Introduction
The spread of drug-resistant clinical tuberculosis
(TB) and its co-infection with HIV have seriously affected TB
prevention and treatment (1).
Inherently, this has meant deterioration in the control of
epidemics. As anti-TB drugs that were identified early for clinical
use, rifampicin (RFP) and isoniazid (INH) have served an important
therapeutic role for TB treatment (2,3).
Statistical data from WHO indicates that the drug resistance of TB
to the first-line drugs INH and RFP is 13.3 and 6.3%, respectively
(4). Research has also suggested
that in recent years TB clinical strains have demonstrated high
levels of RFP (13.3%), INH (24.6%) and multi-drug (10.5%)
resistance (5). At present, there
are a number of methods used to detect Mycobacterium
tuberculosis (MTB) drug resistance, but most are time-consuming
(6). The absolute concentration, or
proportional, method requires cultivation of the bacterium for 2–3
months before the drug sensitivity of a strain is able to be
determined (7). The BACTEC method
(8) only takes 2–4 weeks, but this
remains too slow for clinical requirements.
MTB resistance to RFP is primarily associated with a
mutation in the rpoB gene (9,10). If
the rpoB gene encoding the β subunit of RNA polymerase
mutates, RFP and the β subunit cannot combine to produce the
antibacterial effect. This results in MTB resistance to RFP. At
present, 70 mutation sites have been identified in the rpoB
gene in patients diagnosed with MTB resistance to RFP (9,11,12). Of
these, >95% are associated with drug resistance in the
rpoB gene codons 507–533, which consist of 81 bases; this is
known as the rifampicin resistance-determining region (RRDR). The
531 Ser, 526 His and 516 Asp codons are considered the key mutation
locations for RFP resistance (13,14).
Thus, there is great value in an early clinical diagnosis of
resistance, to establish a rapid detection method of rpoB
gene mutations in this region and the relative likelihood of RFP
resistance.
Mutations of various relevant enzyme-encoding genes
in MTB, which encode the enzymes involved in the INH pathway,
create resistance to INH. The genes involved primarily include
katG, inhA, KasA, ndh and ahpC
(15,16). Among these, katG encodes
catalase-peroxidase, which is involved in the transition of INH
into its active form. inhA encodes enoyl reductase, which is
involved in mycolic acid biosynthesis. katG and inhA
are the primary molecular factors in MTB resistance to INH.
Molecular biological detection techniques may be used to detect the
detailed molecular characteristics of katG and inhA
gene mutations, which may provide rapid early diagnosis for the
treatment of drug-resistant TB. The mutation spectrum and mutation
rates of katG and inhA have regional differences, but
katG315 and inhA-15 are the most common mutation
sites (17,18). Therefore, the detection of mutations
at these two sites has potential application in determination of
INH resistance.
In the present study, a DNA microarray chip was
prepared to detect the mutation sites, based on the gene mutations
associated with the molecular mechanisms of RFP and INH resistance.
The DNA microarray chip technique was used to detect RFP and INH
resistance of clinical TB strains in Soochow City (China) and the
results were compared with drug sensitivity results from the
absolute concentration method, based on Lowenstein-Jensen
cultivation. The value of rapid detection of RFP and INH resistance
in TB clinical strains within Soochow City (by DNA microarray chip)
was considered, in terms of the method's ability to promote the
early diagnosis and treatment of TB, decrease TB morbidity and
increase the recovery rates from TB in Soochow City.
Materials and methods
Research population
From January 2014 to December 2014, the sputum
samples were collected from 42 patients (male/female, 27/15; age
range, 18–75 years) with TB in the Affiliated Hospital of
Infectious Diseases of Soochow University (Soochow, China). All the
patients were negative for acute hepatitis B, antibodies (Abs) to
Hepatitis C virus (HCV), Hepatitis D virus (HDV), human
immunodeficiency virus (HIV), combined tumor and other symptoms of
liver damage such as autoimmune disease, alcoholic liver disease
and drug-induced hepatitis. Our study was approved by the local
ethics committee, and all patients provided their written informed
consent.
Conventional drug sensitivity
test
Following conventional processing (5), the patients' sputum samples were placed
in a BACTEC MGIT-960 MTB culture machine (BD Diagnostics, Sparks,
MD, USA), and acid-fast staining was performed to identify
TB-positive samples (19). Following
cultivation, the MTB was assessed for drug sensitivity using a
Lowenstein-Jensen culture medium (Cell Biotech Co., Ltd., Jinan,
China) (5). The cultivated MTBs were
subjected to three treatments of RFP and INH (Longtai Corporation,
Jilin, China) each, based on concentration (RFP: 0, 50 and 250
µg/ml; INH: 0, 1 and 10 µg/ml). Following a 4-week cultivation
period, the following criteria were used to determine the
sensitivity: Counts >200, drug sensitive; counts ≤200, drug
resistant. ‘Moderate sensitivity’ was determined to indicate drug
resistance.
Detection by DNA microarray chip
This study was based on the designing of
oligonucleotide probes which can specifically detect the mutations
on the promoter of rpoB, KatGandinhA. Briefly, the
DNA microarray chip technique (20)
was used to test mutations in the rpoBgene at the 511, 513,
526, 531 and 533 codons (common mutation sites) to give an
indication of RFP resistance in the RRDR. For INH resistance, the
katG315 and inhA-15 mutation sites were assessed. The
standard protocol of the GeeDom MTB drug detection kits
(18000065.001; CapitalBio Corporation, Beijing, China) were
followed to assess the RFP and INH resistance of the samples. The
nucleic acid was extracted using a GeeDom Extractor 36 rapid
nucleic acid extraction instrument (CapitalBio Corporation). This
was followed by polymerase chain reaction (PCR) amplification. The
sequence of primers is respectively forward AGGCGATCACACCGCAGACGT
and reverse CGAGCCGATCAGACCGATGT for amplifying rpoB
fragment, forward GATCGTCGGCGGTCACACTTTC and reverse
CTCTTCGTCAGCTCCCACTCGTAG for amplifying KatG fragment, and forward
CACCCGCAGCCAGGGCC and CGATCCCCCGGTTTCCTCC for amplifying inhA
fragment. The programme for PCR is as follows: Pre-denaturation
94°C for 4 min; 30 cycle: Denaturation 94°C for 40 sec, annealing
reaction 56°C for 50 sec, elongation reaction 72°C 60 sec; and
elongation reaction 72°C for 10 min. Once combined with a
hybridization buffer (CapitalBio Corporation), the products were
placed in a BioMixer II chip hybridization instrument (Core Life
Sciences, Inc., Irvine, CA, USA) for hybridization. Products were
then put in a lideWasher8 chip dry cleaning instrument (CapitalBio
Corporation) for washing and drying. Finally, the chip was placed
in the TB drug resistance chip identification system (CapitalBio
Corporation) for scanning and interpretation (LuxScanTM 10K/B
software, CapitalBio Corporation). The interpreted results and chip
information were recorded in detail according to the standard
protocol. Every five repeated hybrid grid points correspond to one
cell of specific content. The assay was performed in
triplicate.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc., San Diego, CA, USA)
showing mean ± standard error of the mean. Mann-Whitney U tests of
SSPS 12.0 (SPSS, Inc., Chicago, IL, USA) were used to assess the
difference between different groups. A two-tailed P<0.05 was
considered statistically significant.
Results
RFP and INH resistance of MTB
according to the conventional drug sensitivity test
A conventional drug sensitivity test using the
Lowenstein-Jensen culture medium demonstrated that of the 42
positive TB samples tested, 15 were sensitive and 27 resistant to
RFP. In total, 21 samples were sensitive and 21 samples resistant
to INH.
Detection of MTB rpoB-RRDR relevant
mutation sites according to the DNA microarray chip
The mutations and wild type detection spectrum are
presented in Fig. 1, and Table I summarizes the results. The results
were estimated based on every five repeated hybrid grid points
correspond to one cell of specific content. There were three (7.1%)
mutations of the rpoB gene at RRDR-526 (CAC→TA2.3C; His→Tyr;
H526Y), one mutation (2.4%) at RRDR-526 (CAC→GAC; His→Asp; H526D),
20 mutations (47.6%) at RRDR-531 (TCG→TTG; Ser→Leu; S531L) and one
mutation (2.4%) at RRDR-531 (TCG→TGG; Ser→Trp; S531W). Furthermore,
there were 17 mutations (40.5%) for which the RRDR-511, 513, 526,
531, and 533 mutation sites were all wild type (WT).
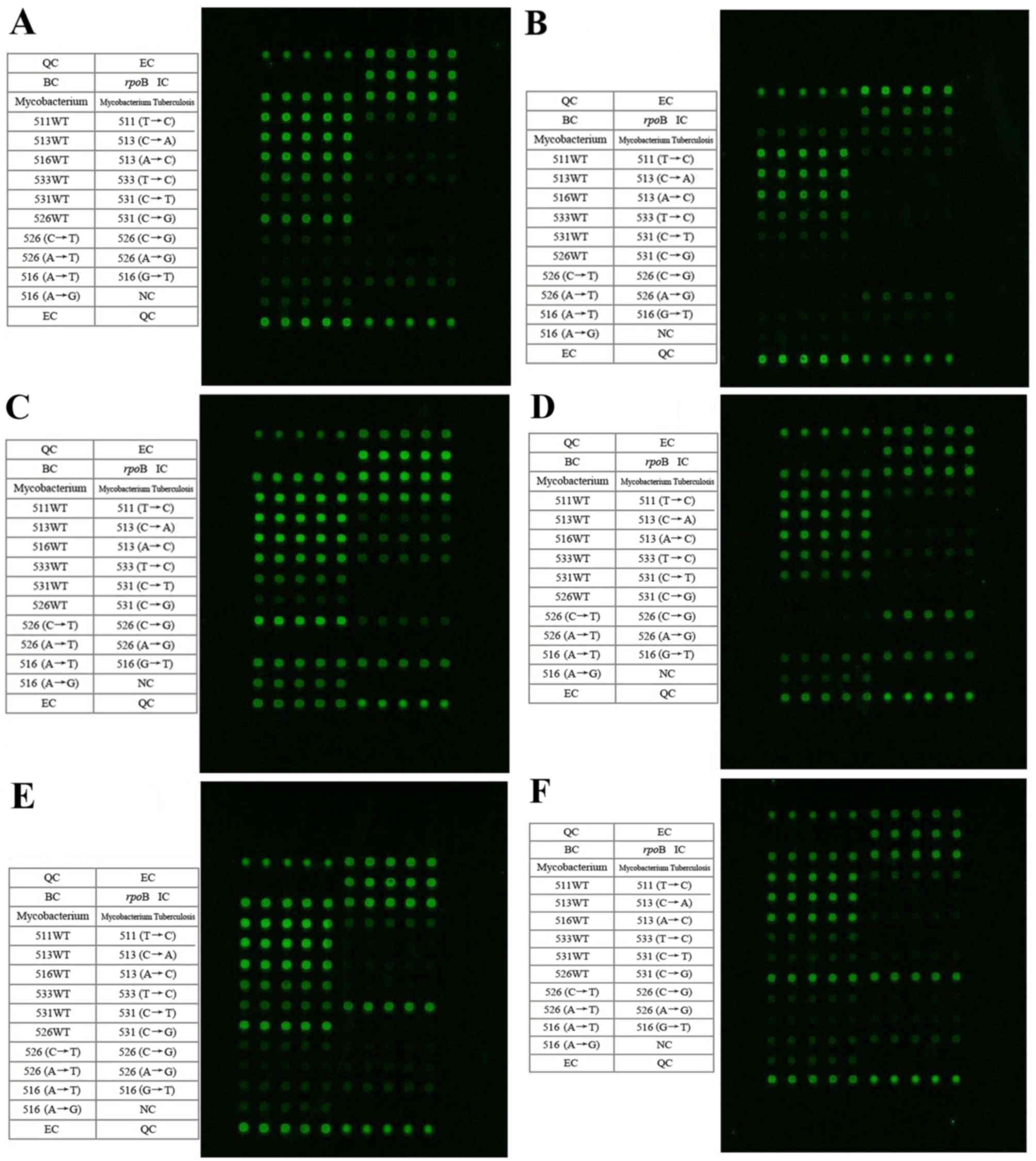 | Figure 1.Detection spectrums of the microarray
chip for the rpoB-RRDR relevant mutation points. The
microarray chip spectrums present the samples with mutation(s) at:
(A) WT MTB rpoB-RRDR 511, 513, 516, 526, 531 and 533, (B)
MTB rpoB-RRDR 526 (CAC→CGC), (C) MTB rpoB-RRDR 526
(CAC→TAC), (D) MTB rpoB-RRDR 526 (CAC→GAC), (E) MTB
rpoB-RRDR 531 (TCG→TTG) and (F) MTB rpoB-RRDR 531
(TCG→TGG). The contents of the table on the left side correspond to
the microarray hybridization dot matrix on the right side in each
figure. Every five repeated hybrid grid points correspond to one
cell of specific content. RRDR, rifampicin resistance-determining
region; MTB, Mycobacterium tuberculosis; QC, chip
preparation quality control; EC, chip hybridization quality
control; BC, blank comparison quality control; IC, targeted gene
amplification quality control; WT, wild type. |
 | Table I.Microarray chip detection of mutations
in Mycobacterium tuberculosis rpoB-RRDR relevant mutation
sites for the 42 samples. |
Table I.
Microarray chip detection of mutations
in Mycobacterium tuberculosis rpoB-RRDR relevant mutation
sites for the 42 samples.
| rpoBcodon
mutation | Codon
variation | Amino acid
variation | Strain no. | Percentage,% |
|---|
| RRDR-531 | CAC→TAC | His→Tyr | 3 |
7.1 |
|
| CAC→GAC | His→Asp | 1 |
2.4 |
|
| TCG→TTG | Ser→Leu | 20 | 47.6 |
|
| TCG→TGG | Ser→Trp | 1 |
2.4 |
| RRDR-511, 513, 516,
526, 531, 533 | No change | No change | 17 | 40.5 |
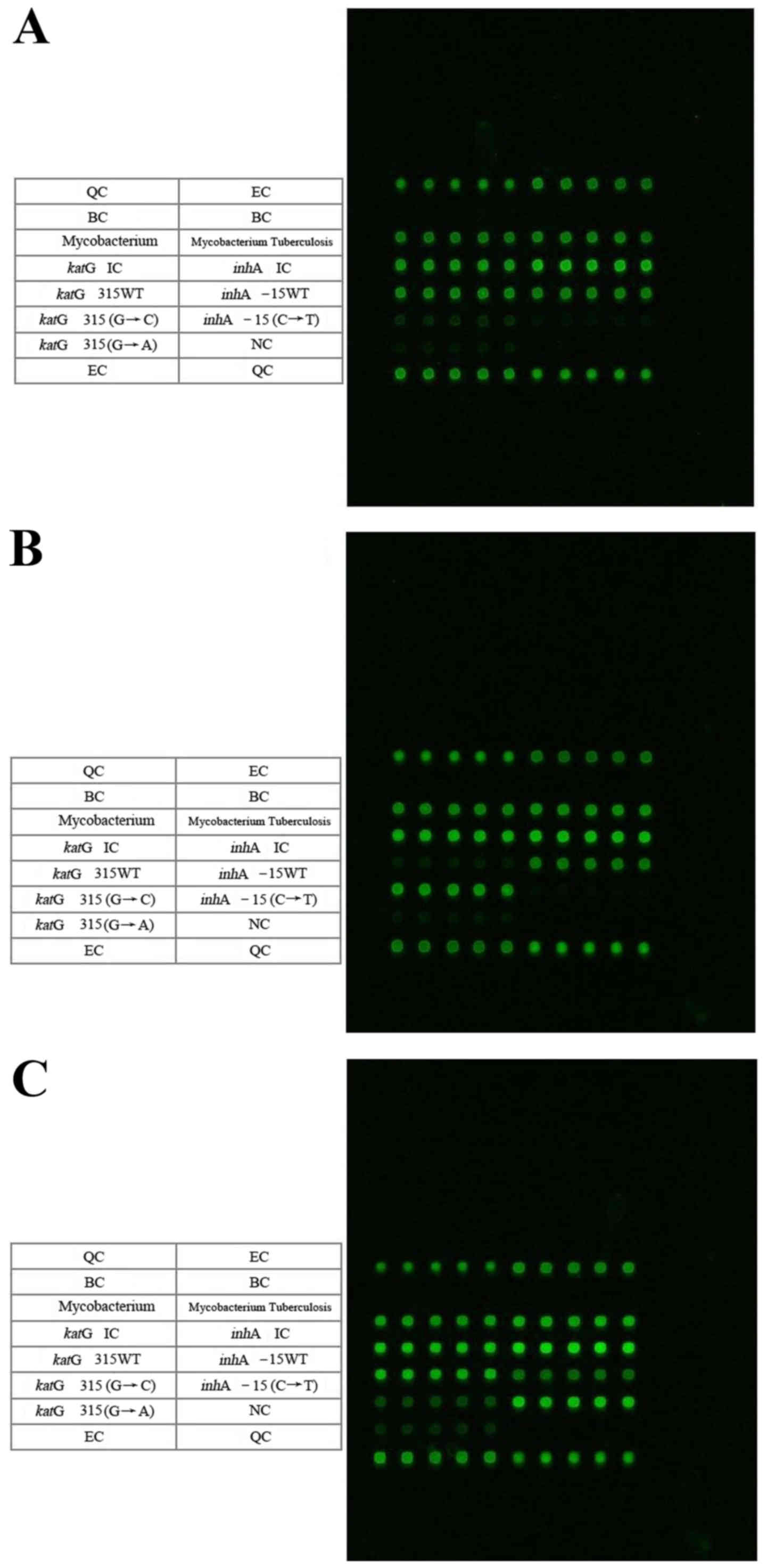
Detection of MTB katG315 and inhA-15
mutation points according to the DNA microarray chip
The microarray detection spectrums for the
KatG315 (G→C) mutation and the inhA (C→T) mutations
are presented in Fig. 2, and
Table II summarizes the results.
Similarly, the results were estimated based on every five repeated
hybrid grid points correspond to one cell of specific content.
There were 26 mutations (61.9%) of katG315 (AGC→ACC;
Ser→Thr; S315T), 16 cases (38.1%) of inhA-15 and
katG315 WTs, 1 mutation (2.4%) of inhA-15 (C→T) and
41 cases (97.6%) of inhA-15 WT (Fig. 2). Thus, the mutations of
katG315 (AGC→ACC) were more common, compared with the
inhA-15 mutation mutations (C→T), and accounted for 96.3% of
all the mutations associated with drug resistance.
 | Table II.Microarray chip detection of
KatG315 and inhA-15 mutation points for the 42
samples. |
Table II.
Microarray chip detection of
KatG315 and inhA-15 mutation points for the 42
samples.
| Codon or gene
locus | Codon or gene locus
variation | Amino acid
variation | Strain no. | Percentage, % |
|---|
| KatG315 | AGC→ACC | Ser→Thr(S315T) | 26 | 61.9 |
|
| WT | – | 16 | 38.1 |
| inhA-15 | C→T | – | 1 |
2.4 |
|
| WT | – | 41 | 97.6 |
Reliability of DNA microarray chip for
drug sensitivity diagnosis
Results from the Lowenstein-Jensen cultivation
method (5) were compared with the
results from the DNA microarray chip detection. The susceptibility
and specificity for RFP resistant strains were 92.3 and 93.8%,
respectively, when diagnosed by DNA microarray chip detection. The
susceptibility and specificity for the INH resistant strains were
66.7 and 81%, respectively (Table
III).
 | Table III.Susceptibility and specificity of DNA
microarray chip on the determination of RFP and INH R/S compared
with the standard method of Lowenstein-Jensen cultivation for the
42 samples. |
Table III.
Susceptibility and specificity of DNA
microarray chip on the determination of RFP and INH R/S compared
with the standard method of Lowenstein-Jensen cultivation for the
42 samples.
|
| Determination by
Lowenstein-Jensen cultivation | Determination by
DNA microarray chip, R/S | Susceptibility,
% | Specificity, % |
|---|
| RFP resistance | 26 | 24/2 |
|
|
| RFP
sensitivity | 16 | 1/15 | 92.3 | 93.8 |
| Total | 42 | 25/17 |
|
|
| INH resistance | 21 | 14/7 |
|
|
| INH
sensitivity | 21 | 4/17 | 66.7 | 81.0 |
| Total | 42 | 18/24 |
|
|
Discussion
RFP and INH are the primary first-line anti-TB
drugs. However, the application and effectiveness of these drugs
has been greatly impacted by the increase in drug resistance.
Statistical data from WHO indicates that the drug resistance of TB
to these first-line drugs was 13.3 and 6.3%, respectively (4). A previous study has also suggested that
in TB clinical strains demonstratehigh levels of RFP (13.3%), INH
(24.6%), and multi-drug (10.5%) resistance (5). The conventional drug sensitivity
assessment takes a long time and is unable to provide prompt
guidance for appropriate clinical treatment (7,8). Thus,
the potential rapid detection of RFP and INH resistance by MTB
would be impactful. Molecular biological techniques like PCR-single
strand confirmation polymorphism (SSCP) are easy, rapid and
inexpensive, but may only detect gene mutations; they are generally
unable to determine the mutation points and the characteristics of
those mutations. A number of the genes associated with
drug-resistance are naturally polymorphic, or their mutations are
not always associated with drug resistance; therefore, this may
result in false positives using PCR-SSCP and other similar
techniques (21).
DNA microarray chips comprise a molecular biological
technique that has been developed since the 1990′s. The technique
has been used widely for multiple applications due to its
rapidness, accuracy, high efficiency and easy operation (20). For different mutations, the
corresponding oligonucleotide probes are designed and immobilized
on a solid support. The gene fragments that contain the mutations
are amplified by PCR and then hybridized with the gene microarray
chips. As a large number of probes may be located on the solid
support at the same time, numerous sample sequences may be detected
and analyzed simultaneously. This technique addresses the
shortcomings of traditional nucleic acid blotting techniques, a low
degree of automation, small number of operating sequences and low
detection efficiency. Along with the subsequent identification of
genes associated with MTB drug-resistance and their relevant
mutation sites, a number of previous studies have considered the
application of DNA microarray chips for the detection of MTB
drug-resistance. A total of 12 genes associated with
drug-resistance for 5 first-line and second-line anti-TB drugs,
including fluoroquinolones, have been detected and analyzed. Based
on the molecular mechanisms of MTB drug-resistance associated with
the gene mutations, gene chips are designed to detect
drug-resistance-related genes and MTB drug-resistance may be judged
by the detected mutations (22,23).
RFP and INH are bactericidal drugs and comprise a
drug combination that has long been effective. Numerous RFP
resistant strains of MTB that result from rpoB gene
mutations are also resistant to INH (24). >95% of RFP resistance related gene
mutations were identified in the rpoB gene-RRDR and ~85% of
the strains resistant to drugs had mutations on the 531 Ser, 526
His and 516 Asp codons in the RRDR. Based on this, the GeeDom
Mycobacterium tuberculosis drug detection kits were used to
analyze the mutations and relevant drug-resistance of the
rpoB gene RRDR 511, 513, 516, 526, 531 and 533 mutation
sites, in the samples from patients in Soochow City, China. The
results were compared with those derived using the absolute
concentration method. The susceptibility and specificity of the DNA
microarray chip to RFP sensitivity was 92.3 and 93.8%,
respectively.
INH is a first-line anti-TB drug that is used
together with RFP. A study previously demonstrated that
multidrug-resistance resulting from double-drug resistance to INH
and RFP was the primary reason for refractory TB (25). Based on the prevalence of
katG315 and inhA-15 mutations in the strains
resistant to the drugs and their relevance to INH resistance, in
addition to previous results (26),
the primary mutation mechanism of INH-resistance in the MTB
katGgene investigated was 315 AGC→ACC, Ser→Thr (S315T).
GeeDom Mycobacterium tuberculosis drug detection kits were
used to detect the INH resistance of the MTB clinical strains, and
the results were compared with those derived using the absolute
concentration method. The susceptibility and specificity of the DNA
microarray chip to INH sensitivity was 66.7 and 81%, respectively.
The susceptibility of the DNA microarray chip to INH sensitivity
was relatively low, which may be due to the INH resistance
mechanisms being associated with multi-gene mutations.
The results of a few of the drug sensitivity tests
were not consistent. This may be because the samples had primary
(natural) drug resistance, rather than drug resistance caused by
gene mutations. Natural drug resistance occurs due to the barrier
mechanism, consisting of an extracellular multilayer coating and an
active multidrug efflux ion pump, disturbing drug transportation by
the MTB (27,28). Alternatively, the inconsistency may
have arisen because gene mutations occurred outside the detection
range, resulting in RFP and INH resistance. For example, a few
mutations have previously been identified in the C II and C III
regions at N-terminal end of the rpoB gene, outside the RRDR
(12,29), resulting in RFP resistance. In
addition, gene mutations other than katG and inhA-15
(like KasA, ndh and ahpC) may result in INH
resistance (and these were not tested for in the present study).
Mutations may have also happened within detection range (RRDR-511,
513, 516, 526, 531 and 533), but may not have consisted of the
detected mutation forms. For example, the detection for the
526-codon mutation point only included four forms, which were
CAC→TAC, GAC, CTC and CGC4; however, there are other known possible
mutations of this point, including CAC→AAC.
In addition, the mutation rate that occurred during
the detection of the strains sensitive to INH and RFP may have been
caused by a difference in the drug concentration. That is, the drug
concentrations in the absolute concentration method were high.
Strains that were detected as drug sensitive through the absolute
concentration method may have been drug resistant under lower drug
concentrations. It is possible that the mutation of drug-resistant
strains only happened at low drug concentrations. If this were the
case, the detection of RFP and INH resistance by the GeeDom
Mycobacterium tuberculosis drug detection kits may not
achieve 100% accuracy, compared with the absolute concentration
method.
Ultimately, the susceptibility and specificity of
the DNA microarray chip to RFP resistance was 92.3 and 93.8%,
respectively, which is high and has great significance for the
rapid detection of resistance in clinical applications. In
conclusion, a DNA microarray chip may be developed as an effective
method for the rapid screening of MTB resistance to RFP and INH,
specifically for RFP resistance. This was effective in Soochow
City, and has great significance for the clinical rapid diagnosis
of MTB and subsequently treating TB patients with effective drugs
against MTB as early as possible.
Acknowledgements
The present study was supported by grants from the
Year 2014 Suzhou City Key Clinical Disease Diagnosis and Treatment
Technology Special Project (grant no. LCZX201314), Year 2014
Jiangsu Province Association of Preventive Medicine Scientific
Research Project (grant no. Y2013023), Year 2015 Suzhou City Key
Clinical Disease Diagnosis and Treatment Technology Special Project
(grant no. LCZX201414) and Suzhou Municipal Health Bureau Hygiene
Project through Science and Education (grant no. KJXW2013033).
References
|
1
|
Maimaiti R, Zhang Y, Pan K, Mijiti P,
Wubili M, Musa M and Andersson R: High prevalence and low cure rate
of tuberculosis among patients with HIV in Xinjiang, China. BMC
Infect Dis. 17:152017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Timmins GS and Deretic V: Mechanisms of
action of isoniazid. Mol Microbiol. 62:1220–1227. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ranguelova K, Suarez J, Magliozzo RS and
Mason RP: Spin trapping investigation of peroxide-and
isoniazid-induced radicals in Mycobacterium tuberculosis
catalase-peroxidase. Biochemistry. 47:11377–11385. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
World Health Organization (WHO), .
Anti-tuberculosis drug resistance in the worldFourth global report.
WHO; Geneva: 2008
|
|
5
|
Yu X, Shen X, Tang P, et al: Analysis of
multi-drug resistance of Mycobacterium tuberculosis derived from
tuberculosis patients in Suzhou city. J Clin Pulmon Med.
17:269–270. 2012.
|
|
6
|
Li G: The study of drug sensitivity for
mycobacteria, Modern phthisiology. Beijing, PLA Medical
Publication; pp. 36–57. 2000
|
|
7
|
Carbonnelle B and Carpentier E:
Bacteriological diagnosis of tuberculosis: Current hieratic
classification of methods. Rev Med Interne. 16:518–523. 1995.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Demers AM, Venter A, Friedrich SO,
Rojas-Ponce G, Mapamba D, Jugheli L, Sasamalo M, Almeida D,
Dorasamy A, Jentsch U, et al: Direct susceptibility testing of
Mycobacterium tuberculosis for pyrazinamide by use of the bactec
MGIT 960 system. J Clin Microbiol. 54:1276–1281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramaswamy S and Musser JM: Molecular
genetic basis of antimicrobial agent resistance in Mycobacterium
tuberculosis 1998 update. Tuber Lung Dis. 79:3–29. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scarpellini P, Braglia S, Carrera P, Cedri
M, Cichero P, Colombo A, Crucianelli R, Gori A, Ferrari M and
Lazzarin A: Detection of rifampin resistance in Mycobacterium
tuberculosis by double gradient-denaturing gradient gel
electrophoresis. Antimicrob Agents Chemother. 43:2550–2254.
1999.PubMed/NCBI
|
|
11
|
Herrera L, Jiménez S, Valverde A,
García-Aranda MA and Sáez-Nieto JA: Molecular analysis of
rifampicin-resistant Mycobacterium tuberculosis isolated in Spain
(1996–2001). Description of new mutations in the rpoB gene and
review of the literature. Int J Antimicrob Agents. 21:403–408.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Der Zanden AG, TeKoppele-Vije EM,
Bhanu N Vijaya, Van Soolingen D and Schouls LM: Use of DNA extracts
from Ziehl-Neelsen-stained slides for molecular detection of
rifampin resistance and spoligotyping of Mycobacterium
tuberculosis. J Clin Microbiol. 41:1101–1118. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harding CV and Boom WH: Regulation of
antigen presentation by Mycobacterium tuberculosis A role for
Toll-like receptors. Nat Rev Microbiol. 8:296–307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kleinnijenhuis J, Oosting M, Joosten LA,
Netea MG and Van Crevel R: Innate immune recognition of
Mycobacterium tuberculosis. Clin Dev Immunol. 21:4053102011.
|
|
15
|
Bostanabad SZ, Nojoumi SA, Jabbarzadeh E,
Shekarabei M, Hoseinaei H, Rahimi MK, Ghalami M, Dizji S Pourazar,
Sagalchyk ER and Titov LP: High level isoniazid resistance
correlates with multiple mutation in the katG encoding catalase
proxidase of pulmonary tuberculosis strains from the frontier
localities of Iran. Tuberk Toraks. 59:27–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cade CE, Dlouhy AC, Medzihradszky KF,
Salas-Castillo SP and Ghiladi RA: Isoniazid-resistanee conferring
mutations in Mvcobacterium tuberculosis katG: Catalase, peroxidase,
and INH-NADH adduct formaion activities. Protein Sci. 19:458–474.
2010.PubMed/NCBI
|
|
17
|
Seifert M, Catanzaro D, Catanzaro A and
Rodwell TC: Genetic mutations associated with isoniazid resistance
in Mycobacterium tuberculosis: A systematic review. PLoS One.
10:e01196282015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khosravi AD, Goodarzi H and Alavi SM:
Detection of genomic mutations in katG, inhA and rpoB genes of
Mycobacterium tuberculosis isolates using polymerase chain reaction
and multiplex allele-specific polymerase chain reaction. Braz J
Infect Dis. 16:57–62. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin SY, Hwang SC, Yang YC, Wang CF, Chen
YH, Chen TC and Lu PL: Early detection of Mycobacterium
tuberculosis complex in BACTEC MGIT cultures using nucleic acid
amplification. Eur J Clin Microbiol Infect Dis. 35:977–984. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Al-Rubaye DS, Henihan G, Al-Abasly AK,
Seagar AL, Al-Attraqchi AA, Schulze H, Hashim DS, Kamil JK,
Laurenson IF and Bachmann TT: Genotypic assessment of
drug-resistant tuberculosis in Baghdad and other Iraqi provinces
using low-cost and low-density DNA microarrays. J Med Microbiol.
65:114–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ali IF, Babak F, Fazlollah MS and
Nematollah JJ: Rapid detection of MDR-Mycobacterium tuberculosis
using modified PCR-SSCP from clinical Specimens. Asian Pac J Trop
Biomed. 4 Suppl 1:S165–S170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Linger Y, Kukhtin A, Golova J, Perov A,
Lambarqui A, Bryant L, Rudy GB, Dionne K, Fisher SL, Parrish N and
Chandler DP: Simplified microarray system for simultaneously
detecting rifampin, isoniazid, ethambutol, and streptomycin
resistance markers in Mycobacterium tuberculosis. J Clin Microbiol.
52:2100–2107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi XC, Liu XQ, Xie XL, Xu YC and Zhao ZX:
Gene chip array for differentiation of mycobacterial species and
detection of drug resistance. Chin Med J (Engl). 125:3292–3297.
2012.PubMed/NCBI
|
|
24
|
Ohno H, Koga H and Kohno S:
Multidrug-resistant tuberculosis. 2. Mechanisms of drug-resistance
in Mycobacterium tuberculosis-genetic mechanisms of
drug-resistance. Kekkaku. 73:657–663. 1998.(In Japanese).
PubMed/NCBI
|
|
25
|
Wang SF, Zhao B, Song YY, Zhou Y, Ou XC,
Li Q, Xia H, Pang Y, Duanmu HJ, Fu Y and Zhao YL: Risk factors for
drug-resistant tuberculosis in China: analysis of the results of
the national drug resistant tuberculosisbaselinesurvey in 2007.
Chin J Antituber. 35:221–226. 2013.
|
|
26
|
van Rie A, Warren R, Mshanga I, Jordaan
AM, van der Spuy GD, Richardson M, Simpson J, Gie RP, Enarson DA,
Beyers N, et al: Analysis for a limited number of gene codons can
predict drug resistance of Mycobacterium tuberculosis in a
high-incidence community. J Clin Microbiol. 39:636–641. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Rossi E, Aínsa JA and Riccardi G: Role
of mycobacterial efflux transporters in drug resistance: An
unresolved question. FEMS Microbiol Rev. 30:36–52. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Danilchanka O, Mailaender C and Niederweis
M: Identification of a novel multidrug efflux pump of Mycobacterium
tuberculosis. Antimicrob Agents Chemother. 52:2503–2511. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Heep M, Brandstätter B, Rieger U, Lehn N,
Richter E, Rüsch-Gerdes S and Niemann S: Frequency of rpoB
mutations inside and outside the cluster I region in
rifampin-resistant clinical Mycobacterium tuberculosis strains. J
Clin Microbiol. 39:107–110. 2001. View Article : Google Scholar : PubMed/NCBI
|