Introduction
Thyroid carcinoma (TC) is the most common endocrine
malignancy accounting for >90% of endocrine gland malignancies
(1). Effective treatment in the
majority of patients with TC includes total thyroidectomy followed
by therapy with radioactive iodide (2). Although the prognosis of patients with
TC is favorable, unfortunately, the capacity for radioactive iodide
accumulation is diminished or lost in a high proportion of patients
with metastases (3). The genetic
events involved in TC consist of numerous gene mutations and gene
dysregulation, and several signaling pathways are activated in
favor of cell growth, survival and angiogenesis (4). However, our understanding of the
detailed molecular mechanism underlying TC tumorigenesis and
development is still not clear.
Accumulating evidence has demonstrated that
noncoding RNAs are involved in various types of human cancers
(5). To date, numerous long
noncoding RNAs (lncRNAs) have been demonstrated to have important
roles in human normal or disease states. Homeobox transcript
antisense RNA (HOTAIR) is a ~2,000 bp lncRNA that is encoded
antisense to the HOXC locus (6).
HOTAIR regulates the transcriptional silencing of genes by binding
to polycomb repressive complex 2 (PRC2) and localizing to the
specific site where H3K27 trimethylation and epigenetic silencing
of gene expression occur (7). In
addition, HOTAIR is able to interact with the lysine-specific
demethylase 1 (LSD1)-CoREST complex to mediate the removal of mono-
and dimethylation from H3K4, a histone marker associated with gene
activation (6,8).
The results from current studies indicate that
HOTAIR is a prognostic factor for the survival of patients with
breast, colon cancer and glioma, and increased HOTAIR expression in
patients has been correlated with increased metastasis (6,9–11). However, the expression and function
of HOTAIR in TC is not well known. The present study demonstrated
that the dysregulation of HOTAIR is correlated with metastasis and
poor prognosis in patients with TC. Through loss-of-function
analysis, the biological function of HOTAIR has been verified in
human thyroid carcinoma cell lines. Collectively, the results of
the present study demonstrate that HOTAIR may act as a novel
biomarker in patients with TC.
Materials and methods
Tissue and plasma samples
Thyroid tissue samples from 35 patients with TC were
collected via surgery from the Department of Thyroid Surgery and
Gastrointestinal Surgery, at the Affiliated Yantai Yuhuangding
Hospital of Qingdao University Medical College (Yantai, China)
between September 2012 and October 2014. Patients involved in the
present study provided written informed consent. Adjacent tissue
and cancerous tissue were reviewed and classified by a pathologist.
Fresh tissue samples were frozen to liquid nitrogen within 30 min
of surgery. Tissue sections from each TC sample.
Whole plasma samples were obtained from the 35
patients with TC and 20 healthy volunteers, and then stored in EDTA
tubes (Zhongyuan Biotech, Beijing, China). The tubes were
centrifuged at 1,200 × g for 10 min at 4°C to spin down the
plasma cells. The supernatants were transferred to microcentrifuge
tubes (Zhongyuan Biotech) and then centrifuged at 12,000 × g
for 10 min at 4°C again to completely remove the cellular
components. The plasma was then carefully collected, aliquoted, and
stored at −80°C until forthputting. Total RNA from 1 ml plasma was
extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's instructions.
Reverse transcription
quantitative-polymerase chain reaction (RT-qPCR)
RT reactions were carried out in 1 µg total RNA
using the PrimeScript RT reagent kit (Takara Bio, Inc., Otsu,
Japan). RT-qPCR was then performed using a SYBR Premix Dimer Eraser
kit (Takara Bio, Inc.). 18S rRNA was evaluated as a housekeeping
gene for the qPCR reactions. The primers used were as follows:
HOTAIR forward, 5′-TCATGATGGAATTGGAGCCTT-3′, and reverse,
5′-CTCTTCCTGGCTTGCAGATTG-3′; 18S rRNA forward,
5′-AGGATCCATTGGAGGGCAAGT-3′, and reverse,
5′-TCCAACTACGAGCTTTTTAACTGCA-3′. All the reactions were carried out
on an ABI7300 real-time PCR system according to the manufacturer's
instructions. Cycling conditions were as follows: 95°C for 10 sec,
one cycle; 95°C for 5 sec, 60°C for 30 sec, 40 cycles; followed by
a 30-min melting curve collection to verify the primer dimers. The
expression levels of HOTAIR in each sample were normalized to that
of the internal control 18S rRNA. The fold change of HOTAIR
expression in the tissue samples and plasma samples compared with
the controls were calculated using the 2−ΔΔCt
method.
Cell lines and culture conditions
The HT-ori3 normal human thyroid cell line and human
thyroid carcinoma cell lines including WRO, TPC-1 and SW579 were
all purchased from Beijing Zhongyuan Ltd. (Beijing, China). All
cells were maintained in a humidified atmosphere containing 10%
CO2 at 37°C.
Small interfering (si)RNA
transfection
Both HOTAIR siRNA and scramble were purchased from
Qiagen (Hilden, Germany). Cells (1×105) were grown on
six-well plates to 70% confluency and transfected using
Lipofectamine™ RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. A total of 48 h
post-transfection, the cells were harvested for RT-qPCR to analyze
HOTAIR knockdown efficiency.
Cell proliferation assay
A cell counting kit-8 (CCK-8) cell proliferation kit
was purchased from Dojindo Laboratories, (Kumamoto, Japan). All the
experimental protocols were conducted in accordance with the
manufacturers' instructions. Briefly, cells were seeded into a
96-well plate at 1×103 cells/well and cultured at 37°C.
CCK-8 solution was added to each well at the indicated times points
and then incubated at 37°C at 0, 12, 24, 36 and 48 h, then for a
futher 2 h. The absorbance at 450 nm was measured with a microplate
reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
experiments were repeated in triplicate and three independent
experiments were performed.
Cell invasion assay
24-well transwell plates (Corning Life Sciences,
Tewksbury, MA, USA) were used for invasion assays. For in
vitro invasion assays, the upper chambers of the transwells (8
µm) were pre-coated with diluted matrigel (BD Biosciences, Franklin
Lakes, NJ, USA). Briefly, 1×105 cells (in serum-free
media) and 10% serum-containing media were plated in the upper
chambers. After 48 hr incubation, the invaded cells were stained
with 0.1% crystal violet, and positively stained cells were counted
with a microplate reader (Bio-Rad Laboratories, Inc.). The
experiments were repeated in triplicate and three independent
experiments were performed.
Statistical analysis
Quantitative variables were expressed as means ±
standard deviations in the statistical analysis. Statistical
significances between groups were determined by two-tailed
Student's t-test. All statistical analyses were carried out with
SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant result. The survival
calculations were illustrated with Kaplan-Meier curve.
Results
Expression levels of HOTAIR are
elevated in TC tissue samples and plasma
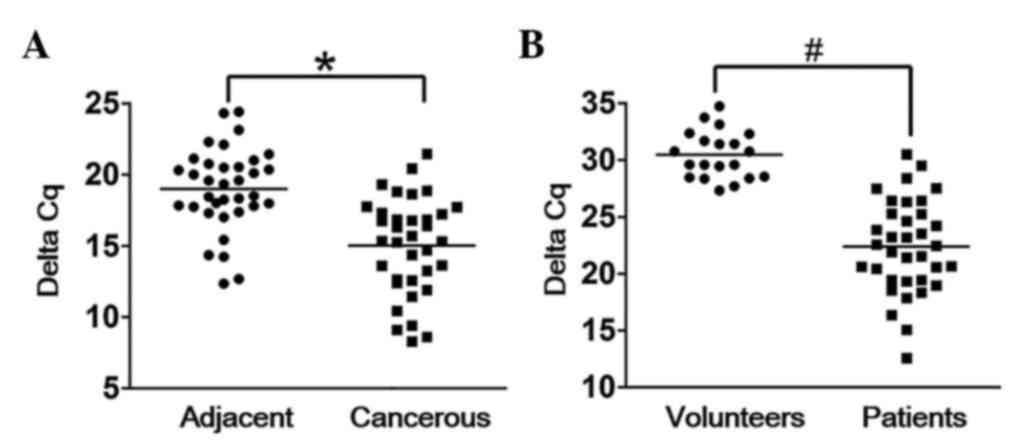
To assess the potential biological function of
HOTAIR, its expression levels in both adjacent tissues and
cancerous tissues were analyzed by RT-qPCR. The results
demonstrated that HOTAIR was significantly upregulated in cancerous
tissues compared with adjacent tissues (Fig. 1A). The expression levels of plasma
HOTAIR were also detected via RT-qPCR, and the results demonstrated
that HOTAIR could be detected in TC patient plasma, whereas there
was almost no HOTAIR expression in the plasma of the healthy
volunteers (Fig. 1B).
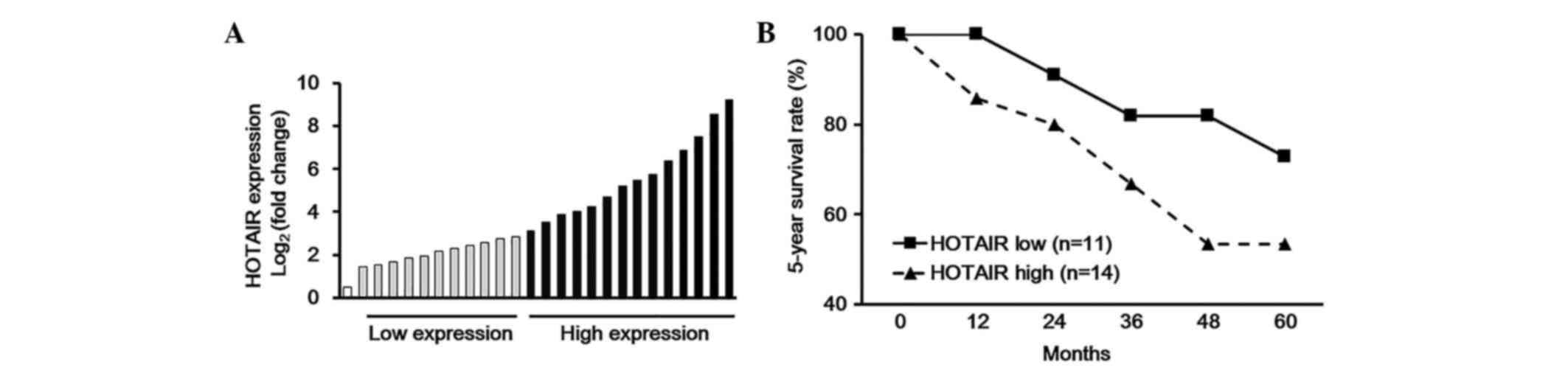
Association between plasma HOTAIR and
5-year survival rates
According to the expression levels of plasma HOTAIR
obtained from the RT-qPCR assay, the 35 TC cases could be divided
into two groups; one group that exhibited high HOTAIR expression
levels, and the other low HOTAIR expression levels (Fig. 2A). Furthermore, the association
between plasma HOTAIR expression levels and 5-year survival rate
was analyzed, and the results demonstrated that higher expression
levels of plasma HOTAIR were positively correlated with worse
5-year survival rates in patients with TC (Fig. 2B).
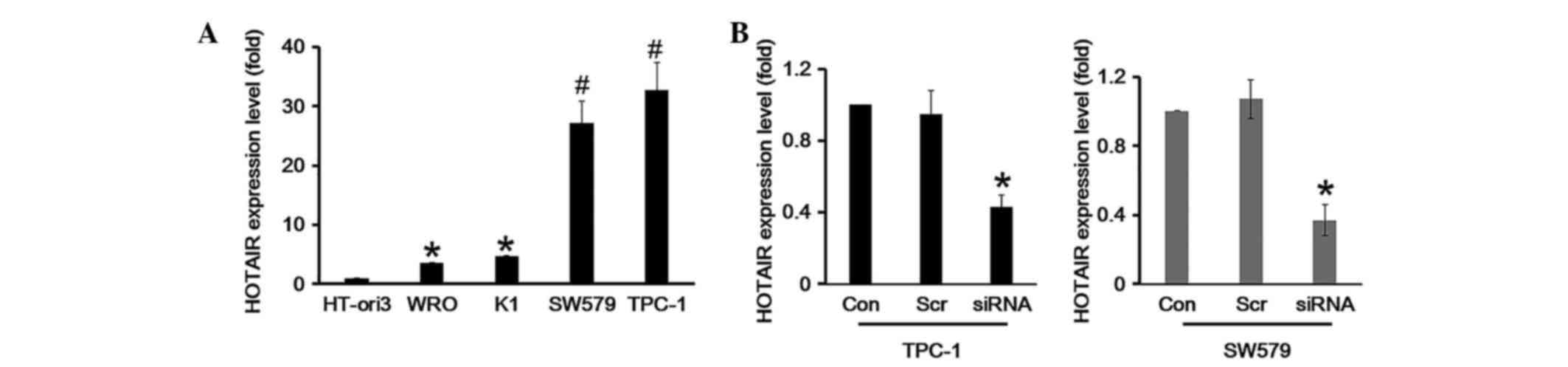
HOTAIR expression is upregulated in TC
cell lines
The detected HOTAIR expression in TC patient plasma
prompted further investigation into the function of HOTAIR. The
HT-ori3 normal human thyroid cell line and three human thyroid
carcinoma cell lines, including WRO, TPC-1 and SW579, were selected
for further study. The results demonstrated that the expression of
HOTAIR in human thyroid carcinoma cell lines showed an aberrant
expression profile. The expression of HOTAIR was markedly
upregulated in TPC-1 and SW579 cell lines (P<0.05), while HOTAIR
was somewhat elevated in the WRO cell line compared with HT-ori3
(P>0.05), as determined by RT-qPCR (Fig. 3A). In order to explore the function
of HOTAIR in TC cells, a siRNA specifically targeting HOTAIR was
designed to knockdown endogenous HOTAIR. The results of the RT-qPCR
demonstrated that HOTAIR was knocked down effectively in TPC-1 and
SW579 cells (Fig. 3B).
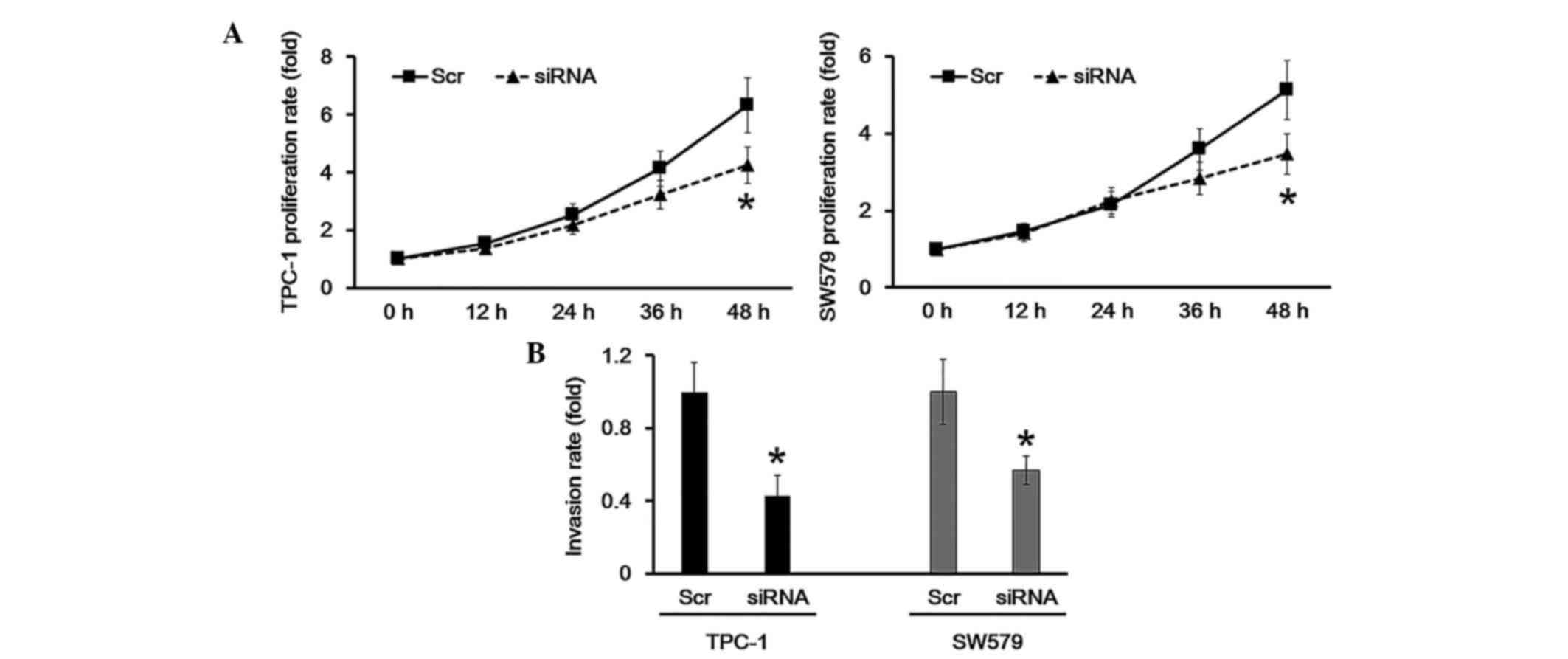
HOTAIR knockdown inhibits cell growth
and invasion
CCK-8 analysis was employed to determine the cell
proliferation rate. The results demonstrated that when HOTAIR was
knocked down in TPC-1 and SW579 cells, the proliferation was
significantly inhibited compared with the scramble group (Fig. 4A). Additionally, a transwell assay
was performed to study whether HOTAIR participated in cell
invasion. The results suggested that HOTAIR knockdown in TPC-1 and
SW579 cells resulted in a decreased invasion ability compared with
the scramble group. These results indicated that HOTAIR knockdown
suppressed cell proliferation and invasion.
Discussion
Significant progress in understanding the molecular
pathogenesis of TC has been made in recent years. Remarkable
knowledge has been accumulated on the role of fundamental signaling
pathways, such as the mitogen-activated protein kinase and the
phosphoinositide 3-kinase/protein kinase B/mammalian target of
rapamycin pathways (4). Though these
signaling pathways provide targets for therapeutic agents, a
further and more detailed investigation of the molecular mechanism
underlying TC tumorigenesis is required.
In recent years, studies on noncoding RNAs has
attracted considerable attention in basic medical research
(12,13). It was reported that miRNAs and
lncRNAs have distinct expression profiles in human plasma in
various types of cancers (14). In
addition, serum miRNAs may serve as important diagnostic biomarkers
for certain cancer types, such as lung cancer (15), breast cancer (16), liver cancer (17) and pancreatic cancer (18). However, lncRNA expression patterns in
plasma have not been investigated for their potential as novel
biomarkers for TC diagnosis or prognosis.
LncRNAs, ranging from 200 to >10,000 nucleotides,
are abundantly transcribed by the mammalian genome (19). LncRNAs have emerged as novel
important regulators of tumorigenesis and development. For
instance, the lncRNA HOXA distal transcript antisense RNA is able
to promote progression and gemcitabine resistance by regulating
HOXA13 in pancreatic cancer (20); a
novel lncRNA AK001796 may act as an oncogene and is involved in
cell growth inhibition by resveratrol in lung cancer (21); lncRNA colon cancer associated
transcript 1 (non-protein coding) is able to promote hepatocellular
carcinoma progression by functioning as a let-7 sponge (22).
LncRNA HOTAIR was first discovered by Howard Chang's
group (5), HOTAIR is overexpressed
in breast, colorectal, liver and nasopharyngeal cancers (23). Tsai et al (8) reported that a 89 bp fragment at the 5′
end of HOTAIR binds to PRC2, and a 646 bp fragment at its 3′ end
binds to the LSD1/CoREST complex (8). HOTAIR has important roles in
tumorigenesis and progression. Briefly, HOTAIR acts as an oncogene
in tumorigenesis, and promotes invasion and metastases in tumor
progression (24). However, whether
HOTARI is involved in TC remains unclear. Therefore, the expression
and function of HOTAIR must be comprehensively understood prior to
the use of HOTAIR in TC treatment.
In the current study, the data suggested that HOTAIR
is differentially expressed in the tissues and plasma of the
patients with TC compared with the controls, and HOTAIR expression
in TC is likely to be associated with the aggressiveness and the
progression of the tumor. The in vitro experiments indicated
that HOTAIR was able to act as an oncogene; knockdown of HOTAIR
inhibited TC cancer cell proliferation and invasion. In conclusion,
to the best of our knowledge, these findings indicate for the first
time that the expression of HOTAIR in plasma can be used as a novel
diagnostic biomarker for TC. The utility of HOTAIR expression could
be established as a prognostic indicator for TC.
References
|
1
|
Fofi C, Festuccia F, Barberi S, Apponi F,
Chiacchiararelli L, Scopinaro F, Punzo G and Menè P: Hemodialysis
in patients requiring 131I treatment for thyroid carcinoma. Int J
Artif Organs. 36:439–443. 2013.PubMed/NCBI
|
|
2
|
Sherman EJ, Su YB, Lyall A, Schöder H,
Fury MG, Ghossein RA, Haque S, Lisa D, Shaha AR, Tuttle RM and
Pfister DG: Evaluation of romidepsin for clinical activity and
radioactive iodine reuptake in radioactive iodine-refractory
thyroid carcinoma. Thyroid. 23:593–599. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wakabayashi H, Taki J, Inaki A, Toratani
A, Kayano D and Kinuya S: Extremity Radioactive Iodine Uptake on
Post-therapeutic Whole Body Scan in Patients with Differentiated
Thyroid Cancer. Asia Ocean J Nucl Med Biol. 3:26–34.
2015.PubMed/NCBI
|
|
4
|
Netea-Maier RT, Klück V, Plantinga TS and
Smit JW: Autophagy in thyroid cancer: Present knowledge and future
perspectives. Front Endocrinol (Lausanne). 6:222015.PubMed/NCBI
|
|
5
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales D Rivea, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thum T and Condorelli G: Long noncoding
RNAs and microRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ling H: Non-coding RNAs: Therapeutic
Strategies and Delivery Systems. Adv Exp Med Biol. 937:229–237.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Latronico MV and Condorelli G: Therapeutic
applications of noncoding RNAs. Curr Opin Cardiol. 30:213–221.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He D, Wang J, Zhang C, Shan B, Deng X, Li
B, Zhou Y, Chen W, Hong J, Gao Y, et al: Down-regulation of
miR-675-5p contributes to tumor progression and development by
targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol
Cancer. 14:732015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Y, Xu K and Yagüe E: MiR-218 targets
survivin and regulates resistance to chemotherapeutics in breast
cancer. Breast Cancer Res Treat. 151:269–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rebucci M, Sermeus A, Leonard E, Delaive
E, Dieu M, Fransolet M, Arnould T and Michiels C: MiRNA-196b
inhibits cell proliferation and induces apoptosis in HepG2 cells by
targeting IGF2BP1. Mol Cancer. 14:792015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin X, Sun Y, Yang H, Li J, Yu S, Chang X,
Lu Z and Chen J: Deregulation of the MiR-193b-KRAS axis contributes
to impaired cell growth in pancreatic cancer. PloS One.
10:e01255152015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kohls K, Schmidt D, Holdenrieder S, Müller
SC and Ellinger J: Detection of cell-free lncRNA in serum of cancer
patients. Urologe A. 54:819–825. 2015.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Q, Xu E, Dai J, Liu B, Han Z, Wu J,
Zhang S, Peng B, Zhang Y and Jiang Y: A novel long noncoding RNA
AK001796 acts as an oncogene and is involved in cell growth
inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol.
285:79–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
24
|
Yu X and Li Z: Long non-coding RNA HOTAIR:
A novel oncogene (Review). Mol Med Rep. 12:5611–5618.
2015.PubMed/NCBI
|