using the Mann-Whitney U-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
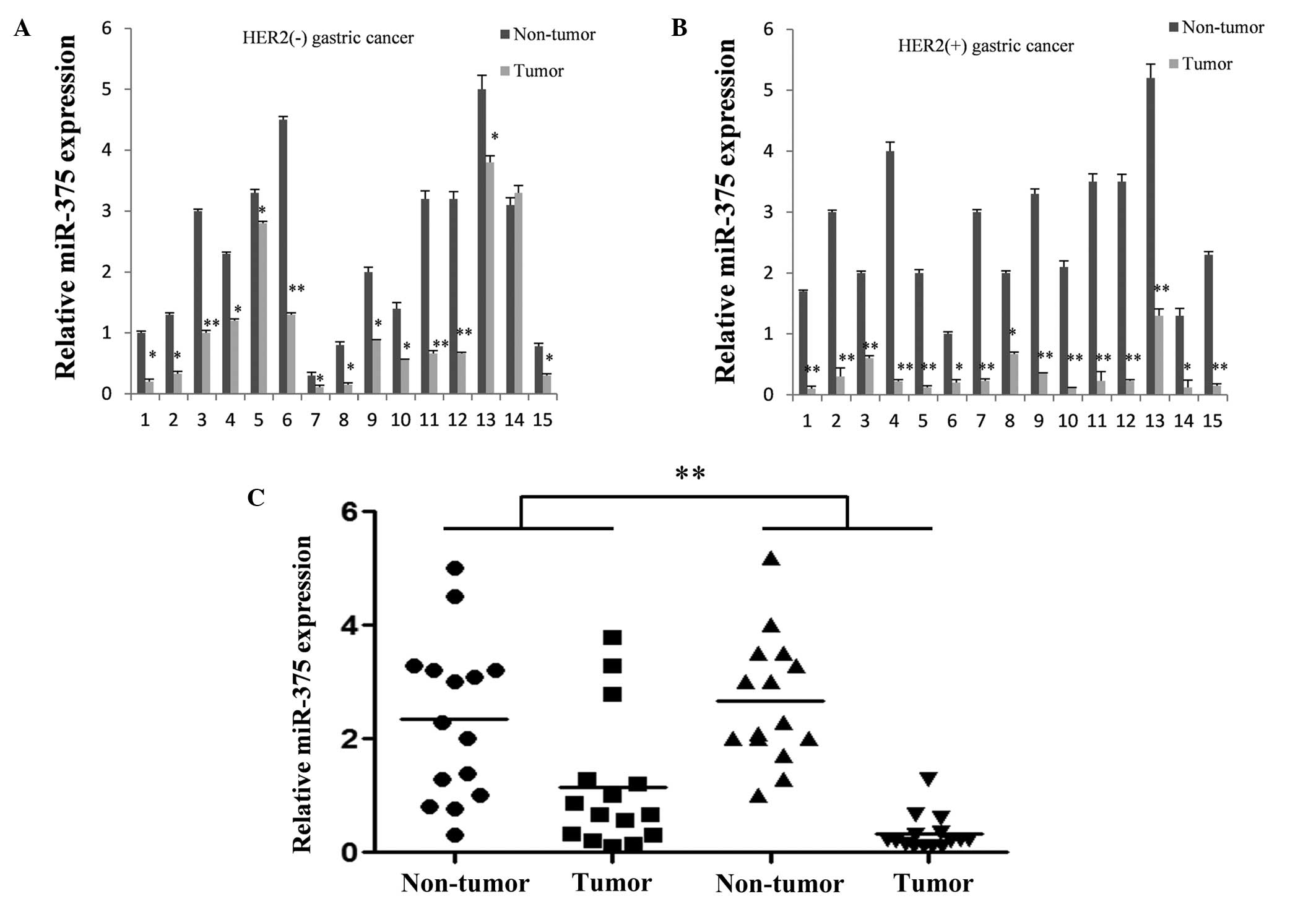
miR-375 is downregulated in gastric
cancer tissues, particularly ERBB2-positive tissues
Quantitative PCR was used to compare the expression
levels of miR-375 among 30 cases of normal and gastric cancer
tissue samples. Each tumor and normal sample was derived from a
single patient specimen. For the majority of cases, the expression
level of miR-375 was significantly decreased in the gastric cancer
tissues when compared with the corresponding non-cancerous tissues
(Fig. 1A and B). In addition,
miR-375 expression levels were markedly reduced in ERBB2-positive
gastric cancer tissues (Fig.
1C).
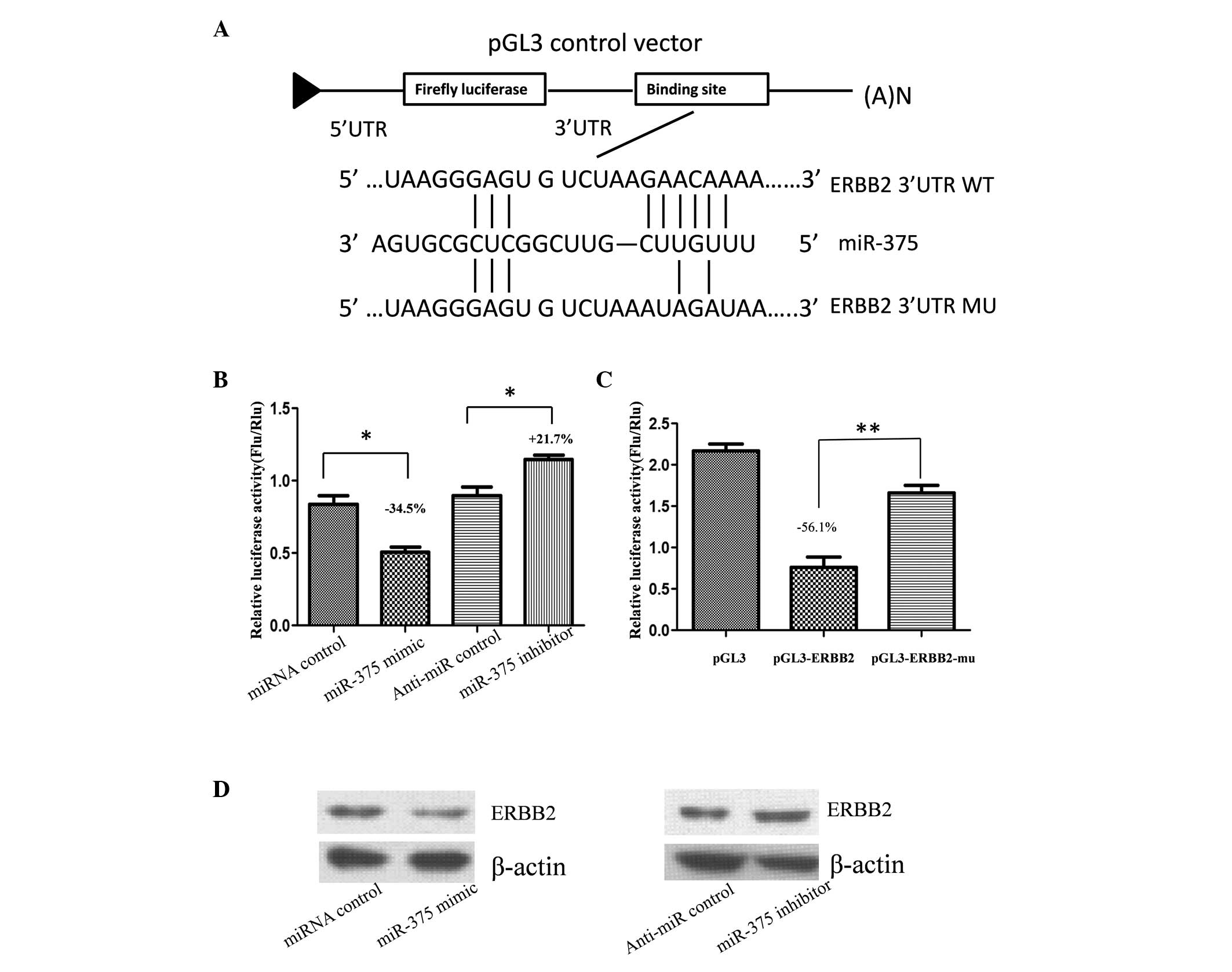
ERBB2 is the target gene of miR-375
miRNA is an important post-transcriptional negative
regulator for protein coding genes. Thus, to explore the
association between reduced miR-375 expression levels and ERBB2
overexpression, miR-375 targets were predicted using the online
bioinformatic tools, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). According
to the results of the online prediction, miR-375 targets ERBB2
directly. Therefore, to validate whether ERBB2 is the target gene
of miR-375, the full length of the 3′-UTR of human ERBB2 was cloned
into the downstream region of the firefly luciferase reporter gene
using the pGL3 control vector (pGL3-ERBB2) for the dual luciferase
assay (Fig. 2A). Human gastric
adenoma BGC-823 cells were cotransfected with pGL3-ERBB2 and
miR-375 mimic or inhibitor (Fig.
2B). Compared with the miRNA control, luciferase activity was
significantly suppressed with miR-375 by ~34.5% (P<0.05).
Furthermore, luciferase activity was significantly upregulated by
~21.7% (P<0.05) with the miR-375 inhibitor, as compared with the
anti-miR control. These results indicate that miR-375 targets the
3′-UTR of ERBB2, leading to a change in firefly luciferase
translation.
A seed sequence mutation clone was also used to
further confirm the binding site for miR-375 (Fig. 2A). A putative miR-375 binding
region in the 3′-UTR of ERBB2 with four mutant nucleotides
(pGL3-ERBB2-Mu) and the pGL3 empty vector were used as controls.
The histogram in Fig. 2C indicates
that enzyme activity was reduced by ~56.1% in cells that had been
cotransfected with miR-375 mimic and pGL3-ERBB2, as compared with
pGL3-ERBB2-Mu (P<0.01). These results indicate that miR-375
suppresses gene expression by binding to the seed sequence at the
3′-UTR of ERBB2, thus, ERBB2 may be a direct target of miR-375.
miR-375 regulates endogenous ERBB2
expression in human gastric cancer cells
Although ERBB2 was identified as a target gene for
miR-375, whether miR-375 regulated endogenous ERBB2 expression was
unknown. Thus, BGC-823 cells were transfected with miR-375 mimic or
inhibitor to determine whether the dysregulation of miR-375
expression affected endogenous ERBB2 expression. Compared with the
corresponding control, the level of ERBB2 protein expression was
significantly suppressed by miR-375 mimic and upregulated by
miR-375 inhibitor (Fig. 2D).
miR-375 overexpression suppresses gastric
cancer cell proliferation
To further investigate whether miR-375 exhibits
tumor-suppressive functions by targeting ERBB2, the effect of ERBB2
on miR-375-mediated cell proliferation was investigated.
BGC-823 cells that had been transfected with miR-375
demonstrated a lower capacity of proliferation compared with cells
that had been transfected with the miRNA control, indicating that
miR-375 suppresses gastric cancer cell proliferation. When the
cells were cotransfected with the ERBB2 expression vector and
miR-375 mimic, the cell proliferation ability was partially
restored, as compared with the control, indicating that miR-375
overexpression mediates the suppression of cell growth through
inhibiting ERBB2 expression.
Discussion
miR-375 was first identified in murine pancreatic
β-cells and the expression was also shown to be enriched in human
pancreatic islet cells. However, miR-375 is not a tissue specific
molecule, as it has also been detected in other tissues, including
the brain and lungs, where it is important for maintaining normal
function.
Low expression levels of miR-375 were first reported
by three studies in 2010 (9,13,14),
which hypothesized that miR-375 was associated with gastric
carcinogenesis. In the present study, the expression levels of
miR-375 were detected in 30-paired cases of normal and gastric
cancer tissue samples. The results demonstrated that miR-375 was
downregulated in almost all the gastric cancer tissues. Notably,
the expression level of miR-375 was significantly lower in
ERBB2-positive gastric cancer tissues as compared with
ERBB2-negative gastric cancer tissues. In addition, miR-375 was
shown to suppress ERBB2 expression by directly targeting the 3′-UTR
of ERBB2, and overexpression of miR-375 was shown to partially
inhibit gastric cancer cell proliferation through the ERBB2
pathway.
ERBB2 is a member of the EGFR family that is
associated with increased proliferation of tumor cells. Between 6
and 35% of patients with gastric and gastroesophageal junction
cancers exhibit ERBB2 gene amplification and protein overexpression
(3). ERBB2 expression is also an
important predictor of gastric cancer patient classification, which
is used for further specific therapies. However, defined by
immunohistochemistry, ERBB2 quantification remains imprecise. The
results of the present study indicate that ERBB2 expression level
detection, associated with quantified miR-375, may be used to
enhance the accuracy of clinical gastric cancer classification
(16).
In conclusion, the present study has partially
clarified the associations between miR-375 and ERBB2-positive
gastric cancer. To the best of our knowledge, this is the first
study to demonstrate that miR-375 is associated with ERBB2-positive
gastric cancer. Therefore, miR-375 is a candidate tumor suppressor
miRNA molecule in gastric cancer and may be a potential clinical
classification marker and therapeutic target for human gastric
cancer.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (nos. 81272743 and 81302094)and
the Shanghai Committee of Science and Technology, China (nos.
11411950800 and 13XD1402500).
References
|
1
|
Danaei G, Vander Hoorn S, Lopez AD, Murray
CJ and Ezzati M; Comparative Risk Assessment collaborating group
(Cancers). Causes of cancer in the world: comparative risk
assessment of nine behavioural and environmental risk factors.
Lancet. 366:1784–1793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delaunoit T: Latest developments and
emerging treatment options in the management of stomach cancer.
Cancer Manag Res. 3:257–266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bang YJ: Advances in the management of
HER2-positive advanced gastric and gastroesophageal junction
cancer. J Clin Gastroenterol. 46:637–648. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hofmann M, Stoss O, Shi D, et al:
Assessment of a HER2 scoring system for gastric cancer: results
from a validation study. Histopathology. 52:797–805. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park YS, Hwang HS, Park HJ, et al:
Comprehensive analysis of HER2 expression and gene amplification in
gastric cancers using immunohistochemistry and in situ
hybridization: which scoring system should we use? Hum Pathol.
43:413–422. 2012. View Article : Google Scholar
|
|
6
|
Fassan M, Mastracci L, Grillo F, et al:
Early HER2 dysregulation in gastric and oesophageal carcinogenesis.
Histopathology. 61:769–776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: a new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ueda T, Volinia S, Okumura H, Shimizu M,
et al: Relation between microRNA expression and progression and
prognosis of gastric cancer: a microRNA expression analysis. Lancet
Oncol. 11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Avissar M, Christensen BC, Kelsey KT and
Marsit CJ: MicroRNA expression ratio is predictive of head and neck
squamous cell carcinoma. Clin Cancer Res. 15:2850–2855. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mathé EA, Nguyen GH, Bowman ED, Zhao Y, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: associations with survival. Clin
Cancer Res. 15:6192–6200. 2009.PubMed/NCBI
|
|
12
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ding L, Xu Y, Zhang W, Deng Y, et al:
MiR-375 frequently downregulated in gastric cancer inhibits cell
proliferation by targeting JAK2. Cell Res. 20:784–793. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, et al: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Washington K: 7th edition of the AJCC
cancer staging manual: stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rüschoff J, Hanna W, Bilous M, Hofmann M,
et al: HER2 testing in gastric cancer: a practical approach. Mod
Pathol. 25:637–650. 2012.PubMed/NCBI
|