Introduction
Ischemic cerebrovascular disease is a disease with a
high incidence worldwide. Progress has been made in the treatment
of ischemic cerebrovascular disease, and the application and
curative effect of stem cells has become an area of increasing
interest. Stromal cell derived factor 1 (SDF-1) is currently the
strongest known chemotactic factor associated with stem cell
mobilization. It can activate a series of signal transduction
pathways through binding to its specific receptor CXC chemokine
receptor 4 (CXCR4), and thus it plays a role in the regulation of
inflammation, mobilization of stem cells and induction of
angiogenesis following ischemia of tissues. Voermans et al
(1) demonstrated that human SDF-1
(hSDF-1α) induced the aggregation of intracellular actin in
CD34+ precursor cells, stimulated the tyrosine
phosphorylation of focal adhesion proteins and changed cytoskeletal
structures, and activated the migration of hematopoietic stem
cells, by rhodamine staining and fluorescence-activated cell
sorting. The combined effects of SDF-1 and vascular endothelial
growth factor can markedly improve the survival time of endothelial
cells. SDF-1 has been found to increase CD34+ cell
proliferation, inhibit apoptosis and promote the differentiation of
CD34+ cells (2,3), which indicates that SDF-1 plays an
important role in the proliferation of hematopoietic stem cells and
the process of homing and mobilization; therefore, studies on SDF-1
have attracted growing attention. SDF-1 was first found in
cytokines secreted by the mouse bone marrow stromal cell line pA6
in 1994 (4). Since four conserved
cysteine residues in the C-terminal and two cysteines in the
N-terminal of its amino acid sequence are separated by another
amino acid, SDF-1 is classified into the CXC subfamily of
chemokines, and is also known as CXCL12 (5). The hSDF-1 gene is located in
chromosome 10q11.1 (6) and encodes
different proteins due to its different splicing modes. The Gen
Bank accession numbers for these different cDNAs and their
associate proteins are SDF-1α, SDF-1β, SDF-1γ, SDF-1δ, SDF-1ε and
SDF-1ϕ. hSDF-1α is an 89-amino-acid protein while SDF-1β, SDF-1γ,
SDF-1δ, SDF-1ε and SDF-ϕ encode 93, 119, 120, 90 and 100-amino-acid
proteins, in all of which the first 89 amino acids are identical to
those of SDF-1α (7,8). The full-length cDNA of hSDF-1α,
which encodes 89 amino acids including the N-terminal signal
peptide, is ∼270 bp. A signal peptide cleavage site exists between
the twentieth and twenty-first amino acids of the N-terminal of
hSDF-1α (9). Crump et al
(10) studied the NMR structure of
SDF-1 in different solution conditions. The results showed that
chemokine tertiary structure consists of a flexible N-terminus
connected by an extended ‘N-loop’ and a single turn of a
310-helix to a three-stranded β-sheet and a C-terminal
α-helix. The functional domain of the C-terminal is important to
maintain SDF-1 conformation and the β-helix to regulate the
activity of the SDF-1 in its interaction with glycosaminoglycans.
The structure of the N-terminal is also important in its
interactions with CXCR4; however the effect of the signal peptide
on the activity of hSDF-1α is unknown.
In the present study, the process and technology of
cloning and expressing the hSDF-1α protein without the N-terminal
signal peptide was reported, and the chemotactic activity of the
recombinant hSDF-1α was identified. This study may help to
understand the technology of the recombinant and purified hSDF-1α,
the diverse physiological functions of hSDF-1α, the activity of
hSDF-1α and the effects of N-terminal signal peptide on the
activity of hSDF-1α.
Materials and methods
HSDF-1α cloning and expression vector
construction
Primers were designed according to the
hSDF-1α cDNA sequence provided by the National Center for
Biotechnology Information (NM_199168). The gene, which had mature
peptide sequences of 213 bp without the signal peptide, was cloned
from the total DNA of the pCMV-SPORT6-SDF1α plasmid (GeneCopoeia,
Guangzhou, China) using polymerase chain reaction (PCR)
amplification with sense primer 5′-GATGCCATGGACGGGAAGCCCGTCAGC-3′ and
antisense primer 5′-CGCGGATCCTTACTTGTTTAAAGCTTTCTCCAGGT-3′
(NcoI and BamHI restriction sites are underlined)
(11). Primers were synthesized by
the Beijing Dingguo Changsheng Biotechnology Co. Ltd. (Beijing,
China). The PCR amplification conditions were as follows: 98°C for
5 min, 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec with a
total of 30 cycles, and 72°C for 10 min for extension. PCR products
were identified by 1.5% agarose gel electrophoresis, retrieved and
purified with a gel extraction kit (Omega Bio-Tek, Inc., Norcross,
GA, USA). The retrieved target fragments and vector pET-15b
(Beijing Dingguo Changsheng Biotechnology Co. Ltd.) were double
digested with NcoI and BamHI restriction enzymes
(Takara Bio Inc., Otsu, Japan) at 37°C for 6 h, respectively.
Digested fragments and plasmids were retrieved with a gel
extraction kit (Takara Bio Inc.) and incubated with T4 ligase
(Takara Bio Inc.) at a constant temperature of 16°C overnight. The
ligation products were then transformed into competent cells of
Escherichia coli (E.coli) DH5α (Beijing Dingguo
Changsheng Biotechnology). The transformed DH5α cells were cultured
on Luria-Bertani (LB) solid medium containing 100 µg/ml ampicillin
sodium salt (Sigma-Aldrich, St. Louis, MO, USA) overnight to screen
the positive clones that were sent to Shanghai Biological
Engineering Co. Ltd. (Shanghai, China) for sequencing. The
recombinant plasmid was verified as correct by sequencing and named
as pET-15b-hSDF-1α.
Inducible expression and analysis of
recombinant protein hSDF-1α
The recombinant hSDF-1α was expressed in the E.
coli cell strain BL21(DE3) (Beijing Dingguo Changsheng
Biotechnology). Bacterial cells transformed with the
pET-15b-hSDF-1α plasmid were grown in LB fluid medium supplied with
100 µg/ml ampicillin sodium salt at 37°C with shaking at 220 rpm
for 12 h (DDHZ-300; TaiCang Experimental Equipment Factory, Suzhou,
China). The following day, 1 ml culture broth was inoculated in 100
ml fresh LB fluid medium supplied with 100 µg/ml ampicillin sodium
salt and grown at 37°C with shaking at 220 rpm for 3 h. Isopropyl
β-D-1-thiogalactopyranoside (IPTG) was added at a final
concentration of 1 mmol/l to induce gene expression for ∼8 h at
30°C with shaking at 220 rpm until the optical density at 600 nm
was 0.7. Bacterial cells were subsequently collected by
centrifugation at 4,444 × g for 20 min at 4°C and frozen at −20°C.
The expression of target proteins was analyzed by
Tris-Tricine-SDS-PAGE electrophoresis (12).
Refolding of recombinant hSDF-1α
protein
The bacterial cell precipitate was mixed with
ultrasound lysis buffer (5 mmol/l EDTA disodium salt and 10 mmol/l
Tris-HCl, pH 8.0) at a ratio of 1:10 (w/v). The precipitate
containing recombinant hSDF-1α was collected following
ultrasonication centrifugation at 8,888 × g for 10 min and
dissolved by denaturing buffer (6 mol/l guanidine hydrochloride, 10
mmol/l β-mercaptoethanol, 5 mmol/l EDTA disodium salt and 10 mmol/l
Tris-HCl, pH 8.0) at a ratio of 1:5 (w/v). The supernatant
containing denatured recombinant hSDF-1α was then collected by
centrifugation at 8,888 × g for 30 min and refolded by dropwise
dilution at 4°C with stirring into refolded buffer (10 mmol/l Tris
HCl, pH 8.0, 5 mmol/l EDTA disodium salt, 1 mmol/l reduced
glutathione and 0.2 mmol/l oxidized glutathione) to yield a final
protein. Following stirring overnight at 4°C the refolded hSDF-1α
was isolated by centrifugation at 8,888 × g for 30 min and the
supernatant containing the refolded recombinant hSDF-1α was
collected.
Separation and purification of
recombinant refolded protein hSDF-1α
Cation exchange was necessary as the next step in
purification (13). Elution buffer A
(0.3 mol/l NaCl, 10 mmol/l Tris HCl, pH 8.5) was first used to
equilibrate the chromatographic column. The supernatant containing
the refolded recombinant hSDF-1α was loaded onto a 5 ml sulfopropyl
(SP) Sepharose fast flow (FF) column (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA) at 2 ml/min and then refolded recombinant
hSDF-1α was eluted from the SP Sepharose FF column in elution
buffer B (1 mol/l NaCl and 10 mmol/l Tris HCl, pH 8.5). The hSDF-1α
was detected by Tris-Tricine-SDS-PAGE electrophoresis.
Purification of recombinant refolded
protein hSDF-1α by size-exclusion chromatography
Size-exclusion chromatography was carried out on a
Sephadex G-75 (GE Healthcare Bio-Sciences) column (1.5/60) with a
flow rate of 1 ml/min, while monitoring the absorbance at 280 nm.
The elution buffer was 10 mmol/l phosphate buffer (pH 7.0) which
was used to equilibrate the Sephadex G-75 column and elute hSDF-1α
protein. The hSDF-1α was detected by Tris-Tricine-SDS-PAGE
electrophoresis.
Determination of recombinant hSDF-1α
activity
A Transwell migration assay with a pore diameter of
8 µm was used to detect the chemotactic migration of THP-1
monocytes to recombinant hSDF-1α (14,15).
THP-1 cells cultured for 3–5 generations were further cultured
using serum-free medium RPMI-1640 (Hyclone, Logan, UT, USA)
containing 0.2% bovine serum albumin (BSA; Sijiqing; Zhejiang
Tianhang Biotechnology Co. Ltd., Beijing, China) for 12 h. The
cells were harvested and resuspended in RPMI-1640 containing 0.5%
fetal bovine serum (FBS) at a concentration of 3×105
cells/ml. hSDF-1α was prepared with concentrations of 0, 10, 100
and 500 ng/ml in 10 mmol/l phosphate buffer (pH 7.0). A 600-µl
amount of hSDF-1α at each of the aforementioned concentrations was
taken and added to 24-well plates (Beyotime Institute of
Biotechnology, Jiangsu, China) in triplicate. A chamber insert was
placed into the 24-well plate and 200 µl cell suspension with a
concentration of 3×105 cells/ml was added to each
chamber. The cells were then incubated in a CO2
incubator for ∼10 h and the cell numbers in each well were counted
and calculated under a microscope (magnification, ×200; TE2000-U;
Nikon Corporation, Tokyo, Japan).
Determination of protein
concentration
Standard protein assay procedures using the Rio-Rad
Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and BSA protein standards were used to determine the hSDF-1α
protein concentration.
Results
Construction and identification of
pET-15b-hSDF-1α
The six known SDF-1 isoforms, SDF-1α, SDF-1β,
SDF-1γ, SDF-1δ, SDF-1ε and SDF-1ϕ share the same first three exons
but contain different fourth exons and have the same signal peptide
at their 5′-end; therefore, the SDF-1 gene was amplified
without the N-terminal signal peptide sequences. Sense and
antisense primers were designed to clone the hSDF-1α gene.
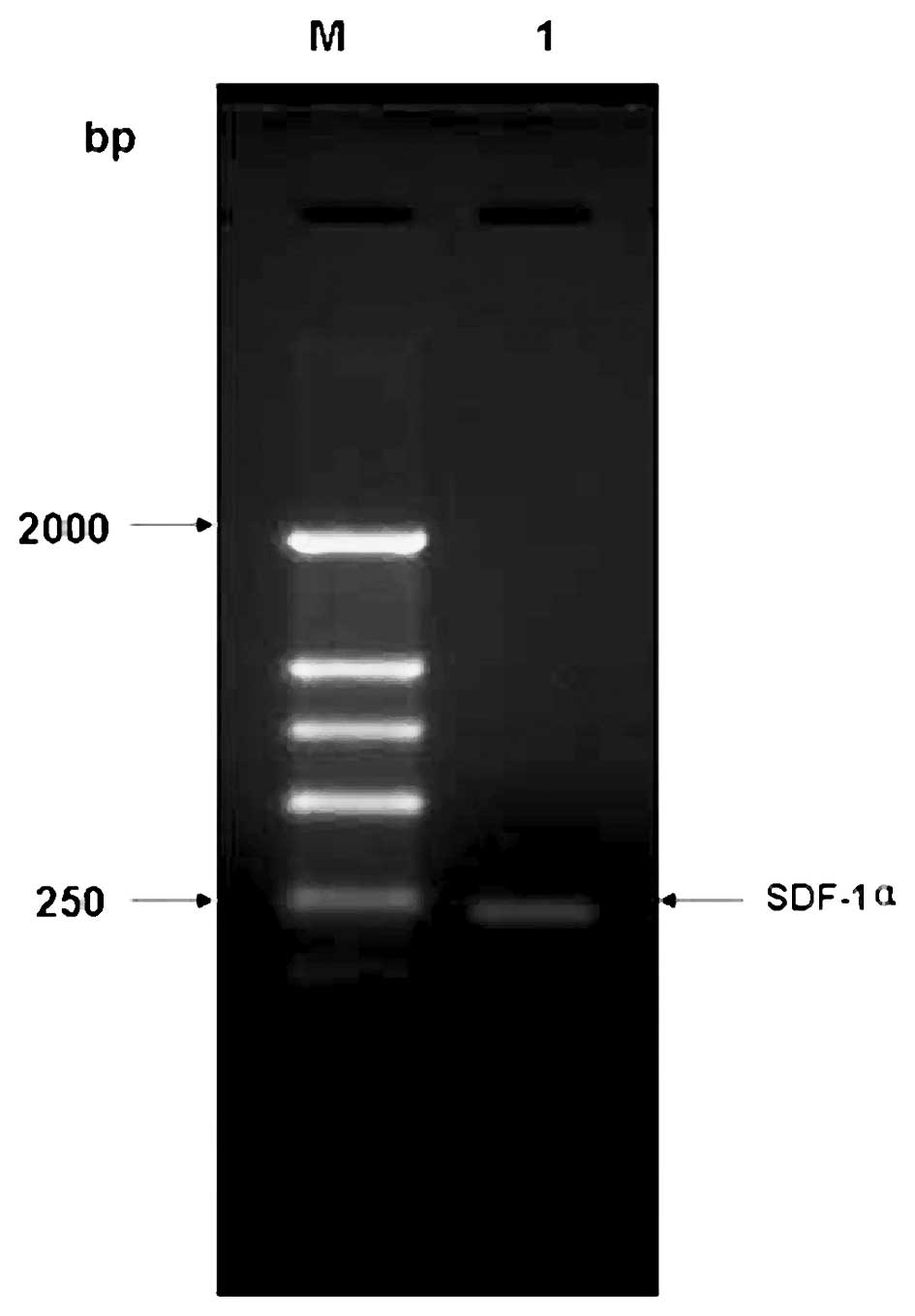
The amplified target fragment identified by 1.5% agarose gel
electrophoresis was ∼213 bp (Fig.
1), indicating that the hSDF-1α gene was successfully
amplified, and was consistent with the theory. The amplified target
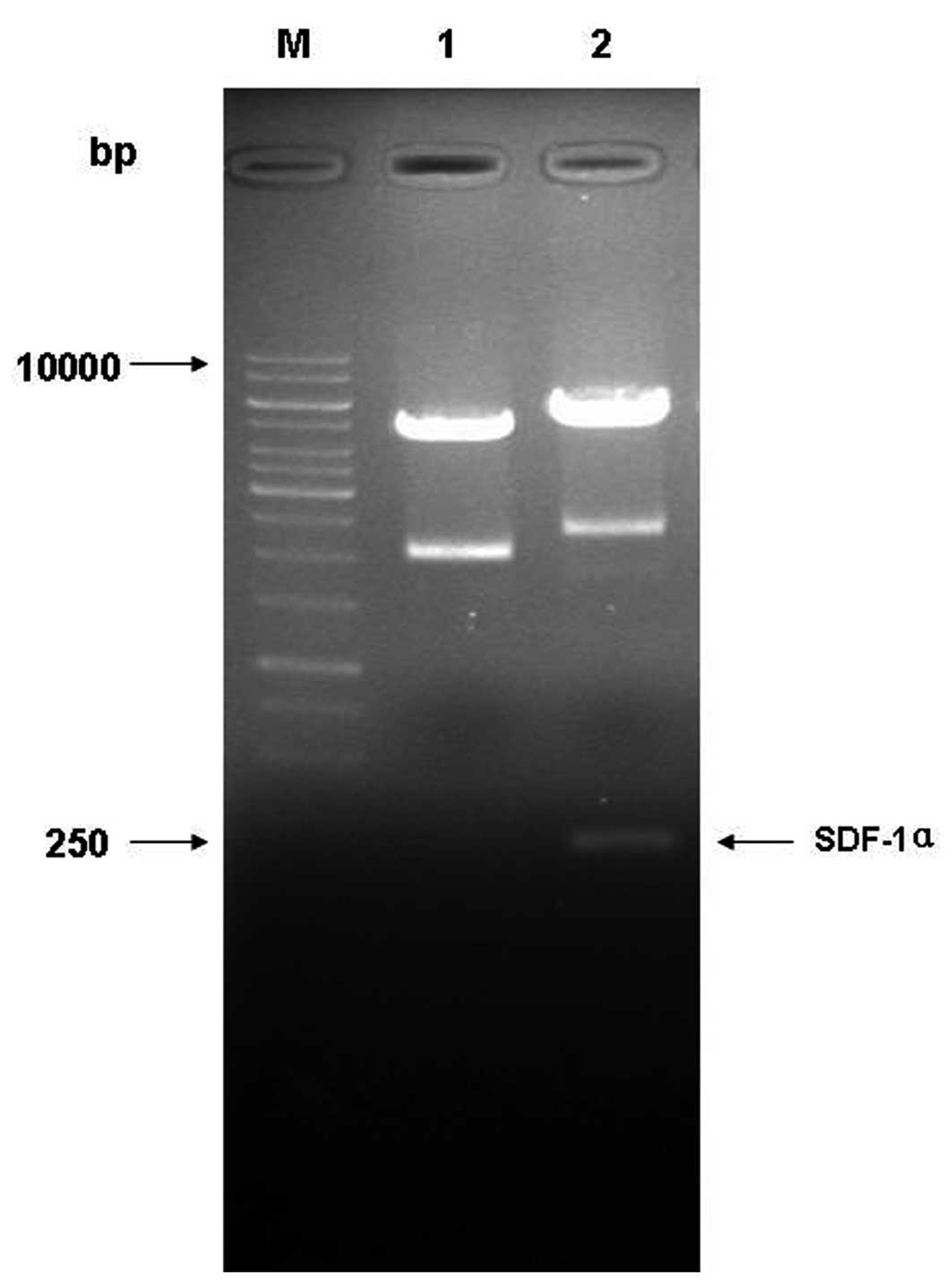
fragment was cloned into the pET-15b vector. The double-enzyme
digestion results demonstrated a band of ∼213 bp in lane 2,
suggesting that the target gene was successfully inserted into
vector pET-15b (Fig. 2). The
positive recombinants identified by double-enzyme digestion were
further sequenced, and the alignment results revealed that the
sequence of the constructed recombinant vector and reading frame
were correct, indicating that the recombinant prokaryotic
expression vector named pET-15b-hSDF-1α was successfully
constructed.
Expression of recombinant protein
hSDF-1α
The recombinant vector pET-15b-hSDF-1α was
transformed into E. coli BL21(DE3) to express the
hSDF-1α gene. The hSDF-1α protein was analyzed with
Tris-Tricine-SDS-PAGE following induction. A band was clearly
observed with a molecular weight corresponding to that estimated
from the deduced amino acid sequence, and the recombinant hSDF-1α
appeared in the precipitate of cell lysates (Fig. 3), suggesting that the recombinant
hSDF-1α was successfully expressed in the form of an inclusion body
in E. coli BL21(DE3). Due to the inactivity of the
aggregated form of hSDF-1α, it was necessary for the hSDF-1α to be
dissolved and refolded in vitro, to provide biologically
active hSDF-1α. Optimal refolding efficiency of hSDF-1α required
optimal conditions of refolding; thus the inactive hSDF-1α was
refolded under optimized refolding conditions. Bradford method
analysis following refolding revealed a refolding rate of 27%.
Cation exchange chromatography required preparation of the refolded
hSDF-1α sample in a low-salt-containing buffer prior to loading on
the cation exchange column to remove any salt or contaminating
materials which would inhibit binding of hSDF-1α to the column. The
refolding process only dilutes the concentration of salt and does
not remove other impurities.
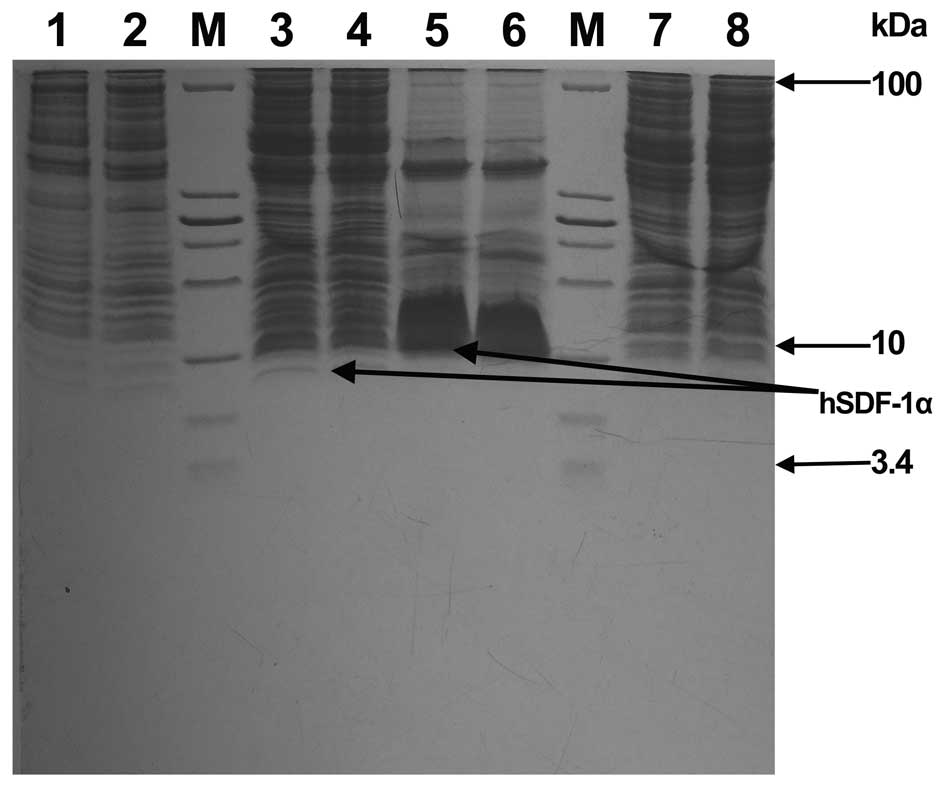 | Figure 3.Expression of recombinant bacteria
E.coli BL21(DE3)/pET15b-SDF1α. Lane M, low protein molecular
weight marker (from top to bottom the molecular weights are 100,
30, 25, 20, 15, 10, 5 and 3.4 kDa); Lanes 1 and 2, bacteria were
cultured at 37°C with shaking at 220 rpm for 2.5 h and then
cultured at 30°C with shaking at 220 rpm for 6 h. 1 ml bacteria was
boiled and analyzed by Tris-Tricine-SDS-PAGE electrophoresis; Lanes
3 and 4, bacteria were cultured at 37°C with shaking at 220 rpm for
2.5 h and then cultured at 30°C with shaking at 220 rpm for 6 h
following the addition of isopropyl β-D-1-thiogalactopyranoside. 1
ml bacteria was boiled and analyzed by Tris-Tricine-SDS-PAGE
electrophoresis; Lanes 5 and 6, the precipitation of bacterial
lysates was analyzed by Tris-Tricine-SDS-PAGE electrophoresis;
Lanes 7 and 8, The supernatant of the bacterial lysates was
analyzed by Tris-Tricine-SDS-PAGE electrophoresis. hSDF, human
stromal cell-derived factor. |
Separation and purification of the
recombinant refolded protein hSDF-1α
A low concentration of refolded hSDF-1α protein with
a high level of impurity necessitated strong cation exchange and
size-exclusion chromatography. Strong cation exchange
chromatography under refolding conditions was employed to remove
the majority of the truncated product, accomplished by thorough
washing with cation exchange buffer (elution buffer A). hSDF-1α was
eluted from the strong cation exchange column in cation exchange
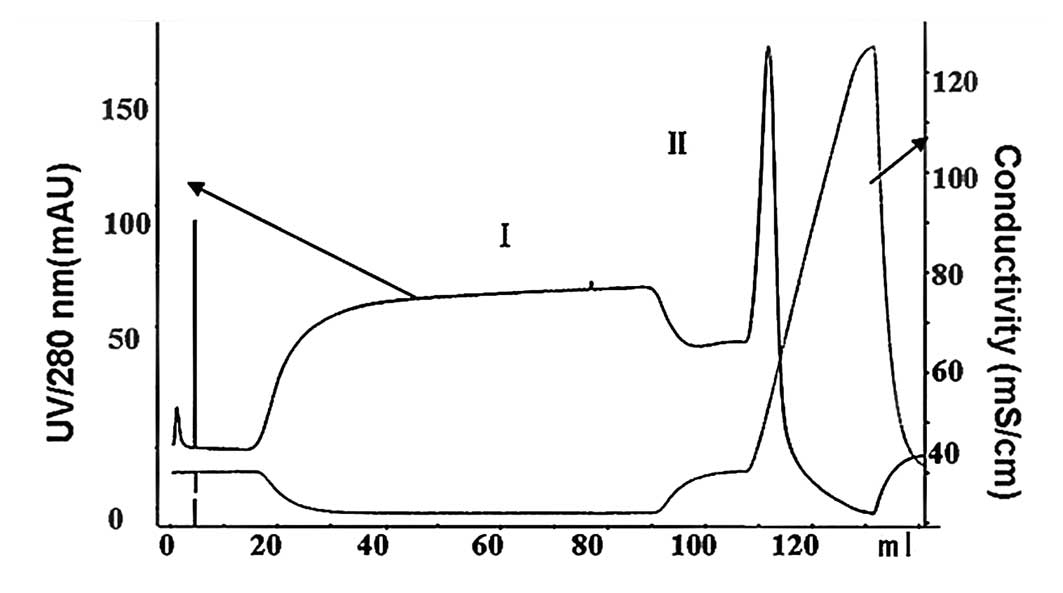
buffer (elution buffer B). During the elution process of 0–100% B,
peaks I and II were observed in the chromatogram of the refolding
solution. Tris-Tricine-SDS-PAGE analysis following strong cation
exchange showed that hSDF-1α existed in peak II with an impurity at
∼15% abundance (Figs. 4 and 5). Strong cation exchange was followed by
size-exclusion chromatography under refolding conditions. This step
removed higher and lower molecular mass impurities.
Tris-Tricine-SDS-PAGE analysis following size-exclusion
chromatography showed that there were few impurities, indicating
that the target protein could be obtained by purification using
strong cation exchange and size-exclusion chromatography (Fig. 6).
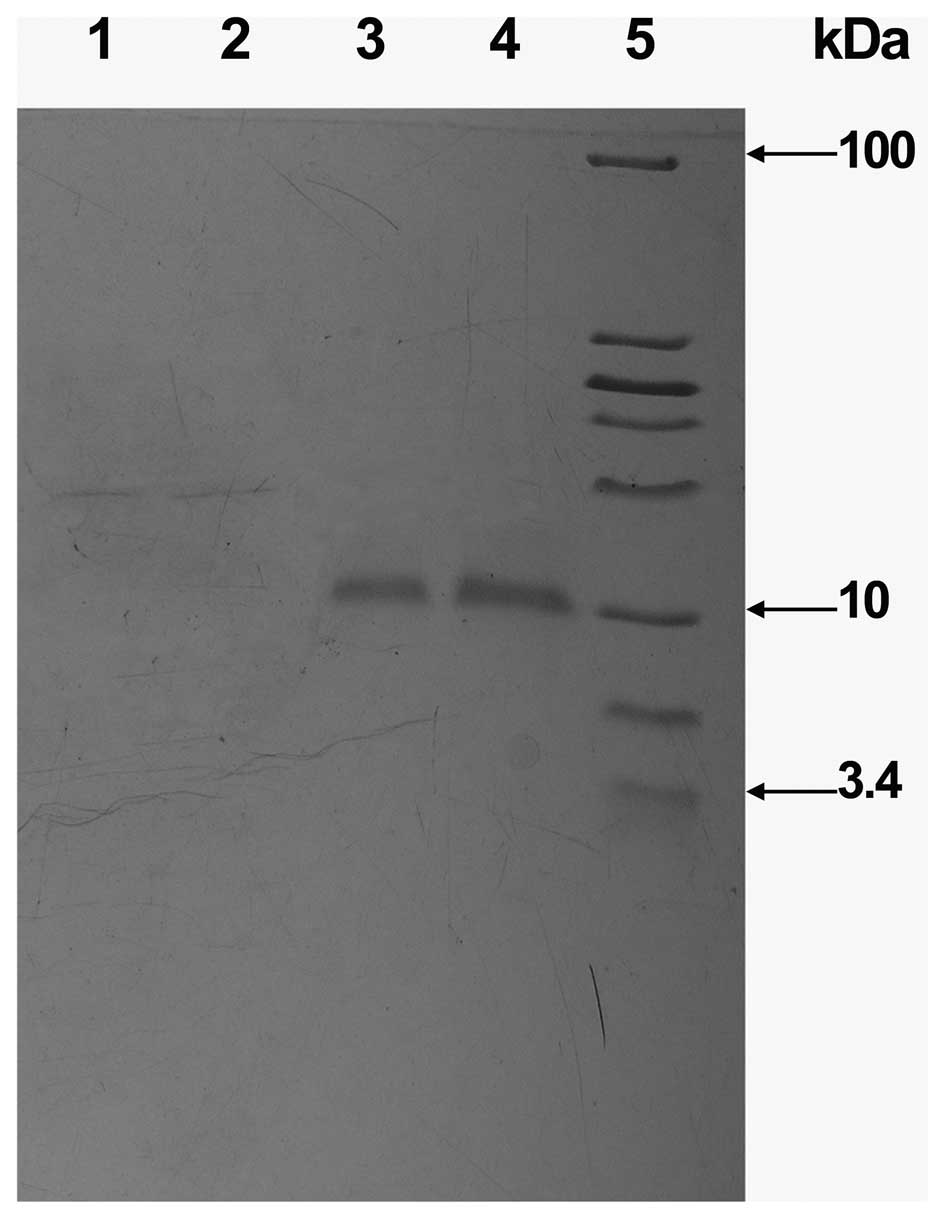 | Figure 5.SDS-PAGE analysis of the protein
fractions collected from the SP Sepharose FF column. Lanes 1 and 2,
proteins which did not bind to the column (Peak I in Fig. 4); Lanes 3 and 4, protein eluted by
elution liquid B (Peak II in Fig.
4); Lane 5, molecular weight marker (from top to bottom the
molecular weights are 100, 30, 25, 20, 15, 10, 5 and 3.4 kDa,
respectively). |
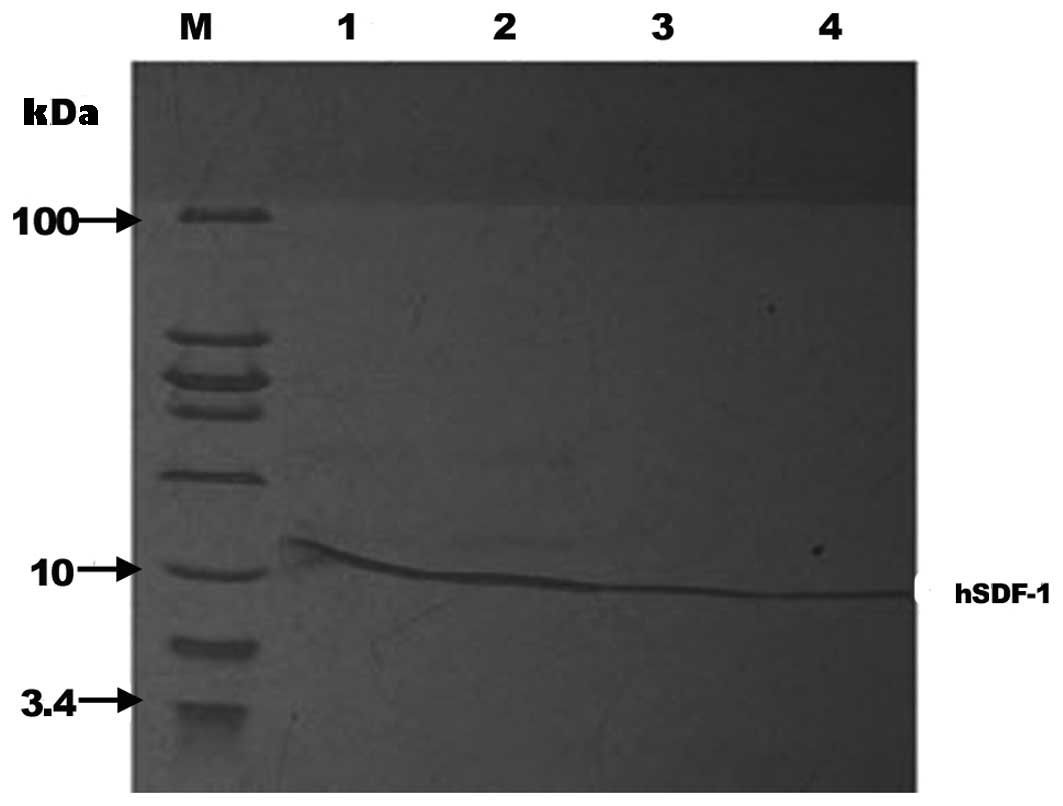 | Figure 6.SDS-PAGE analysis of the protein
fractions collected from strong cation exchange and size-exclusion
chromatography. Lanes 1 and 2, proteins collected from strong
cation exchange chromatography; Lanes 3 and 4, proteins collected
from size-exclusion chromatography; Lane M, molecular weight marker
(from top to bottom the molecular weights are 100, 30, 25, 20, 15,
10, 5 and 3.4 kDa, respectively). hSDF, human stromal cell-derived
factor. |
Determination of recombinant hSDF-1α
activity
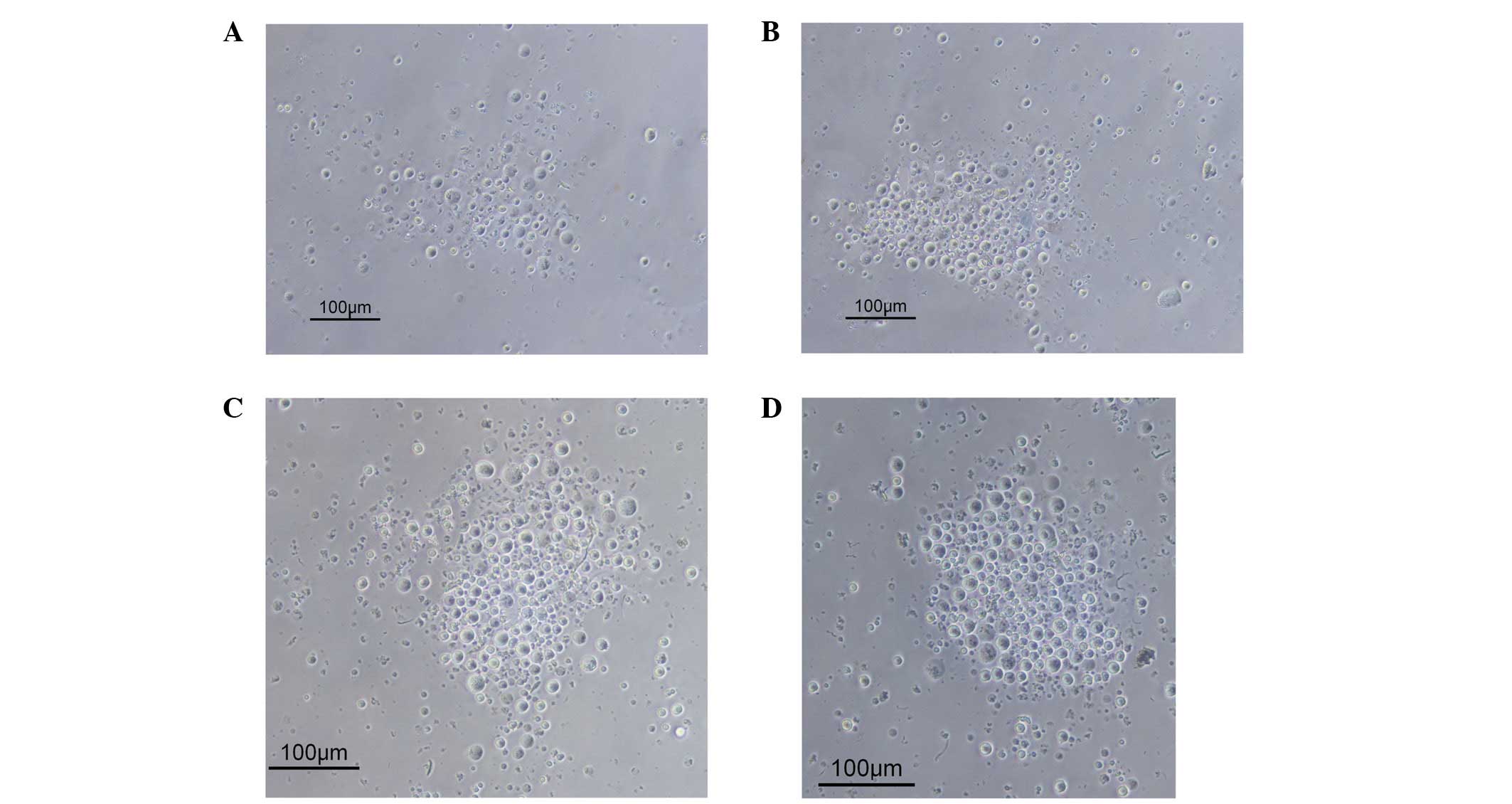
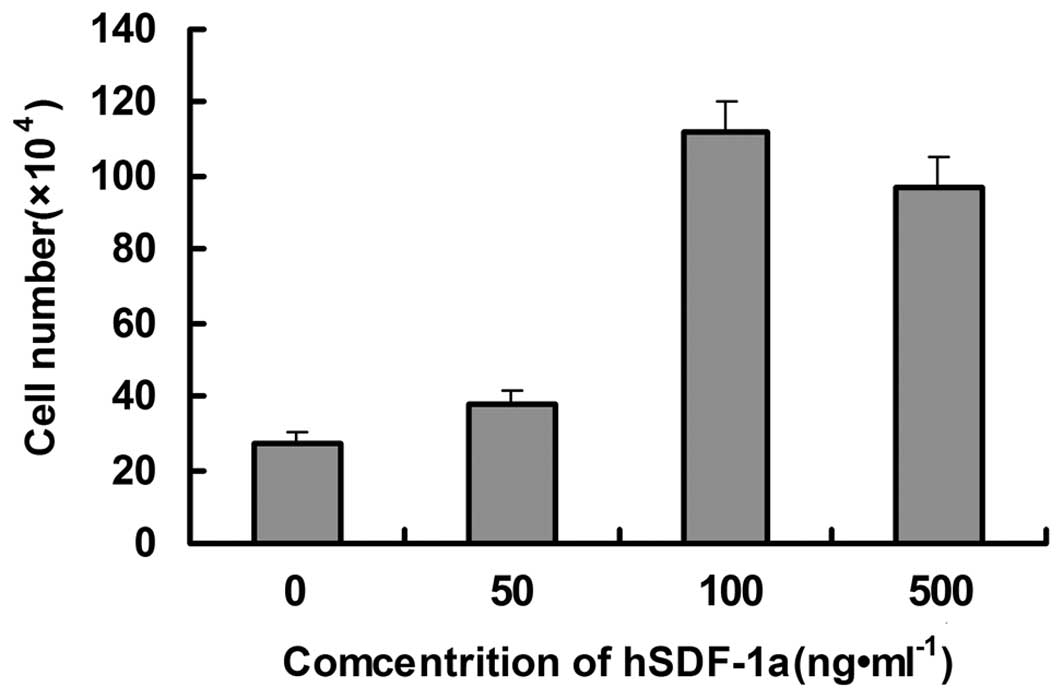
The effects of recombinant hSDF-1α on the
chemotactic activity of THP-1 cells were investigated by Transwell
cell migration assay, with RPMI-1640 containing 0.5% FBS as a
control. The number of cells that migrated to the lower chamber
with recombinant hSDF-1α protein concentrations of 10, 100 and 500
ng/ml was significantly higher than that of the control group,
indicating that a chemotactic effect of the purified recombinant
mutant hSDF-1α protein occurred in the THP-1 cells (Figs. 7 and 8), demonstrating that the cloned,
expressed, refolded and purified recombinant mutant hSDF-1α in the
present study had high biological activity.
Discussion
SDF-1α is a protein that plays an important role in
the migration, proliferation, differentiation and adhesion of
endothelial progenitor cells (EPCs) in vivo. CXCR4 is a
specific receptor of SDF-1, and is a G protein-coupled receptor
composed of 352 amino acids and containing a seven transmembrane
helical structure. CXCR4 is widely expressed in bone marrow
mononuclear cells and stem cells (16). The interaction between CXCR4 and
SDF-1 not only regulates the release of stem cells from bone marrow
into the peripheral circulation but also regulates the recruitment
and residence of stem cells in ischemic tissues (17,18),
which plays an important role in the application of stem cell
therapy for hypoxia, ischemic heart disease and brain injury
(19,20). Crystal structure and NMR studies have
demonstrated that hSDF-1α binds to CXCR4 in its monomeric form, and
the eight amino acids of the N-terminus form an important receptor
binding region, whereas the C-terminus of SDF-1 is not involved in
receptor binding (7). Yu et
al (8) studied NEK293 cells
transfected with SDF-1δ, which has an additional 50 amino acids on
the C-terminals, and therefore is >50% longer than hSDF-1α. The
study showed the presence of secreted protein at ∼14 kDa, the
correct molecular size to be the intact protein, suggesting that
the C-terminals of SDF-1δ were not cleaved. This demonstrated that
SDF-1δ was also able to stimulate CXCR4-mediated clathrin-mediated
endocytosis cell migration in a similar manner to hSDF-1α. This
research showed that the activity of hSDF-1α was associated with
its structure.
In the present study, hSDF-1α without a signal
peptide sequence was created and an efficient protocol for cloning,
expression and purification of the recombinant protein was
developed. The hSDF-1α cDNA sequence which did not contain
the N-terminal signal peptide sequence was successfully constructed
in a pET-15b vector, and hSDF-1α was efficiently expressed in E.
coli BL21(DE3) cells in the form of an inclusion body. Prior to
being separated and purified by strong cation exchange
chromatography, the recombinant hSDF-1α was refolded with oxidized
glutathione and reduced glutathione. This process not only diluted
the concentration of salt but also removed certain impurities and
improved the subsequent separation efficiency. The purity of the
recombinant hSDF-1α reached >85% following cation exchange
chromatography, which meets the requirements of general protein
experiments. Size-exclusion chromatography was then used to
separate out the hSDF-1α. Tris-Tricine-SDS-PAGE analysis showed
that the purity of the recombinant hSDF-1α reached >95%, which
meets the requirements of all protein experiments. To determine if
the recombinant hSDF-1α was functional, a chemotaxis assay
evaluating the ability of hSDF-1α to stimulate the migration of
cells expressing the CXCR4 receptor, was performed using THP-1
cells. The recombinant hSDF-1α stimulated THP-1 cell migration,
showing that the recombinant hSDF-1α had bioactivity, and
indicating that the N-terminal signal peptide of hSDF-1α had little
effect on the activity of hSDF-1α. Further studies are required to
determine if there are quantitative differences in chemotaxis
activities; however the present study laid a good foundation for
further study of the hSDF-1α gene, the function of the
recombinant hSDF-1α protein and the mechanism of interaction of
hSDF-1α with its specific receptor CXCR4.
Acknowledgements
The authors would like to thank the National Basic
Research Program of China (no. 21075097).
References
|
1
|
Voermans C, Anthony EC, Mul E, van der
Schoot E and Hordijk P: SDF-1-induced actin polymerization and
migration in human hematopoietic progenitor cells. Exp Hematol.
29:1456–1464. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shi HW, Zhang P, Wang CM and Han H:
Prokaryotic expression and purification of fusion protein
GST-SDF-1α. Ke Xue Ji Shu Yu Gong Cheng. 7:3365–3367. 2007.[(In
Chinese)].
|
|
3
|
Yao F, Zhou JM, Wang Z, Wei DH, Jiang ZS,
Liu LS and Tong ZY: Cloning and sequence analysis of SDF-1α gene in
rats. Zhongguo Shi Yan Dong Wu Xue Bao. 16:14–18. 2008.[(In
Chinese)].
|
|
4
|
Nagasawa T, Kikutani H and Kishimoto T:
Molecular cloning and structure of a pre-B-cell growth-stimulating
factor. Proc Natl Acad Sci USA. 91:2305–2309. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Psenák O: Stromal cell-derived factor-1
(SDF-1). Its structure and function. Cas Lek Cesk. 140:355–363.
2001.[(In Czech)]. PubMed/NCBI
|
|
6
|
Shirozu M, Nakano T, Inazawa J, Tashiro K,
et al: Structure and chromosomal localization of the human stromal
cell derived factor-1 (SDF-1) gene. Genomics. 28:495–500. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gleichmann M, Gillen C, Czardybon M, Bosse
F, et al: Cloning and characterization of SDF-1γ, a novel SDF-1
chemokine transcript with developmentally regulated expression in
the nervous system. Eur J Neurosci. 12:1857–1866. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu L, Cecil J, Peng SB, Schrementi J,
Kovacevic S, Paul D, Su EW and Wang J: Identification and
expression of novel isoforms of human stromal cell-derived factor
1. Gene. 374:174–179. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng Y: Cloning of SDF-1α gene and effect
on migration of THP-1 (PhD thesis)Nanhua University; 2006
|
|
10
|
Crump MP, Gong JH, Loetscher P,
Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, et al:
Solution structure and basis for functional activity of stromal
cell-derived factor-1; dissociation of CXCR4 activation from
binding and inhibition of HIV-1. EMBO J. 16:6996–7007. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu M, Liang Q, Lu HL, Mao WW, Wu MY, Wang
Q and Han W: Expression and purification of human SDF-1α in
prokarotic cells and its regulative role in murine bone marrow
hematopoiesis. Xian Dai Sheng Wu Yi Xue Jin Zhan. 7:836–839.
2007.[(In Chinese)].
|
|
12
|
Cao ZW: An effective method of
Tricine-SDS-PAGE for separating the 1kDa peptide. Zhong Guo Sheng
Wu Gong Cheng Za Zhi. 38:74–76. 2003.[(In Chinese)].
|
|
13
|
Miao L, Xu LH, Ji X and Bian LJ: Cloning,
expression, purification and its biological activity study of the
soluble tripolymer recombinant angiogenesis inhibitor kringle 5.
Sheng Wu Gong Cheng Jin Zhan. 31:18–22. 2011.[(In Chinese)].
|
|
14
|
Kodali RB, Kim WJ, Galaria II, Miller C,
Schecter AD, Lira SA and Taubman MB: CCL11 (Eotaxin) induces
CCR3-dependent smooth muscle cell migration. Arterioscler Thromb
Vasc Biol. 24:1211–1216. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv YC, Wang Z, Wei DH, et al: The
chemotactic activity of stromal cell derived factor 1α-CXCR4 to the
migration of THP-1 cell and enhanced effect of oxidized low density
lipoprotein. Zhongguo Dong Mai Ying Hua Za Zhi. 15:15–18. 2007.[(In
Chinese)].
|
|
16
|
Wu B, Chien EY, Mol CD, Fenalti G, Liu W,
Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al: Structures
of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide
antagonists. Science. 330:1066–1071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Penn MS: Importance of the SDF-1:CXCR4
axis in myocardial repair. Circ Res. 104:1133–1135. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jujo K, Hamada H, Iwakura A, Thorne T,
Sekiguchi H, Clarke T, Ito A, Misener S, Tanaka T, Klyachko E, et
al: CXCR4 blockade augments bone marrow progenitor cell recruitment
to the neovasculature and reduces mortality after myocardial
infarction. Proc Natl Acad Sci USA. 107:11008–11013. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li YJ, Cai XX, Ma GS, Chen ZH, Tang CC and
Feng Y: The short-term prognostic value of stromal cell-derived
factor-1α in patients with acute myocardial infarction. Nanjing
Gongye Daxue Xxuebao: Ziran Kexue Ban. 31:1781–1784. 2011.[(In
Chinese)].
|
|
20
|
Stumm RK, Rummel J, Junker V, Culmsee C,
Pfeiffer M, Krieglstein J, Höllt V and Schulz S: A dual role for
the SDF-1/CXCR4 chemokine receptor system in adult brain:
isoform-selective regulation of SDF-1 expression modulates
CXCR4-dependent neuronal plasticity and cerebral leukocyte
recruitment after focal ischemia. J Neurosci. 22:5865–5878.
2002.PubMed/NCBI
|