Introduction
Endometriosis manifests as ectopic endometrial cells
outside the uterus. It is an intractable disease that causes
infertility, dysmenorrhea and pelvic pain. Endometriosis occurs in
10% of women of childbearing age. Notably, the incidence of
endometriosis has been rising in recent years (1). The pathogenesis of endometriosis
remains to be elucidated.
Published reports indicate that endometriosis is a
polygenic/multifactorial disease caused by interactions between
multiple genes and the environment (2,3). In
particular, a correlation has been identified between endometriosis
and exposure to environmental toxins such as dioxin (4); dioxin and dioxin-like compounds have
been implicated in the development of endometriosis (5,6).
The phase II conjugation enzymes usually function to
inactivate environmental toxins. Among these, glutathione
S-transferase (GST) may be critical for the detoxification of
dioxins. Human GSTs are classified into two distinct categories:
Soluble or cytosolic and membrane-bound microsomal. The soluble or
cytosolic GSTs are subdivided into seven families named α, µ, ω, π,
σ, θ and ζ (7). Genes in several of
these families are polymorphic, including: GSTA2 in the α
family, GSTM1 and GSTM3 in the µ family, GSTP1
in the π family, GSTO, GSTT1, and GSTT2 in the
θ family, and GSTZ1 in the ζ family. Heritable allelic
differences in GSTM1, GSTM3, GSTT1 and
GSTP1 may have marked relevance for individual
susceptibility to disease.
GSTM1 and GSTT1 are two candidate
genes that may play an important role in the development of
endometriosis. GSTM1 and GSTT1 are located on
chromosomes 1p13.3 and 22q11.23, respectively. They are critical in
the detoxification of the products of oxidative stress produced
during the repair of the ovarian epithelium. GSTM1 and
GSTT1 null alleles have reduced enzyme activity, a state
that may contribute to inefficient detoxification of intermediates
produced during stress. This may increase damage to various host
genes and contribute to the pathogenesis of endometriosis (8,9).
A meta-analysis summarizing the literature up to the
year 2005 suggested that the GSTT1 null genotype, but not
the GSTM1 null genotype, was associated with an increased
risk for endometriosis (7). In the
years since 2005, additional reports investigating this topic have
been published. The objective of the present study was to update
the existing meta-analysis and reevaluate the possible associations
between GSTM1, GSTT1 and combined
GSTM1/GSTT1 (null genotype vs. wild-type) gene
polymorphisms and susceptibility to endometriosis.
Materials and methods
Searches
For this systematic review and meta-analysis, PubMed
(from January 1996 to January 2014), Embase (from January 1996 to
January 2014), Chinese BioMedical Literature database (from January
1996 to January 2014) and Google Scholar (from January 1996 to
January 2014) were searched. The following keywords were used:
‘endometriosis’, ‘polymorphisms’, ‘glutathione S-transferases’,
‘GSTM1’ and ‘GSTT1’ or their combinations.
Reference lists from articles identified by the
electronic search were searched by hand. This process was performed
iteratively until no additional articles could be identified.
Inclusion and exclusion criteria
Articles published in English or Chinese were
included if they reported quantitative outcomes from case-control
genetic association studies on GSTM1, GSTT1 or
combined GSTM1/GSTT1 (null genotype vs. wild-type)
gene polymorphisms and endometriosis versus non-endometriosis or
healthy controls.
Studies were excluded if they were case reports,
case-only studies, letters, reviews or meta-analyses; included
subjects who were related; included cases of adenomyosis, which has
unknown etiology (10); reported
insufficient data; or were duplicate studies.
Selection of studies
Two reviewers (XYX and ZSJ) independently examined
titles and abstracts to select eligible studies. Records were
removed that were ongoing or unpublished studies, or were published
as abstracts or conference proceedings. Where data sets were
overlapping or duplicated, only the most recent information was
included. The full text of potentially relevant studies was
retrieved. Two reviewers (XYX and HJG) independently examined the
full text records to determine which studies met the inclusion
criteria. Disagreement about the selection of studies was resolved
by discussion and consensus.
Data extraction and management
Two reviewers (XYX and ZSJ) independently extracted
data from eligible studies including the first author's last name,
publication year, study location, ethnicity, matching variability,
diagnostic criteria, stages of disease, source of controls, numbers
of cases and controls, and numbers and/or percentages of null
genotypes. Disagreement about data extraction was resolved by
discussion and consensus.
Assessment of quality of evidence in
included studies
Two reviewers (YYL and HJG) independently assessed
quality of evidence in the included studies using the 9-star
Newcastle-Ottawa Scale, which considers selection, comparability
and outcome evaluation criteria.
Assessment of heterogeneity
Heterogeneity was assessed using the χ2 test and I2
test. The I2 statistic was interpreted as follows: I2=0–40%,
heterogeneity may not be important; I2= 30–60%, heterogeneity may
be moderate; I2=50–90%, heterogeneity may be substantial; and
I2=75–100%, considerable heterogeneity (11). If heterogeneity was present,
meta-regression was used to find the source.
Assessment of reporting biases
A funnel plot of effect estimates against their
standard errors (SEs) was created to assess possible reporting bias
between studies. Funnel plot asymmetry was assessed using Egger's
linear regression test and Begg's rank correlation test; P<0.05
suggested publication bias.
GSTM1/GSTT1 and risk for
endometriosis
Two reviewers (XYX and HJG) independently combined
data from trials using a fixed-effect model (DerSimonian and Laird
method) when there was no significant heterogeneity in populations
(I2<50%) and a random-effect model (Mantel-Haenszel method) when
there was considerable heterogeneity. Variables were synthesized
using odds ratios (ORs). A P-value of 0.05 was used as the cut-off
value to determine statistical significance, and data are presented
as the estimated OR with 95% confidence intervals (CIs). All
statistical analyses were performed using STATA software, version
12.0 (StataCorp, College Station, TX, USA). Inconsistencies in data
analysis were resolved through consensus and discussion with a
third reviewer (ZSJ).
Sensitivity and subgroup analyses
Sensitivity analyses were performed to explore the
impact of excluding outlying results. Subgroup analyses were
performed by stratifying patients according to ethnicity
(Caucasian, Asian or mixed), characteristics of controls (hospital
patients or healthy individuals), and quality of evidence
(high-quality or low-quality).
Results
Screening and selection
The searches identified 120 articles. Titles and
abstracts were screened, and 36 studies were identified as
potentially eligible for inclusion. The full text articles for
these studies were retrieved. Following analysis of the full text
articles, four studies were excluded and 32 studies were found to
be eligible for inclusion according to the criteria used for
considering studies in this review (Fig.
1).
Included studies
The characteristics of the included studies are
shown in Table I. There were 32
case-control genetic association studies involving 3,990 cases of
endometriosis and 4,625 controls. One publication addressed two
groups of subjects with different ethnicities and was considered as
two case-control genetic association studies (12); thus, the total number of studies was
considered to be 33. Studies included data relevant to the
GSTM1 genotype, GSTT1 genotype or the combined
GSTM1/GSTT1 genotype. Of the 32 eligible studies, 20
were conducted in Asia (12–31), eight in Europe (32–39), two
in North America (40,41), and two in South America (42,43). The
evidence reported in 23 studies was identified as high-quality, and
that in 10 studies was identified as low-quality.
 | Table I.Characteristics of included studies
on the GSTM1, GSTT1 and combined GSTM1/GSTT1 gene
polymorphisms. |
Table I.
Characteristics of included studies
on the GSTM1, GSTT1 and combined GSTM1/GSTT1 gene
polymorphisms.
|
|
|
|
|
| GSTM1
(n) | GSTT1
(n) | GSTM1+GSTT1
(n) |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Ethnicity | Countries | Source of
controls | Quality | Cases/null | Controls/null | Cases/null | Controls/null | Cases/null | Controls/null | Refs. |
|---|
| Baranov, 1996 | Caucasian | Russia | Healthy
individuals | Low | 42/34 | 67/26 |
|
|
|
| (33) |
| Baranova, 1999 | Caucasian | Russia, France | Hospital
patients | High | 65/50 | 72/33 | 65/13 | 72/7 |
|
| (32) |
| Baranov, 1999 | Caucasian | Russia | Healthy
individuals | Low | 150/88 | 99/42 |
|
|
|
| (34) |
| Hadfield, 2001 | Caucasian | UK | Hospital
patients | High | 132/59 | 52/27 | 116/29 | 50/14 |
|
| (35) |
| Baxter, 2001 | Caucasian | England | Healthy
individuals | High | 84/40 | 219/107 |
|
|
|
| (36) |
| Bischoff, 2002 | Caucasian | USA | Hospital
patients | Low | 62/13 | 36/20 |
|
|
|
| (40) |
| Ivaschenko,
2003 | Caucasian | Russia | Hospital
patients | High | 74/42 | 40/17 | 74/27 | 40/6 | 74/16 | 40/2 | (37) |
| Arvanitis,
2003 | Caucasian | Greece | Healthy
individuals | High | 275/161 | 346/181 | 275/24 | 346/31 | 275/11 | 346/16 | (38) |
| Peng, 2003 | Asian | China | Hospital
patients | High | 76/50 | 80/37 |
|
|
|
| (15) |
| Lin, 2003 | Asian | China | Hospital
patients | High | 68/49 | 28/12 | 68/53 | 28/9 |
|
| (16) |
| Morizane, 2004 | Asian | Japan | Healthy
individuals | Low | 108/57 | 173/89 | 108/52 | 173/71 | 108/30 | 173/43 | (17) |
| Hsieh, 2004 | Asian | China | Hospital
patients | High | 150/95 | 159/8 |
|
|
|
| (18) |
| De Carvalho,
2004 | Mixed | Brazil | Hospital
patients | Low | 61/21 | 32/17 |
|
|
|
| (42) |
| Ding, 2004 | Asian | China | Healthy
individuals | High | 80/46 | 105/55 | 80/59 | 105/47 | 80/34 | 105/24 | (12) |
| Ding, 2004 | Asian | China | Healthy
individuals | High | 41/21 | 107/57 | 41/15 | 107/32 | 41/10 | 107/14 | (12) |
| Babu, 2005 | Caucasian | India | Hospital
patients | High | 310/121 | 215/64 | 310/42 | 215/34 | 310/14 | 215/11 | (19) |
| Hur, 2005 | Asian | Korea | Hospital
patients | Low | 194/112 | 259/145 | 194/104 | 259/125 |
|
| (20) |
| Aban, 2007 | Caucasian | Turkey | Hospital
patients | High | 150/88 | 150/65 | 150/59 | 150/44 |
|
| (21) |
| Chang, 2007 | Asian | China | Hospital
patients | High | 74/48 | 65/30 | 74/46 | 65/32 | 74/27 | 65/13 | (30) |
| Kim, 2007 | Asian | Korea | Hospital
patients | High | 316/183 | 256/146 | 316/178 | 256/124 |
|
| (22) |
| Rozati, 2009 | Caucasian | India | Hospital
patients | High | 97/26 | 102/15 |
|
|
|
| (13) |
| Yang, 2009 | Asian | China | Hospital
patients | Low | 216/134 | 216/100 |
|
|
|
| (14) |
| Cao, 2009 | Asian | China | Hospital
patients | High | 51/33 | 102/61 | 51/22 | 102/39 |
|
| (23) |
| Wu, 2009 | Asian | China | Hospital
patients | High | 96/63 | 85/40 |
|
|
|
| (24) |
| Huang, 2010 | Asian | China | Hospital
patients | High | 28/12 | 29/10 |
|
|
|
| (25) |
| Trabert, 2011 | Caucasian | USA | Healthy
individuals | High | 254/137 | 567/268 |
|
|
|
| (41) |
| Hosseinzadeh,
2011 | Caucasian | Iran | Healthy
individuals | High | 120/87 | 200/80 |
|
|
|
| (26) |
| Wu, 2012 | Asian | China | Healthy
individuals | Low | 121/57 | 171/52 | 121/40 | 171/33 | 121/23 | 171/15 | (27) |
| Seifati, 2012 | Caucasian | Iran | Hospital
patients | High | 101/51 | 142/74 |
|
|
|
| (28) |
| Vichi, 2012 | Caucasian | Italy | Hospital
patients | High | 181/104 | 162/85 | 181/20 | 162/32 |
|
| (39) |
| Matsuzaka,
2012 | Asian | Japan | Hospital
patients | High | 97/43 | 143/67 | 97/38 | 143/56 |
|
| (29) |
| Frare, 2013 | Mixed | Brazil | Healthy
individuals | Low | 50/25 | 46/34 | 50/16 | 46/27 |
|
| (43) |
| Sachan, 2013 | Caucasian | Iran | Healthy people | Low | 66/27 | 100/16 |
|
|
|
| (31) |
Excluded studies
Of the 36 studies that were relevant to the
GSTM1/GSTT1 genotype and endometriosis, four were
excluded. Of these, three were duplicates (13,14,32), and
one included subjects who were related (44).
GSTM1/GSTT1 and risk for
endometriosis
GSTM1genotype
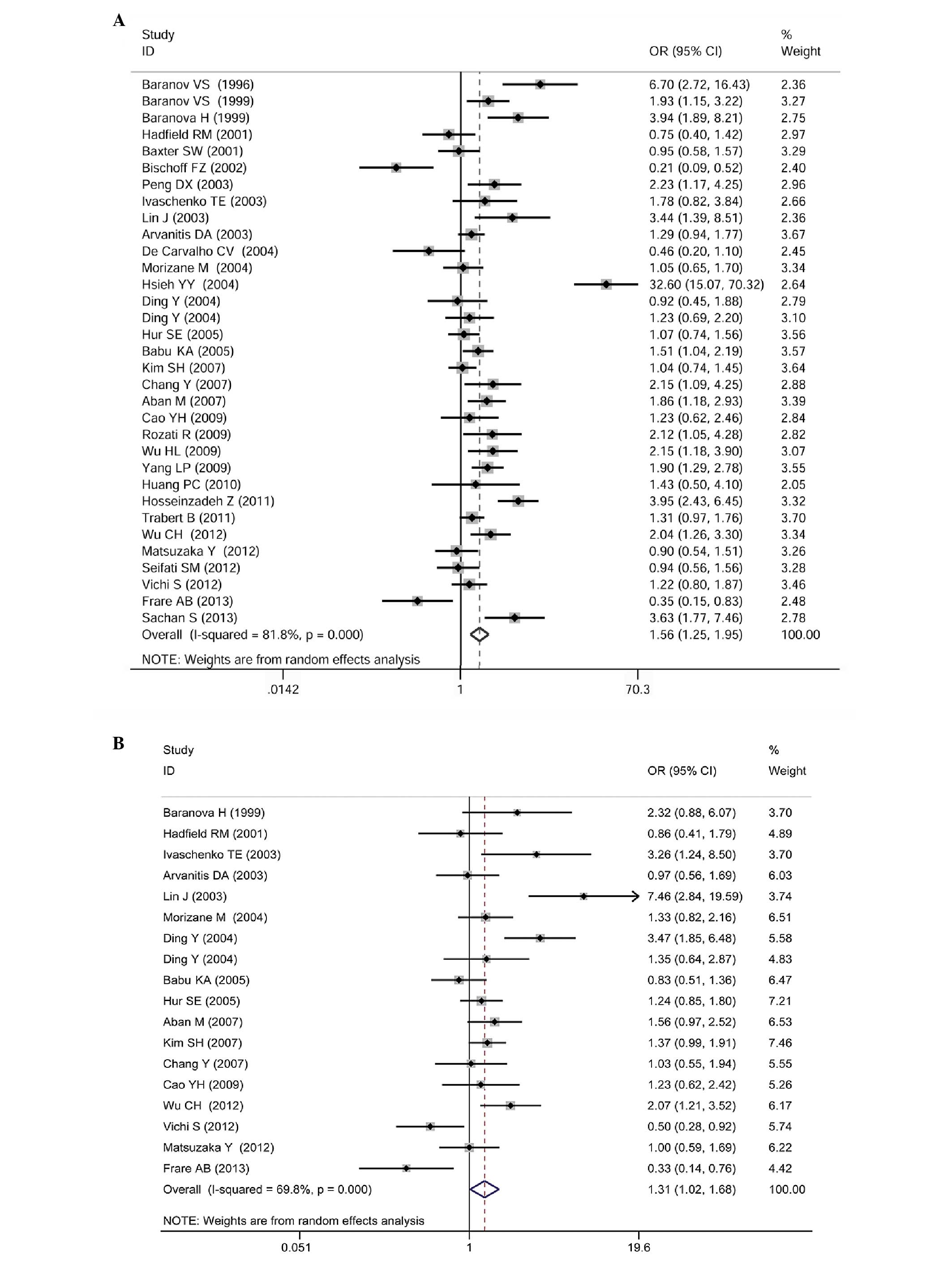
Data reporting on the GSTM1 gene polymorphism
are described in 33 case-control studies (3,990 cases of
endometriosis and 4,625 controls). The meta-analysis demonstrated
that there was a significant association between the GSTM1
null genotype and an increased risk for endometriosis (OR=1.56; 95%
CI: 1.25–1.95; P<0.0001; Fig.
2A).
Subgroup analyses stratified by ethnicity
(Caucasian: OR=1.599; 95% CI: 1.205–2.122; P=0.001; Asian:
OR=1.772; 95% CI: 1.242–2.528, P=0.002), source of controls
(hospital patients: OR=1.561; 95% CI: 1.151–2.117; P=0.004; healthy
individuals: OR=1.569; 95% CI: 1.131–2.176; P=0.007), and quality
of evidence (high-quality: OR=1.563; 95% CI: 1.253–1.949;
P<0.0001) confirmed this finding.
Subgroup analysis stratified for mixed ethnicity
(two case control studies involving 111 cases of endometriosis and
78 controls) demonstrated a significant association between the
GSTM1 null genotype and a decreased risk for endometriosis
(OR=0.404; 95% CI: 0.219–0.745; P=0.004; Table II). Compared with individual
Caucasian and Asian populations, the difference was statistically
significant (P<0.001; data shown in Table III).
 | Table II.Meta-analysis of the association
between GSTM1, GSTT1 and combined
GSTM1/GSTT1 (null genotype vs. wild-type) gene
polymorphisms and susceptibility to endometriosis. |
Table II.
Meta-analysis of the association
between GSTM1, GSTT1 and combined
GSTM1/GSTT1 (null genotype vs. wild-type) gene
polymorphisms and susceptibility to endometriosis.
| Group | No. of studies | No. of subjects
(cases/controls) | OR [95%CI] | P-value |
|---|
| Total studies |
|
|
|
|
|
GSTM1 genotype | 33 | 3,990/4,625 | 1.563
[1.253–1.949] | <0.001 |
|
GSTT1 genotype | 18 | 2,371/2,490 | 1.345
[1.044–1.733] | 0.022 |
|
GSTM1+GSTT1
genotype | 8 | 1,083/1,222 | 1.672
[1.291–2.166] | 0.005 |
| Caucasian |
|
|
|
|
|
GSTM1 genotype | 16 | 2,163/2,569 | 1.599
[1.205–2.122] | 0.001 |
|
GSTT1 genotype | 7 | 1,171/1,035 | 1.124
[0.745–1.697] | 0.577 |
|
GSTM1+GSTT1
genotype | 3 | 659/601 | 1.185
[0.717–1.961] | 0.508 |
| Asian |
|
|
|
|
|
GSTM1 genotype | 15 | 1,716/1,978 | 1.772
[1.242–2.528] | 0.002 |
|
GSTT1 genotype | 10 | 1,150/1,409 | 1.573
[1.186–2.085] | 0.002 |
|
GSTM1+GSTT1
genotype | 5 | 424/621 | 1.898
[1.404–2.565] | <0.001 |
| Mixed |
|
|
|
|
|
GSTM1 genotype | 2 | 111/78 | 0.404
[0.219–0.745] | 0.004 |
|
GSTT1 genotype | 1 | 50/46 |
|
GSTM1+GSTT1
genotype | 0 |
|
|
|
| Controls from
hospital patients |
|
|
|
|
|
GSTM1 genotype | 21 | 2,599/2,425 | 1.561
[1.151–2.117] | 0.004 |
|
GSTT1 genotype | 12 | 1,696/1,542 | 1.284
[0.963–1.712] | 0.089 |
|
GSTM1+GSTT1
genotype | 3 | 458/320 | 1.797
[1.081–2.989] | 0.024 |
| Controls from
healthy individuals |
|
|
|
|
|
GSTM1 genotype | 12 | 1,391/2,200 | 1.569
[1.131–2.176] | 0.007 |
|
GSTT1 genotype | 6 | 675/948 | 1.315
[0.767–2.254] | 0.320 |
|
GSTM1+GSTT1
genotype | 5 | 625/902 | 1.657
[1.085–2.532] | 0.001 |
| High quality |
|
|
|
|
|
GSTM1 genotype | 23 | 2,920/3,426 | 1.563
[1.253–1.949] | <0.001 |
|
GSTT1 genotype | 14 | 1,898/1,841 | 1.376
[1.020–1.858] | 0.037 |
|
GSTM1+GSTT1
genotype | 6 | 854/878 | 1.753
[1.265–2.430] | 0.001 |
| Low quality |
|
|
|
|
|
GSTM1 genotype | 10 | 1,070/1,199 | 1.259
[0.785–2.020] | 0.340 |
|
GSTT1 genotype | 4 | 473/649 | 1.121
[0.646–1.944] | 0.684 |
|
GSTM1+GSTT1
genotype | 2 | 229/344 | 1.542
[1.009–2.356] | 0.045 |
 | Table III.Comparisons of subgroup analyses for
GSTM1, GSTT1 and combined GSTM1/GSTT1
studies. |
Table III.
Comparisons of subgroup analyses for
GSTM1, GSTT1 and combined GSTM1/GSTT1
studies.
| A, Analysis of the
GSTM1 gene |
|---|
|
|---|
|
|
| GSTM1
(n) |
|
|
|---|
|
|
|
|
|
|
|---|
| Subgroup | Subjects | Null | Normal | χ2 | P-value |
|---|
| Ethnicity |
|
|
| 3.245 | 0.72a |
|
Caucasian | Cases | 1,128 | 1,035 |
|
|
|
|
Controls | 1,120 | 1,449 |
|
|
|
Asian | Cases | 1,003 | 713 |
|
|
|
|
Controls | 909 | 1,069 |
|
|
|
Mixed | Cases | 46 | 65 | 18.737 |
<0.001b |
|
| Controls | 51 | 27 | 23.467 |
<0.001c |
| Source of
controls |
|
|
| 0.130 | 0.718 |
|
Hospital patients | Cases | 1,397 | 1,202 |
|
|
|
| Controls | 1,073 | 1,352 |
|
|
| Healthy
individuals | Cases | 780 | 611 |
|
|
|
| Controls | 1,007 | 1,193 |
|
|
| Quality |
|
|
| 0.825 | 0.364 |
| High
quality | Cases | 1,609 | 1,311 |
|
|
|
| Controls | 1,539 | 1,887 |
|
|
| Low
quality | Cases | 568 | 502 |
|
|
|
| Controls | 541 | 658 |
|
|
|
| B, Analysis of the
GSTT1 gene |
|
|
|
| GSTT1
(n) |
|
|
|
|
|
|
|
|
| Subgroup | Subjects | Null | Normal | χ2 | P-value |
|
| Ethnicity |
|
|
| 6.766 | 0.009 |
|
Caucasian | Cases | 214 | 957 |
|
|
|
| Controls | 168 | 867 |
|
|
|
Asian | Cases | 607 | 543 |
|
|
|
| Controls | 568 | 841 |
|
|
| Source of
controls |
|
|
| 0.638 | 0.425 |
|
Hospital patients | Cases | 631 | 1,065 |
|
|
|
| Controls | 522 | 1,020 |
|
|
| Healthy
individuals | Cases | 206 | 469 |
|
|
|
| Controls | 241 | 707 |
|
|
| Quality |
|
|
| 0.062 | 0.803 |
| High
quality | Cases | 625 | 1,273 |
|
|
|
|
Controls | 507 | 1,334 |
|
|
| Low
quality | Cases | 212 | 261 |
|
|
|
| Controls | 256 | 393 |
|
|
|
| C, Analysis of
GSTM1+GSTT1 genes |
|
|
|
| GSTM1+GSTT1
(n) |
|
|
|
|
|
|
|
|
| Subgroup | Subjects | Null | Normal | χ2 | P-value |
|
| Ethnicity |
|
|
| 7.642 | 0.006 |
|
Caucasian | Cases | 41 | 618 |
|
|
|
| Controls | 29 | 572 |
|
|
|
Asian | Cases | 124 | 300 |
|
|
|
| Controls | 109 | 152 |
|
|
| Source of
controls |
|
|
| 0.091 | 0.763 |
|
Hospital patients | Cases | 57 | 401 |
|
|
|
| Controls | 26 | 294 |
|
|
| Healthy
individuals | Cases | 108 | 517 |
|
|
|
| Controls | 112 | 790 |
|
|
| Quality |
|
|
| 0.022 | 0.882 |
| High
quality | Cases | 112 | 542 |
|
|
|
| Controls | 80 | 598 |
|
|
| Low
quality | Cases | 53 | 176 |
|
|
|
| Controls | 58 | 286 |
|
|
GSTT1 genotype
Data reporting on the GSTT1 gene polymorphism
are described in 18 case-control studies (2,371 cases of
endometriosis and 2,490 controls). The meta-analysis demonstrated a
significant association between the GSTT1 null genotype and
an increased risk for endometriosis (OR=1.31; 95% CI: 1.02–1.68;
P=0.037; Fig. 2B).
Subgroup analysis stratified by ethnicity
demonstrated a significant association between the GSTT1
null genotype and an increased risk for endometriosis among Asians
(OR=1.573; 95% CI: 1.186–2.085; P=0.002), but not among Caucasians
(OR=1.124; 95% CI: 0.745–1.697; P=0.577).
Subgroup analyses stratified by the source of
controls found no significant association between the GSTT1
null genotype and an increased risk for endometriosis among
hospital-based studies (OR=1.284; 95% CI: 0.963–1.712; P=0.089) or
among healthy individuals (OR=1.315; 95% CI: 0.767–2.254;
P=0.320).
Subgroup analyses stratified by quality of evidence
demonstrated a significant association between the GSTT1
null genotype and an increased risk for endometriosis among studies
considered high-quality evidence (OR=1.376; 95% CI: 1.020–1.858;
P=0.037), but not among studies considered low-quality evidence
(OR=1.121, 95% CI: 0.646–1.944; P=0.684; Table II).
Combined GSTM1/GSTT1 genotype
Data reporting on the combined
GSTM1/GSTT1 gene polymorphism are described in eight
case-control studies (1,083 cases of endometriosis and 1,222
controls). The meta-analysis demonstrated a significant association
between the combined GSTM1/GSTT1 null genotype and an
increased risk for endometriosis (OR=1.68, 95% CI: 1.29–2.17;
P<0.0001; Fig. 2C).
This association was unchanged by subgroup analyses
stratified by source of controls (hospital-based studies: OR=1.797;
95% CI: 1.081–2.989; P=0.024; healthy individuals: OR=1.657; 95%
CI: 1.085–2.532; P=0.001) or quality of evidence (high-quality
evidence: OR=1.753; 95% CI: 1.265–2.430; P=0.001; low-quality
evidence: OR=1.542; 95% CI: 1.009–2.356, P=0.045; Table II).
Subgroup analysis stratified by ethnicity
demonstrated a significant association between the combined
GSTM1/GSTT1 null genotype and an increased risk for
endometriosis among Asian populations (OR=1.898; 95% CI:
1.404–2.565; P<0.001), but not among Caucasian populations
(OR=1.185; 95% CI: 0.717–1.961; P=0.508).
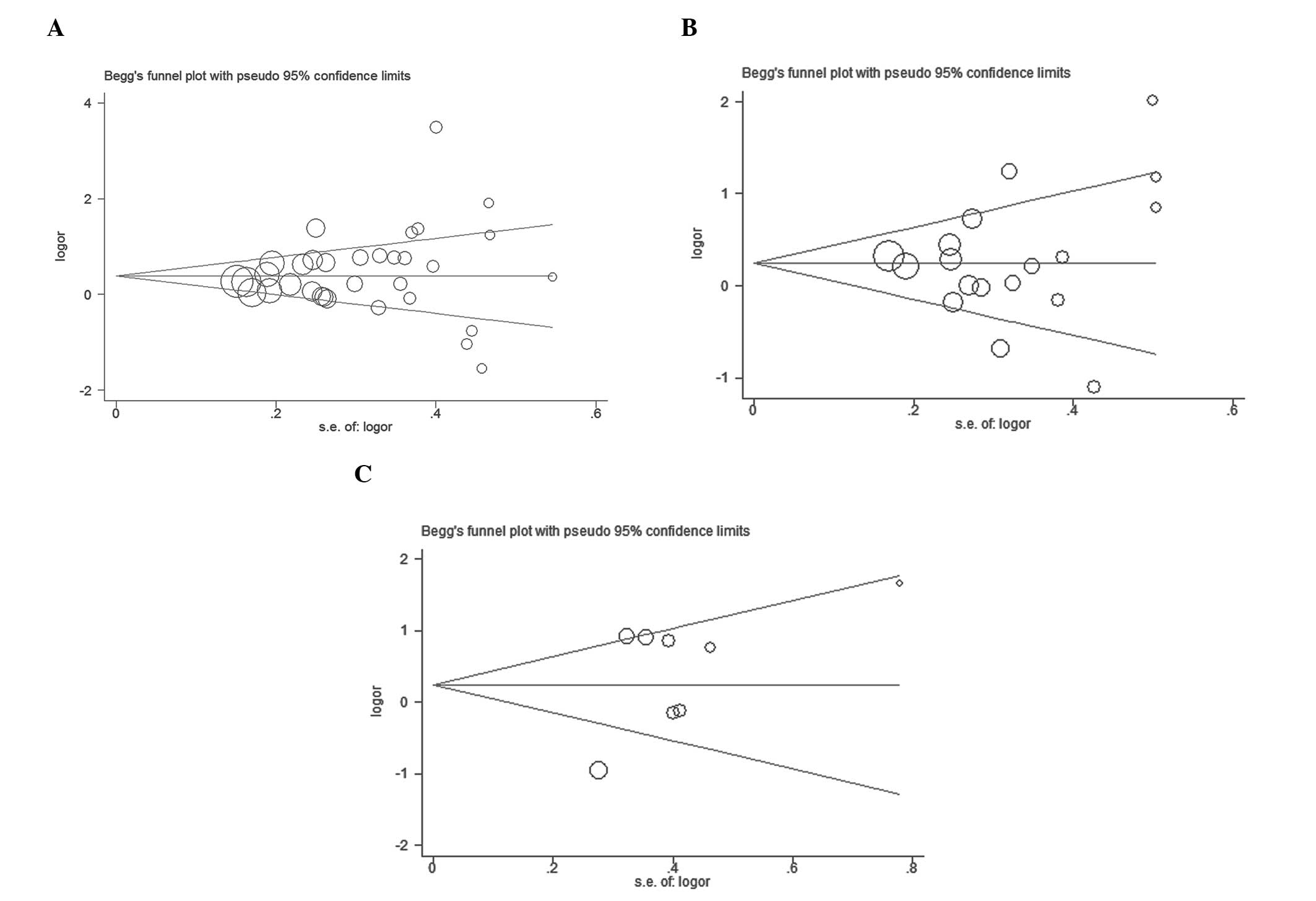
Publication bias
Visual inspection of a Funnel plot, Egger's test and
Begg's rank correlation test revealed no significant publication
bias for the GSTM1, GSTT1 and combined
GSTM1/GSTT1 studies (Fig.
3; Table IV).
 | Table IV.Heterogeneity and publication bias of
GSTM1, GSTT1 and combined GSTM1/GSTT1
studies. |
Table IV.
Heterogeneity and publication bias of
GSTM1, GSTT1 and combined GSTM1/GSTT1
studies.
|
| Heterogeneity | Publication bias
(P-value) |
|---|
|
|
|
|
|---|
| Group | I2 value
(%) | P-value | Egger's test | Begg's funnel
plot |
|---|
| Total studies |
|
|
|
|
|
GSTM1 genotype | 81.8 | <0.001 | 0.313 | 0.412 |
|
GSTT1 genotype | 69.9 | <0.001 | 0.557 | 0.705 |
|
GSTM1+GSTT1
genotype | 44.7 | 0.081 | 0.170 | 1.000 |
| Caucasian |
|
|
|
|
|
GSTM1 genotype | 79.2 | <0.001 | 0.454 | 0.322 |
|
GSTT1 genotype | 64.7 | 0.009 | 0.339 | 0.764 |
|
GSTM1+GSTT1
genotype | 58.5 | 0.090 | 0.021 | 0.296 |
| Asian |
|
|
|
|
|
GSTM1 genotype | 83.8 | <0.001 | 0.098 | 0.083 |
|
GSTT1 genotype | 62.4 | 0.004 | 0.160 | 0.210 |
|
GSTM1+GSTT1
genotype | 13.6 | 0.081 | 0.340 | 0.806 |
| Mixed |
|
|
|
|
|
GSTM1 genotype | 0.0 | 0.664 | <0.001 | 0.317 |
| Controls from
hospital patients |
|
|
|
|
|
GSTM1 genotype | 83.6 | <0.001 | 0.390 | 0.506 |
|
GSTT1 genotype | 65.9 | 0.001 | 0.335 | 0.451 |
|
GSTM1+GSTT1
genotype | 62.2 | 0.071 | 0.585 | 1.000 |
| Controls from
healthy individuals |
|
|
|
|
|
GSTM1 genotype | 79.4 | <0.001 | 0.598 | 0.784 |
|
GSTT1 genotype | 78.4 | <0.001 | 0.431 | 0.707 |
|
GSTM1+GSTT1
genotype | 45.9 | 0.116 | 0.531 | 1.000 |
| High quality |
|
|
|
|
|
GSTM1 genotype | 80.9 | <0.001 | 0.042 | 0.068 |
|
GSTT1 genotype | 69.9 | <0.001 | 0.530 | 0.189 |
|
GSTM1+GSTT1
genotype | 49.0 | 0.081 | 0.641 | 1.000 |
| Low quality |
|
|
|
|
|
GSTM1 genotype | 85.1 | <0.001 | 0.788 | 0.516 |
|
GSTT1 genotype | 77.2 | 0.004 | 0.347 | 1.000 |
|
GSTM1+GSTT1
genotype | 62.9 | 0.101 |
|
|
Heterogeneity analysis
There was evidence of significant heterogeneity
(I2>50%) between studies of GSTM1 and GSTT1, and
those used in subgroup analyses, although not among studies of
GSTM1/GSTT1 combined (Table IV). Therefore, the random-effect
model was used in all analyses with the exception of the analysis
of combined GSTM1/GSTT1 gene polymorphisms. For the
GSTM1 and GSTT1 gene polymorphisms, a meta-regression
was conducted in which publication year, ethnicity, source of
controls, sample size, and quality of evidence were covariates. All
the covariates were entered into the meta-regression model
simultaneously, and the covariates that had the highest P-values
were omitted one at a time in order to identify any sources of
heterogeneity among them. However, the meta-regression analysis did
not identify any of these covariates as a significant source of
heterogeneity (Figs. 4 and 5).
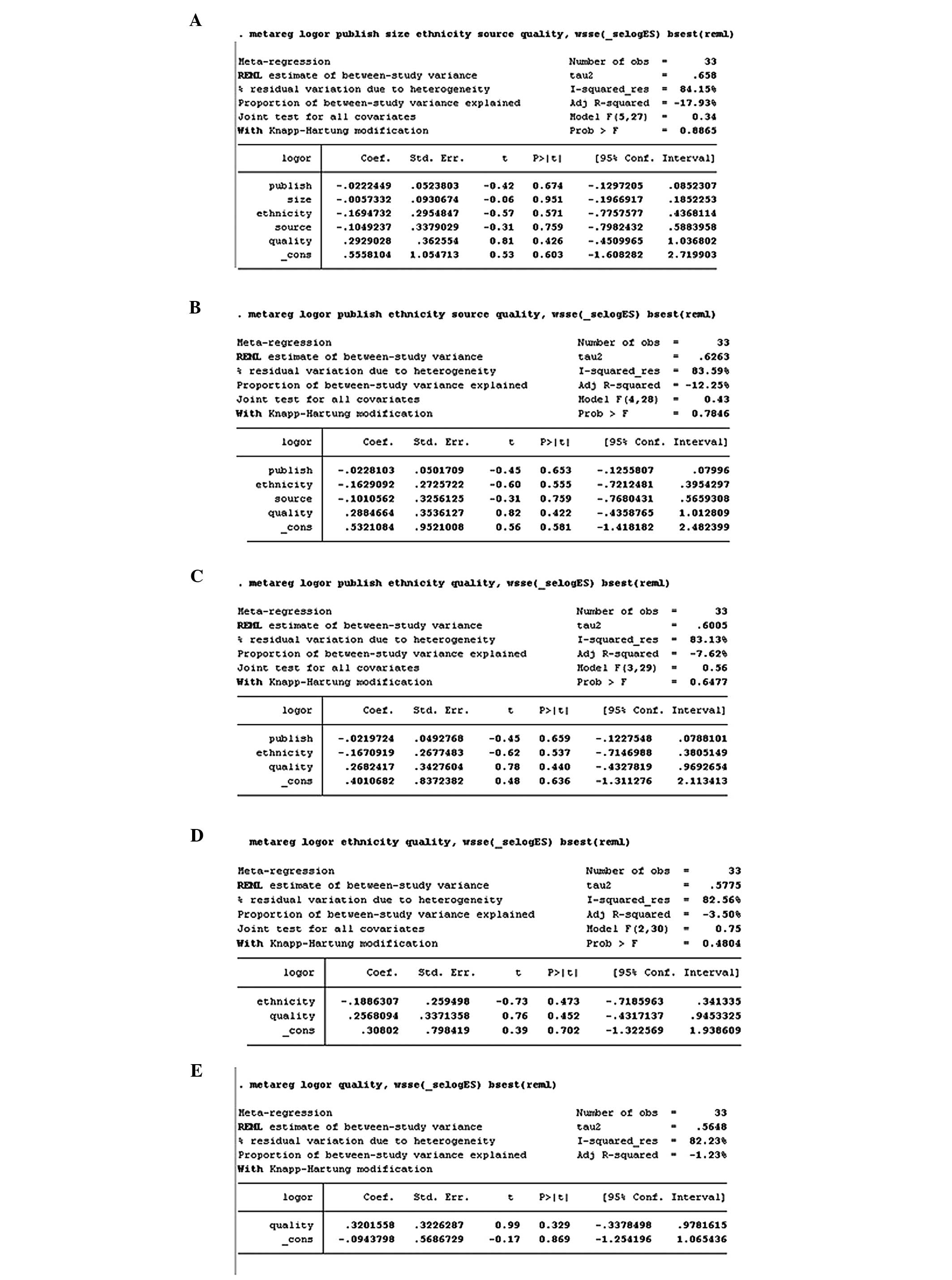 | Figure 4.Meta-regression for GSTM1
studies, with publication year, ethnicity, source of controls,
sample size, and quality of evidence as covariates. All covariates
were entered into the meta-regression model simultaneously, and the
covariates with the highest P-values were omitted one at a time to
identify sources of heterogeneity. The meta-regression did not
identify any of these covariates as a significant source of
heterogeneity. Variables were omitted in the following order: Size
(A→B), source (B→C), publication year (C→D), ethnicity (D→E).
GSTM1, glutathione S-transferase µ1. |
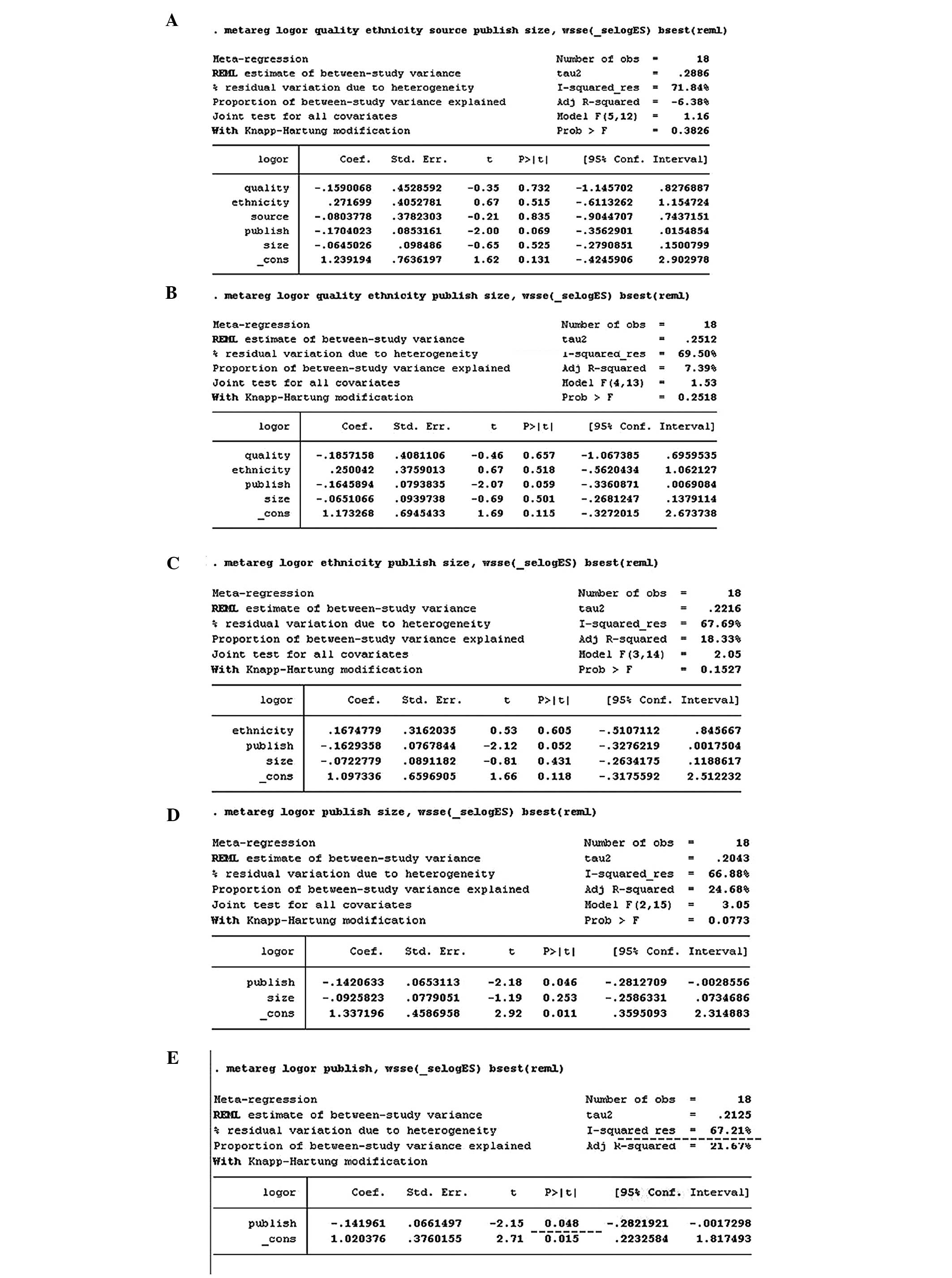 | Figure 5.Meta-regression for GSTT1
studies, with publication year, ethnicity, source of controls,
sample size, and quality of evidence as covariates. All covariates
were entered into the meta-regression model simultaneously, and
covariates with the highest P-values were omitted one at a time to
identify sources of heterogeneity. Meta-regression identified
publication year as a significant source of heterogeneity
(P=0.048), but after omitting this covariate heterogeneity remained
substantial (I2=67.21%) Variables were omitted in the
order: Source (A→B), quality (B→C), ethnicity (C→D), size (D→E).
GSTT1, glutathione S-transferase θ1 |
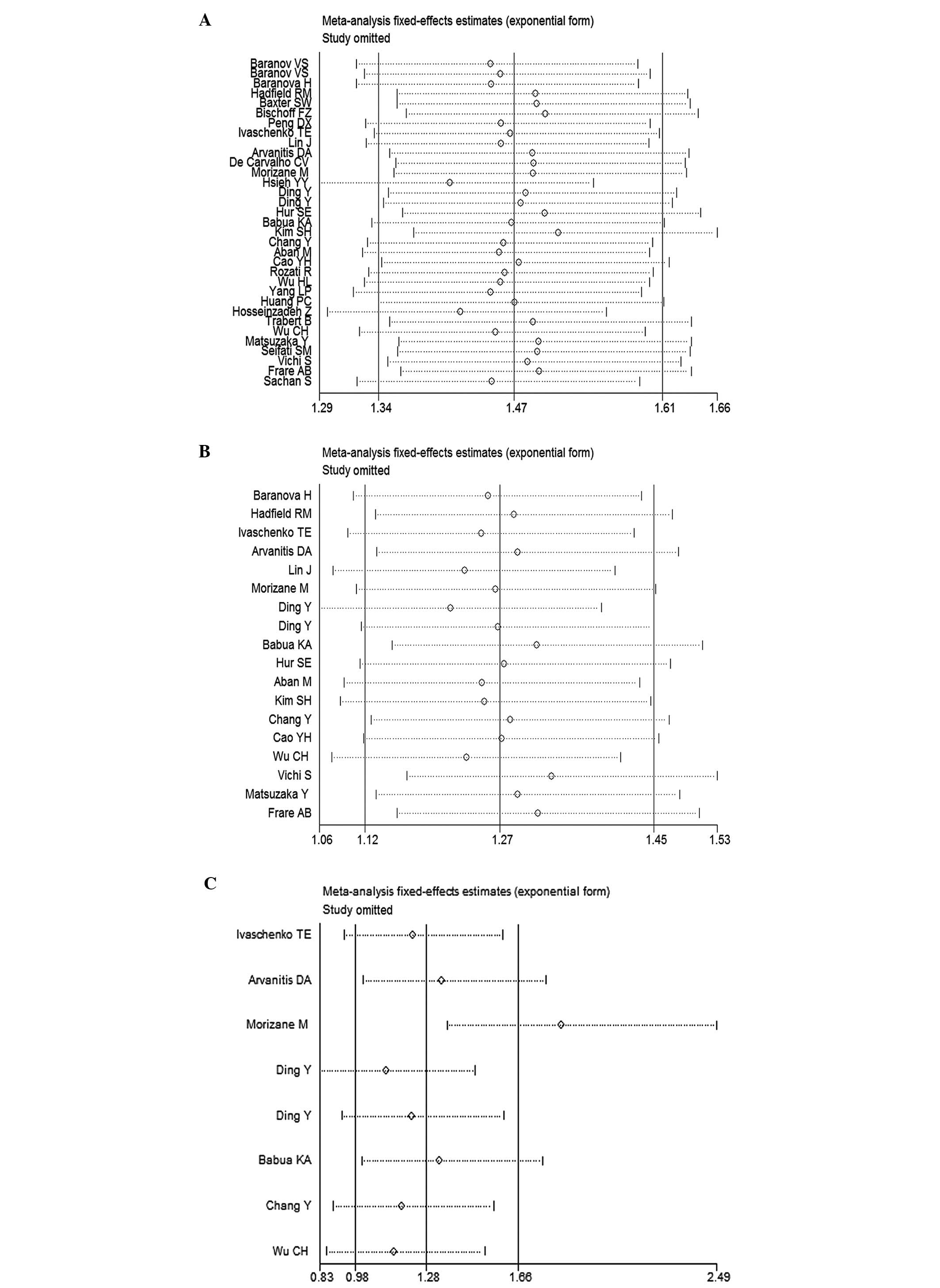
Sensitivity analysis
To explore the effects of individual studies on the
pooled OR estimates, a sensitivity analysis was performed, with the
omission of one study at a time. The OR estimates for the
GSTM1 polymorphism were not notably altered (Fig. 6A). The OR estimates for the
GSTT1 and combined GSTM1/GSTT1 polymorphisms
were altered when studies were excluded (Fig. 6B and C).
Discussion
In the present study, a meta-analysis of data from
33 studies was conducted to examine the associations between the
GSTM1, GSTT1 and combined GSTM1/GSTT1
null genotypes and susceptibility to endometriosis. The risk for
endometriosis was significantly increased in the presence of the
GSTM1, GSTT1 and combined GSTM1/GSTT1
null genotypes compared with the wild-type. Subgroup analyses
stratified by ethnicity, source of controls and quality of evidence
confirmed this finding among several subgroups, but particularly
among studies considered high-quality evidence. Notably, among
patients of mixed ethnicity, the GSTM1 null genotype was
significantly associated with a decreased risk for endometriosis
compared with the wild-type.
A similar meta-analysis of 23 studies performed in
2005 demonstrated an increased risk for endometriosis in women with
the GSTT1 null genotype (8).
However, the authors requested that their findings be interpreted
with caution as asymmetry in the funnel plot was evident, which was
likely due to publication bias (8).
This previous study did not include subgroup analyses or an
evaluation of the combined GSTM1/GSTT1 null
genotype-endometriosis association.
Previous meta-analyses have found that the
GSTM1/GSTT1 gene polymorphism is associated with
cervical cancer (45), breast cancer
(46), bladder cancer (47), gastric cancer (48,49) and
acute leukemia (50). In accordance
with the observations of the present study, several studies have
shown that the GSTM1 (OR=32.6, 95% CI: 15.07–70.32,
P<0.0001) (18) and GSTT1
(OR>3; P<0.0001) null genotypes (12,16) are
associated with an increased risk for endometriosis. However, other
reports suggest the GSTM1 (OR=0.21, 95% CI: 0.09–0.52,
P<0.0001; OR=0.35, 95% CI: 0.15–0.83, P<0.0001) (40,43),
GSTT1 (OR≥5; P<0.0001) (16) and combined GSTM1/GSTT1
(OR=0.38; P<0.001) (17) null
genotypes are associated with a decreased risk for endometriosis.
These divergent results may be explained by differences in
GSTM1/GSTT1 null genotype frequencies and study
locations. The frequency of the GSTM1/GSTT1 null
genotype may vary from 10 to 65% depending on the region and
population studied (51). Different
study locations may introduce confounding variables associated with
variations in lifestyles and exposures to toxic substances of the
study populations.
The results of the present study must be interpreted
with caution due to the presence of substantial heterogeneity.
Among analyses of the studies of GSTM1 and GSTT1, the
cause of heterogeneity remains unclear, despite meta-regression
analyses being conducted. Among the analyses of combined
GSTM1/GSTT1 studies, subgroup and sensitivity
analyses suggested that studies that included patients with
advanced stage endometriosis caused most of the variability.
Publication bias was unlikely to have influenced the findings.
In addition to the heterogeneity, there were several
limitations to this study. Firstly, the composition of the
endometriosis patient and control populations varied between
studies. For instance, some studies included only patients with
advanced endometriosis (17–20,22,27,35),
while control populations consisted of a mixture of infertile
(29), postmenopausal (43) and premenopausal (18,35)
women, and newborn babies that had not been exposed to the
environment (17). Furthermore,
patients and controls were not always accurately matched by age or
environmental exposures. Secondly, gene-gene or gene-environment
interactions may jointly increase the risk for endometriosis;
therefore, different lifestyle and environmental factors may
contribute to differential genotypic frequencies in cases and
controls. Attempts were made to mitigate inaccuracies associated
with this limitation through a subgroup analysis stratified
according to ethnicity. Thirdly, this study was based on published
articles. As a positive result is more likely to be published,
publication bias is an inherent limitation of all meta-analyses
irrespective of the outcomes of the Egger's linear regression test
and Begg's rank correlation test.
In conclusion, the present meta-analysis shows the
GSTM1, GSTT1 and combined GSTM1/GSTT1
null genotypes are likely associated with increased susceptibility
to endometriosis. These data are in contrast to those reported
previously. Therefore, further studies reporting higher quality
evidence are necessary to verify these conclusions.
Acknowledgements
The study was supported by Hubei Provincial Natural
Science Foundation of China (Grant No.2013060602010236).
References
|
1
|
Viganò P, Parazzini F, Somigliana E and
Vercellini P: Endometriosis: Epidemiology and aetiological factors.
Best Pract Res Clin Obstet Gynaecol. 18:177–200. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giudice LC, Tazuke SI and Swiersz L:
Status of current research on endometriosis. J Reprod Med.
43(Suppl): 252–262. 1998.PubMed/NCBI
|
|
3
|
Kennedy S: Is there a genetic basis to
endometriosis? Semin Reprod Endocrinol. 15:309–318. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mayani A, Barel S, Soback S and Almagor M:
Dioxin concentrations in women with endometriosis. Hum Reprod.
12:373–375. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eskenazi B, Mocarelli P, Warner M, Samuels
S, Vercellini P, Olive D, Needham LL, Patterson DG Jr, Brambilla P,
Gavoni N, et al: Serum dioxin concentrations and endometriosis: A
cohort study in Seveso, Italy. Environ Health Perspect.
110:629–634. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heilier JF, Nackers F, Verougstraete V,
Tonglet R, Lison D and Donnez J: Increased dioxin-like compounds in
the serum of women with peritoneal endometriosis and deep
endometriotic (adenomyotic) nodules. Fertil Steril. 84:305–312.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nebert DW and Vasiliou V: Analysis of the
glutathione S-transferase (GST) gene family. Hum Genomics.
1:460–464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo SW: Glutathione S-transferases M1/T1
gene polymorphisms and endometriosis: A meta-analysis of genetic
association studies. Mol Hum Reprod. 11:729–743. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo SW: The association of endometriosis
risk and genetic polymorphisms involving dioxin detoxification
enzymes: A systematic review. Eur J Obstet Gynecol Reprod Biol.
124:134–143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hudelist G, Keckstein J and Wright JT: The
migrating adenomyoma: Past views on the etiology of adenomyosis and
endometriosis. Fertil Steril. 92:1536–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding Y, Chen ZF, Lin RY, Wang XF, Ding JB,
Ai XZ and Wen H: Relationship between endometriosis and glutathione
S-transferase M1, T1 genes of the Uygurs and Hans in Xinjiang.
Zhonghua Fu Chan Ke Za Zhi. 39:101–104. 2004.(In Chinese).
PubMed/NCBI
|
|
13
|
Rozati R, Baludu GS and Reddy BS: Possible
aggravating impact of gene polymorphism in women with
endometriosis. Indian J Med Res. 129:395–400. 2009.PubMed/NCBI
|
|
14
|
Yang LP and An XF: Analysis on the
relationship between polymorphism of Exon7 situs in CYP1A1 gene,
GSTM1 gene and susceptibility of endometriosis in women of
Han nationality in Jilin City. Zhongguo Fu You Bao Jian.
24:2556–2564. 2009.(In Chinese).
|
|
15
|
Peng DX, He YL, Qiu LW, Yang F and Lin JM:
Association between glutathione S-transferase M1 gene deletion and
genetic susceptibility to endometriosis. Di Yi Jun Yi Da Xue Xue
Bao. 23:458–459, 462. 2003.(In Chinese). PubMed/NCBI
|
|
16
|
Lin J, Zhang X, Qian Y, et al: Glutathione
S-transferase M1 and T1 genotypes and endometriosis risk: A
case-controlled study. Chin Med J (Engl). 116:777–780.
2003.PubMed/NCBI
|
|
17
|
Morizane M, Yoshida S, Nakago S, Hamana S,
Maruo T and Kennedy S: No association of endometriosis with
glutathione S-transferase M1 and T1 null mutations in a Japanese
population. J Soc Gynecol Investig. 11:118–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsieh YY, Chang CC, Tsai FJ, Lin CC, Chen
JM and Tsai CH: Glutathione S-transferase M1*null genotype but not
myeloperoxidase promoter G-463A polymorphism is associated with
higher susceptibility to endometriosis. Mol Hum Reprod. 10:713–717.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Babu KA, Reddy NG, Deendayal M, Kennedy S
and Shivaji S: GSTM1, GSTT1 and CYP1A1 detoxification
gene polymorphisms and their relationship with advanced stages of
endometriosis in South Indian women. Pharmacogenet Genomics.
15:167–172. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hur SE, Lee JY, Moon HS and Chung HW:
Polymorphisms of the genes encoding the GSTM1, GSTT1
and GSTP1 in Korean women: No association with
endometriosis. Mol Hum Reprod. 11:15–19. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aban M, Ertunc D, Tok EC, Tamer L, Arslan
M and Dilek S: Modulating interaction of glutathione-S-transferase
polymorphisms with smoking in endometriosis. J Reprod Med.
52:715–721. 2007.PubMed/NCBI
|
|
22
|
Kim SH, Choi YM, Lee GH, Hong MA, Lee KS,
Lee BS, Kim JG and Moon SY: Association between susceptibility to
advanced stage endometriosis and the genetic polymorphisms of aryl
hydrocarbon receptor repressor and glutathione-S-transferase T1
genes. Hum Reprod. 22:1866–1870. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao YH, Yao L, Wang D and Han P:
Relationship between endometriosis and glutathione S-transferase
Nl, T1 and P1 genetic polymorphism. Maternal and Child Health Care
of China. 24:805–814. 2009.(In Chinese).
|
|
24
|
Wu HL, Feng D and Liu PX: Relationship
between glutathione S-transferase M1 gene polymorphism and
susceptibility to endometriosis. Yi Xue Lin Chuang Yan Jiu.
26:764–766. 2009.(In Chinese).
|
|
25
|
Huang PC, Tsai EM, Li WF, Liao PC, Chung
MC, Wang YH and Wang SL: Association between phthalate exposure and
glutathione S-transferase M1 polymorphism in adenomyosis, leiomyoma
and endometriosis. Hum Reprod. 25:986–994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hosseinzadeh Z, Mashayekhi F and Sorouri
ZZ: Association between GSTM1 gene polymorphism in Iranian
patients with endometriosis. Gynecol Endocrinol. 27:185–189. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu CH, Guo CY, Yang JG, Tsai HD, Chang YJ,
Tsai PC, Hsu CC and Kuo PL: Polymorphisms of dioxin receptor
complex components and detoxification-related genes jointly confer
susceptibility to advanced-stage endometriosis in the Taiwanese Han
population. Am J Reprod Immunol. 67:160–168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seifati SM, Parivar K, Aflatoonian A,
Firouzabadi Dehghani R and Sheikhha MH: No association of
GSTM1 null polymorphism with endometriosis in women from
central and southern Iran. Iran J Reprod Med. 10:23–28.
2012.PubMed/NCBI
|
|
29
|
Matsuzaka Y, Kikuti YY, Goya K, Suzuki T,
Cai LY, Oka A, Inoko H, Kulski JK, Izumi S and Kimura M: Lack of an
association human dioxin detoxification gene polymorphisms with
endometriosis in Japanese women: Results of a pilot study. Environ
Health Prev Med. 17:512–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang Y: Association studies of the
relationship between GSTM1, GSTT1 gene polymorphism
and susceptibility to endometriosis in women of Han nationality in
Hunan province. Masters thesis, Central South University.
2007.http://cdmd.cnki.com.cn/Article/CDMD-10533-2007172085.htm(In
Chinese).
|
|
31
|
Sachan S, Nair RR, Khanna A and Singh K:
CYP1A1 and GSTM1 genes polymorphism and its association with
endometriosis: A pilot study. Asian Pacific Journal of
Reproduction. 2:297–300. 2013. View Article : Google Scholar
|
|
32
|
Baranova H, Canis M, Ivaschenko T,
Albuisson E, Bothorishvilli R, Baranov V, Malet P and Bruhat MA:
Possible involvement of arylamine N-acetyltransferase 2,
glutathione S-transferases M1 and T1 genes in the development of
endometriosis. Mol Hum Reprod. 5:636–641. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baranov VS, Ivaschenko T, Bakay B, Aseev
M, Belotserkovskaya R, Baranova H, Malet P, Perriot J, Mouraire P
and Baskakov VN: Proportion of the GSTM1 0/0 genotype in
some Slavic populations and its correlation with cystic fibrosis
and some multifactorial diseases. Hum Genet. 97:516–520. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baranov VS, Ivashchenko TE, Shved NIU,
Iarmolinskata MI, Sel'kov SA, Gorbushin SM, Savitskiĭ GA, Malet P,
Kanis M, Bruhat M and Baranova E: Genetic factors of predisposition
to endometriosis and response to its treatment. Genetika.
35:243–248. 1999.(In Russian). PubMed/NCBI
|
|
35
|
Hadfield RM, Manek S, Weeks DE, Mardon HJ,
Barlow DH and Kennedy SH: OXEGENE Collaborative Group: Linkage and
association studies of the relationship between endometriosis and
genes encoding the detoxification enzymes GSTM1,
GSTT1 and CYP1A1. Mol Hum Reprod. 7:1073–1078. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Baxter SW, Thomas EJ and Campbell IG:
GSTM1 null polymorphism and susceptibility to endometriosis
and ovarian cancer. Carcinogenesis. 22:63–65. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ivashchenko TE, Shved NIU, Kramareva NA,
Aĭlamazian EK and Baranov VS: Analysis of the polymorphic alleles
of genes encoding phase 1 and phase 2 detoxication enzymes in
patients with endometriosis. Genetika. 39:525–529. 2003.(In
Russian). PubMed/NCBI
|
|
38
|
Arvanitis DA, Koumantakis GE, Goumenou AG,
Matalliotakis IM, Koumantakis EE and Spandidos DA: CYP1A1, CYP19,
and GSTM1 polymorphisms increase the risk of endometriosis.
Fertil Steril. 79(Suppl 1): 702–709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vichi S, Medda E, Ingelido AM, Ferro A,
Resta S, Porpora MG, Abballe A, Nisticò L, De Felip E, Gemma S and
Testai E: Glutathione transferase polymorphisms and risk of
endometriosis associated with polychlorinated biphenyls exposure in
Italian women: A gene-environment interaction. Fertil Steril.
97:1143–1151.e3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bischoff FZ, Marquez-Do D, Dang D, Carson
SA, Buster JE and Simpson JL: NAT2 and GSTM1 DNA
polymorphisms: Increased GSTM1 (active) genotypes in
endometriosis. Fertil Steril. 77(Suppl 1): S172002. View Article : Google Scholar
|
|
41
|
Trabert B, Schwartz SM, Peters U, De Roos
AJ, Chen C, Scholes D and Holt VL: Genetic variation in the sex
hormone metabolic pathway and endometriosis risk: An evaluation of
candidate genes. Fertil Steril. 96:1401–1406.e3. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
De Carvalho CV, D'Amora P, Schor E,
Baracat EC, Girão MJC and Da Silva IDG: GSTM1 polymorphism
analysis in patients with mild and severe endometriosis. Fertil
Steril. 82(Suppl 2): S2912004. View Article : Google Scholar
|
|
43
|
Frare AB, Barbosa AM, Costa IR, Souza SR,
Silva RC, Bordin BM, Ribeiro Júnior CL and Moura KK: GSTM1
and GSTT1 polymorphisms in endometriosis in women from
Goiás, Brazil. Genet Mol Res. 12:2764–2770. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Arvanitis DA, Goumenou AG, Matalliotakis
IM, Koumantakis EE and Spandidos DA: Low-penetrance genes are
associated with increased susceptibility to endometriosis. Fertil
Steril. 76:1202–1206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu Y and Xu LZ: Meta-analysis of
association between GSTM1 gene polymorphism and cervical
cancer. Asian Pac J Trop Med. 5:480–484. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sergentanis TN and Economopoulos KP:
GSTT1 and GSTP1 polymorphisms and breast cancer risk:
A meta-analysis. Breast Cancer Res Treat. 121:195–202. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
García-Closas M, Malats N, Silverman D,
Dosemeci M, Kogevinas M, Hein DW, Tardón A, Serra C, Carrato A,
García-Closas R, et al: NAT2 slow acetylation, GSTM1 null
genotype, and risk of bladder cancer: results from the Spanish
Bladder Cancer Study and meta-analyses. Lancet. 366:649–659. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen B, Cao L, Zhou Y, Yang P, Wan HW, Jia
GQ, Liu L and Wu XT: Glutathione S-transferase T1 (GSTT1)
gene polymorphism and gastric cancer susceptibility: A
meta-analysis of epidemiologic studies. Dig Dis Sci. 55:1831–1838.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang H, Zhou Y, Zhuang W, Yin YQ, Liu GJ,
Wu TX, Yao X, Du L, Wei ML and Wu XT: Glutathione S-transferase M1
null genotype associated with gastric cancer among Asians. Dig Dis
Sci. 55:1824–1830. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Das P, Shaik AP and Bammidi VK:
Meta-analysis study of glutathione-S-transferases (GSTM1,
GSTP1, and GSTT1) gene polymorphisms and risk of
acute myeloid leukemia. Leuk Lymphoma. 50:1345–1351. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nelson HH, Wiencke JK, Christiani DC,
Cheng TJ, Zuo ZF, Schwartz BS, Lee BK, Spitz MR, Wang M, Xu X, et
al: Ethnic differences in the prevalence of the homozygous deleted
genotype of glutathione S-transferase theta. Carcinogenesis.
16:l243–1245. 1995. View Article : Google Scholar
|