Introduction
Intervertebral disc degeneration (IDD) is the main
cause of low back pain and ~20–40% of middle and old-aged patients
can suffer from nerve root pain to different degrees (1). The microRNA chip technique was used to
screen differentially expressed microRNAs in degenerative nucleus
pulposus tissues (2) and it was
found that the abnormal expression of microRNA-21 is positively
correlated with the degenerative Pfirrmann grade. The occurrence of
IDD is related to aging, cell dehydration, inflammatory response,
cell apoptosis and autophagy (3).
Herniation of the nucleus pulposus activates the body's immune
system, leading to leukocyte activation, release of a variety of
inflammatory factors and nerve cell degeneration, resulting in
neuropathic pain (4). Rat models
have confirmed (5) the prolonged
inflammatory response with significant increase of IL-6 and TNF-α.
Decreased contents of type II collagen (Col II) and aggrecan are
typical pathological changes of IDD, and autophagy is an important
way to regulate the extracellular matrix (ECM) metabolism of the
intervertebral disc (6). Studies
(7) have confirmed that the
autophagy-related gene 7 (ATG7) is closely related to the formation
of autophagic vacuoles, and it is also the target gene of
microRNA-21. Based on this, our study further analyzed the
degenerative nucleus pulposus tissue obtained in vivo to
investigate the mechanism of microRNA-21 expression of regulating
IL-6 inflammatory response and cell autophagy so as to provide new
treatment ideas.
Materials and methods
Patient data
A total of 10 patients diagnosed with lumbar disc
herniation accompanied by nerve root pain from May to October 2016
in our hospital (observation group) and 10 patients with lumbar
burst fractures (control group) were continuously selected. In the
observation group, there were 6 males and 4 females aged 52–67 with
an average age of 56.9±7.2 years. In terms of sampling site, there
were 2 cases of L2-3, 1 case of L3-4, 4 cases of L4-5 and 3 cases
of L5-S1. In the control group, there were 5 males and 5 females
aged 48–68 with an average age of 55.7±6.9 years. In terms of
sampling site, there were 3 cases of L2-3, 2 cases of L3-4, 3 cases
of L4-5 and 2 cases of L5-S1. Baseline data of the two groups was
comparable. This study was approved by the Ethics Committee of
Clinical College of Maanshan, and signed written informed consents
were obtained from the patients.
Research methods
The nucleus pulposus tissues of the lesions were
obtained for cell culture during the operation. Real-time
quantitative polymerase chain reaction (PCR) was used to detect the
expression of microRNA-21. ELISA method was used to detect the
levels of IL-6, Col II and aggrecan and western blotting was used
to detect ATG7 and LC3-II/−I.
Cell culture and identification
The tissue samples were washed with PBS and placed
in a DMEM/F12 (1:1) (both from Biosharp, Hefei, China) sterile
culture flask containing 10% fetal bovine serum and stored at low
temperature. The residual components in nucleus pulposus tissues
were removed using ophthalmic forceps on a sterile operation desk,
and nucleus pulposus tissues were washed using 1% double resistant
flushing fluid, and about 1 mm3 tissue was cut into
pieces. A total of 1.5-time-volume of 0.25% trypsin (Biosharp) was
added into the centrifuge tube for digestion for 30 min, followed
by centrifugation at 2,000 × g for 5 min; isopyknic 0.2% Col II was
added into the sediment and mixed for about 4 h and then isopyknic
DMEM/F12 complete culture solution was added to terminate the
digestion, followed by screening via cell strainer and
centrifugation at 1,000 × g for 5 min; 4 ml DMEM/F12 (1:1) complete
culture solution containing 15% fetal calf serum was added to blow
away cells; re-suspension density was 1×105/ml; and
cells were incubated in an incubator containing 5% CO2
at 37°C. The solution was changed the next day for subculture. The
cell adherence, growth and morphological changes were observed
under inverted microscope (BX-42; Olympus, Tokyo, Japan). The
second generation of cells was taken for immunocytochemical
staining to identify Col II expression, confirmed to be nucleus
pulposus cells.
Real-time quantitative PCR
The total RNA in cells was extracted using the
conventional TRIzol reagent, and its concentration and purity were
determined by ultraviolet spectrophotometer. cDNA was synthesized
using a reverse transcription kit, and primer sequences were
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) according
to GenBank sequence. MicroRNA-21: forward,
5-GGTTTCATCCAGGATCGAGCAGG-3 and reverse,
5-ACAAAGATGGTCACGGTCTGCC-3, 445 bp; GAPDH forward,
5-CGCGAGAAGATGACCCAGAT-3 and reverse, 5-GCACTGTGTTGGCGTACAGG-3, 225
bp. Reaction system: 2 µl cDNA + 3 µl upper primer and 3 µl lower
primer + 0.5 µl Taq polymerase + 1 µl dNTPs + 3 µl MgCl2
+ 5 µl 10X buffer, and water was added until the total volume was
20 µl. Reaction conditions: 95°C for 5 min, 95°C for 30 sec, 58°C
for 30 sec, 72°C for 60 sec, a total of 30 cycles, ending at 72°C
for 10 min. PCR products were identified via 2% agarose gel
electrophoresis, followed by imaging via gel imaging analysis
system and gray value analysis via digital photograph. The results
are presented using 2−ΔΔCq method.
ELISA method
IL-6, Col II and aggrecan reagents were purchased
from Jiangsu Beyotime Technology Co. Ltd., (Jiangsu, China); cell
sap was detected three times via a microplate reader after being
centrifuged at 3,000 × g for 20 min, and the average was taken.
Western blotting
RIPA lysate was added to extract the total cell
protein, followed by rough quantification via Coomassie brilliant
blue method and dose standardization via β-actin antibody. A total
of 30 µg total protein was taken and separated via 8% SDS-PAGE, and
the separated zone was transferred to a PVDF membrane and mouse
anti-human ATG7, LC3-II and LC3-I monoclonal antibodies (1:2000;
Sigma, St. Louis, MO, USA) was added overnight; then rabbit
anti-mouse polyclonal secondary antibody (1:500; Sigma) was added
to incubate at room temperature for 4 h, followed by washing via
PBS and development via ECL. Results were scanned and saved, and
semi-quantitative analysis was performed using Lab Works 4.5 gel
imaging software (Invitrogen, Carlsbad, CA, USA).
Statistical analysis
SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis; measurement data was presented
as mean ± standard deviation, and independent sample t-test was
used for intergroup comparison; enumeration data were presented as
case or percentage (%), and Chi-square test was used for intergroup
comparison. P<0.05 indicates that the difference was
statistically significant.
Results
Identification of nucleus pulposus
cells
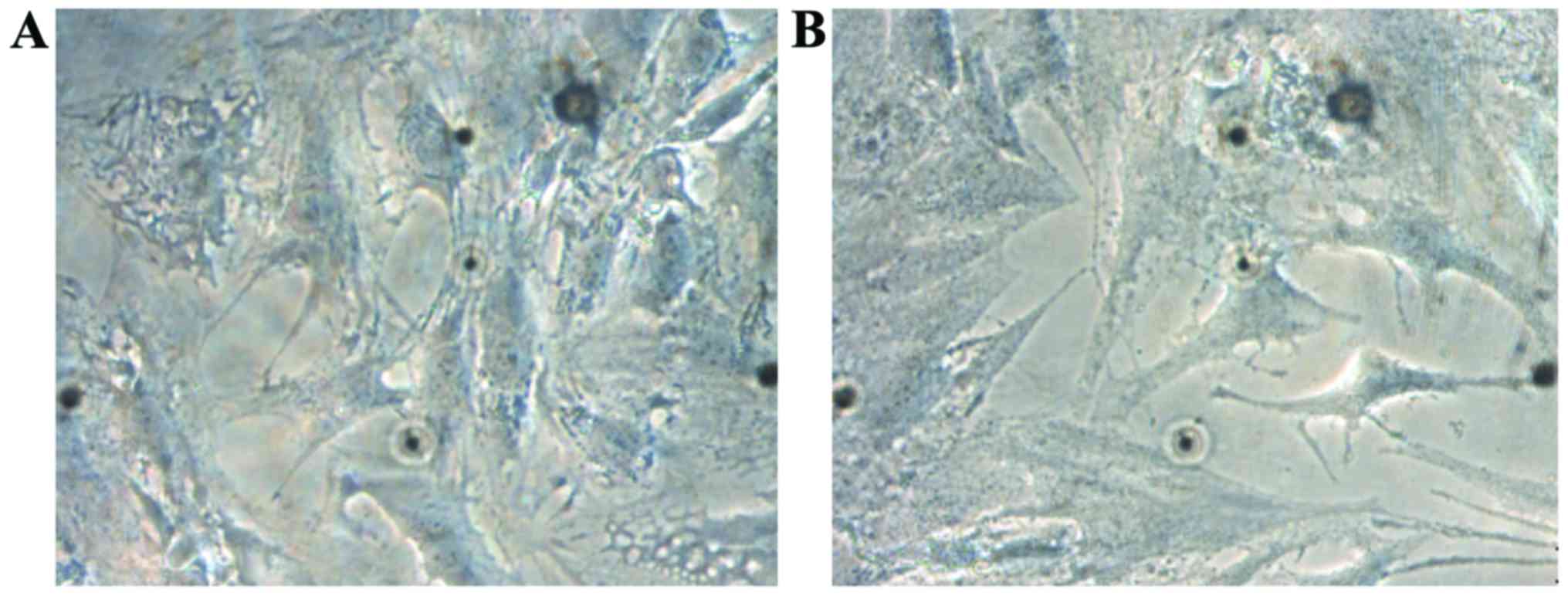
The second generation of cells was observed under
the inverted phase contrast microscope; in both groups, cells were
mainly short fusiform with good refractivity, and short and thick
protrusions spread all around in arborescent type, and there were
secondary protrusions. The number of cells in the observation group
was decreased and that of protrusions was increased (Fig. 1).
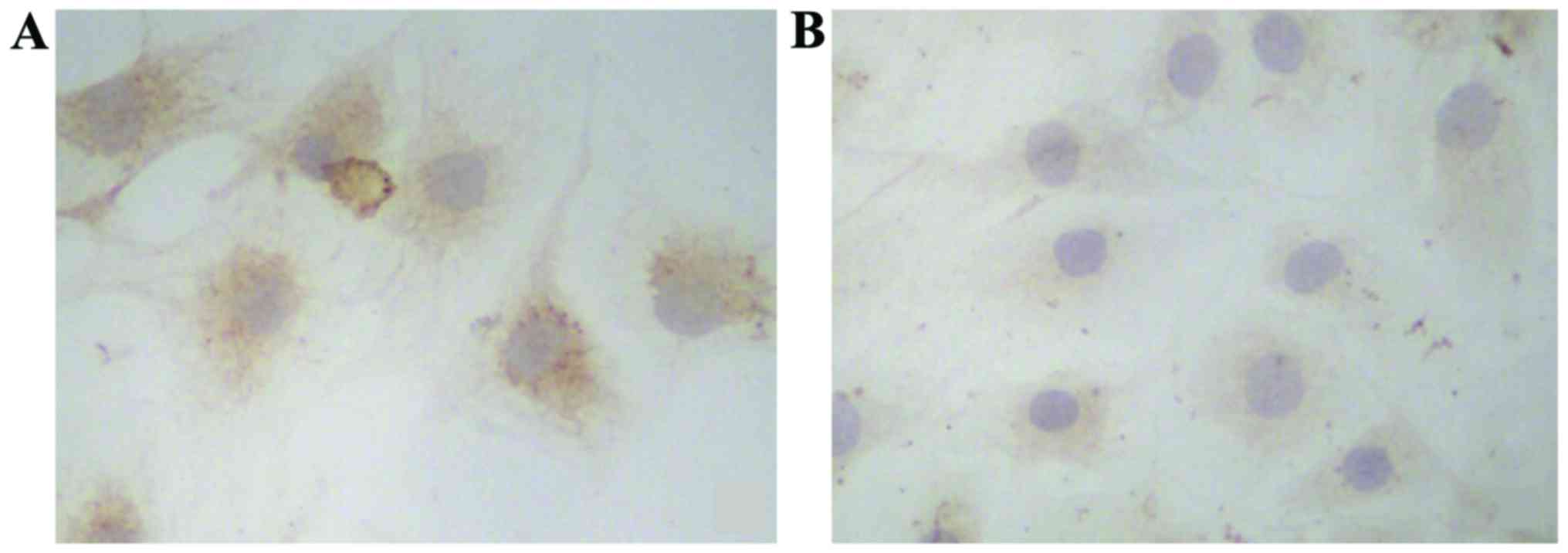
Immunostaining showed that the cytoplasm of the
control group was stained brown yellow and that the control group
was stained pale yellow (Fig.
2).
Results of real-time quantitative
PCR
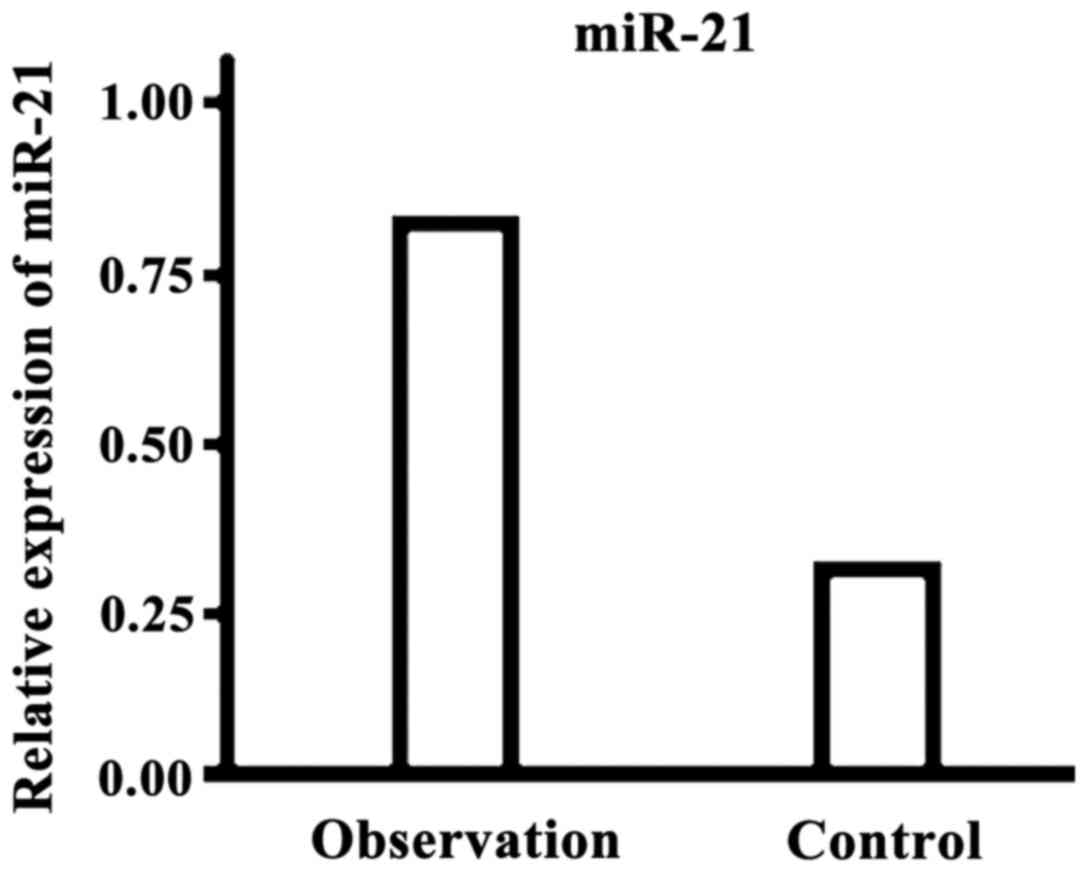
The level of microRNA-21 in the observation group
was significantly higher than that in the control group and the
difference was statistically significant (P<0.05, Fig. 3).
Results of ELISA method
The level of IL-6 in the observation group was
significantly higher than that of the control group, but the levels
of Col II and aggrecan were significantly lower than those in the
control group; the differences were statistically significant
(P<0.05, Table I).
 | Table I.Results of ELISA method (µmol/l). |
Table I.
Results of ELISA method (µmol/l).
| Group | IL-6 | Col II | Aggrecan |
|---|
| Observation
group | 125.6±34.9 | 223.1±65.9 | 125.8±54.2 |
| Control group | 64.7±22.5 | 352.8±82.7 | 264.7±72.9 |
| t-test | 12.635 | 10.234 | 15.285 |
| P-value | <0.001 | <0.001 | <0.001 |
Results of western blotting
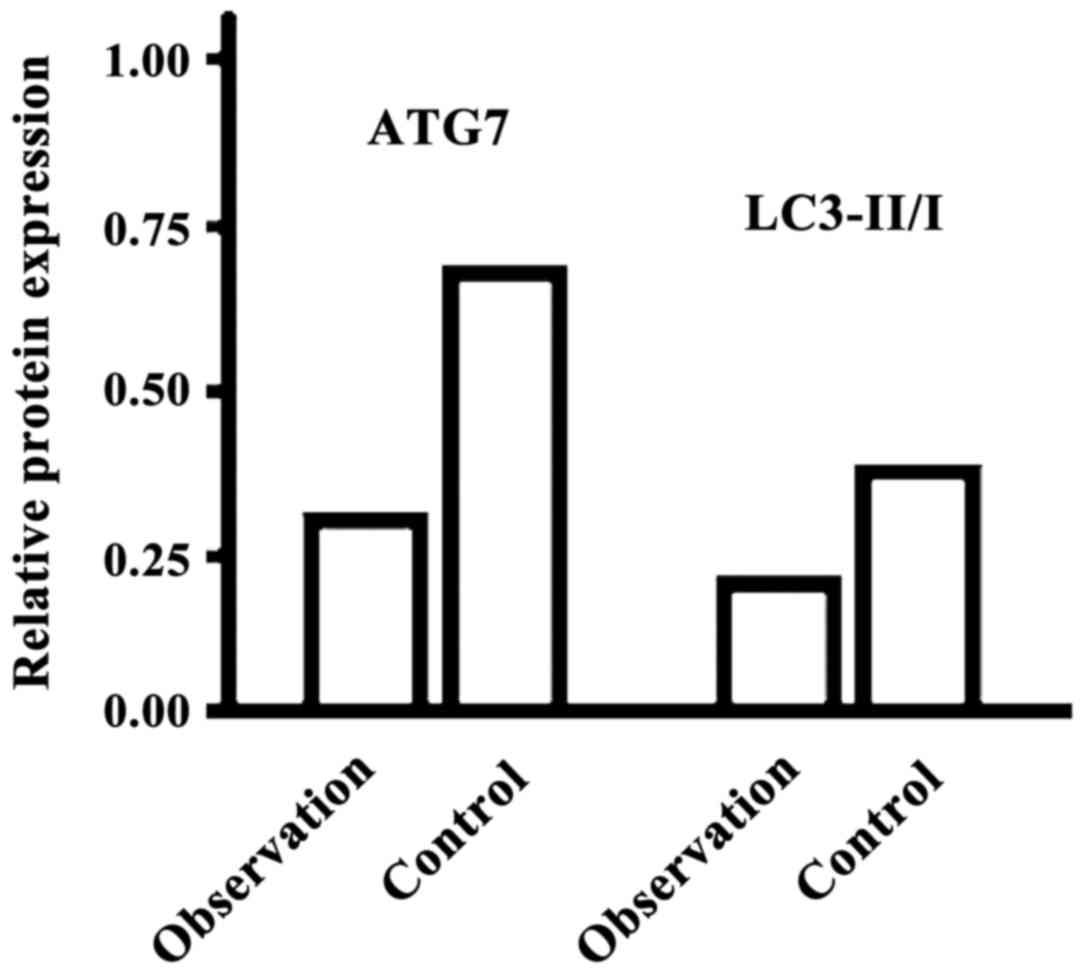
The levels of ATG7 and LC3-II/−I in the observation
group were significantly lower than those of the control group, and
the differences were statistically significant (P<0.05, Fig. 4).
Discussion
It is agreed that IDD is a physiological and
pathological process related to ECM synthesis and catabolic
imbalance, cell apoptosis, inflammatory response and vascular
proliferation. Some biological treatments, such as active
substances of intervertebral disc regeneration, stem cell
transplantation, autologous chondrocyte transplantation and gene
therapy can promote intervertebral disc regeneration or repair in
different degrees and reverse the degeneration process, but most of
these results are in animal experiments or in vitro animal
model stages (8,9).
This study showed that the levels of microRNA-21 and
IL-6 in the observation group were significantly higher than those
of the control group, but the levels of Col II and aggrecan were
significantly lower than those in the control group, and the levels
of ATG7 and LC3-II/−I in cells were significantly decreased,
suggesting that the expression of microRNA-21 is abnormally high in
lumbar intervertebral disc nerve root pain with increased IL-6
inflammatory response and reduced cell autophagy. MicroRNAs are
endogenous non-coding RNAs widely distributed in the human body and
mainly bind to the 3′ untranslated region (3′UTR) in target mRNA,
inhibiting the translation process or increasing the target mRNA
degradation, which affects the expressions of target genes and 30%
genome coding protein, which plays an important role in a variety
of pathological and physiological processes. Liu et al
(10) confirmed that miR-21
expression is significantly increased in the degenerative nucleus
pulposus tissues and miR-21-transfected human nucleus pulposus
cells can stimulate cell proliferation. Studies on overexpression
of miR-21 or silent miR-21 expression showed that the
overexpression of miR-21 can aggravate cell degeneration, increase
the expression of MMP and ECM degradation, upregulate the
inflammatory responses, such as IL-6, and promote cell apoptosis
and autophagy (11). Therefore, it
is thought that miR-21 expression is closely related to the
occurrence of IDD.
This study found that (12) IL-6 and TNF-α inflammatory factors in
intervertebral disc tissue play important roles in the occurrence
of IDD damage and neuropathic pain. They can be significantly
increased at an early stage. The injection of recombinant IL-6 in
dorsal root ganglion can cause hyperalgesia, and IL-6 inhibitors
can significantly reduce the pain of rats in a chronic compression
model, and the application of IL-6 gene silencing treatment can
significantly reduce the mechanical hyperalgesia in rats in a
spinal nerve abruption model (13).
At the same time, anti-inflammatory factors, such as IL-10, have
significant analgesic effects (14).
Jiang et al (15) pointed out
that the number of autophagosomes in human degenerative nucleus
pulposus cells was significantly reduced, and LC3-II/−I and
beclin-1 expression levels were decreased. Autophagy inhibitor
3-methyladenine can significantly reduce the number of autophagic
vacuole (16). Wang et al
(17) pointed out that resveratrol
can increase the capacity of cell autophagy, and inhibit
TNF-α-induced MMP-3 expression. ATG7 is an important target of
autophagy. ATG7 mRNA 3′UTR has complementary sequences with miR-21.
A dual-luciferase reporter gene test showed that miR-210 mimic
significantly reduces the activity of wild-type ATG, but has no
effect on mutant type, confirming that ATG7 is the target gene of
miR-21 (10).
The innovation of this study is that it obtained the
degenerative nucleus pulposus tissue from the human body and
established a cell culture method, providing an important basis for
follow-up studies. The deficiency of this study is that it failed
to further analyze the mechanism of miR-21 expression for
IL-6-mediated inflammatory response and cell autophagy, and whether
miR-21 is a potential target for intervention treatments remains to
be verified.
Acknowledgements
The authors would like to acknowledge the assistance
of Dr Yvonne Opalinski in the preparation of this article.
References
|
1
|
Wang HQ and Samartzis D: Clarifying the
nomenclature of intervertebral disc degeneration and displacement:
From bench to bedside. Int J Clin Exp Pathol. 7:1293–1298.
2014.PubMed/NCBI
|
|
2
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50.
2014.PubMed/NCBI
|
|
3
|
Deng X, Zhao F, Kang B and Zhang X:
Elevated interleukin-6 expression levels are associated with
intervertebral disc degeneration. Exp Ther Med. 11:1425–1432.
2016.PubMed/NCBI
|
|
4
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson ZI, Schoepflin ZR, Choi H, Shapiro
IM and Risbud MV: Disc in flames: Roles of TNF-α and IL-1β in
intervertebral disc degeneration. Eur Cell Mater. 30:104–116;
discussion 116–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu K, Chen W, Wang X, Peng Y, Liang A,
Huang D, Li C and Ye W: Autophagy attenuates the catabolic effect
during inflammatory conditions in nucleus pulposus cells, as
sustained by NF-κB and JNK inhibition. Int J Mol Med. 36:661–668.
2015.PubMed/NCBI
|
|
7
|
Yang Y and Liang C: MicroRNAs: An emerging
player in autophagy. ScienceOpen Res. 2015:102015.
|
|
8
|
Daly C, Ghosh P, Jenkin G, Oehme D and
Goldschlager T: A Review of animal models of intervertebral disc
degeneration: Pathophysiology, regeneration, and translation to the
clinic. BioMed Res Int. 2016:59521652016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bian Z and Sun J: Development of a KLD-12
polypeptide/TGF-β1-tissue scaffold promoting the differentiation of
mesenchymal stem cell into nucleus pulposus-like cells for
treatment of intervertebral disc degeneration. Int J Clin Exp
Pathol. 8:1093–1103. 2015.PubMed/NCBI
|
|
10
|
Liu H, Huang X, Liu X, Xiao S, Zhang Y,
Xiang T, Shen X, Wang G and Sheng B: miR-21 promotes human nucleus
pulposus cell proliferation through PTEN/AKT signaling. Int J Mol
Sci. 15:4007–4018. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen B, Huang SG, Ju L, Li M, Nie FF,
Zhang Y, Zhang YH, Chen X and Gao F: Effect of microRNA-21 on the
proliferation of human degenerated nucleus pulposus by targeting
programmed cell death 4. Braz J Med Biol Res. 49:e50202016.
View Article : Google Scholar
|
|
12
|
Molinos M, Almeida CR, Caldeira J, Cunha
C, Gonçalves RM and Barbosa MA: Inflammation in intervertebral disc
degeneration and regeneration. J R Soc Interface. 12:201411912015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weber KT, Alipui DO, Sison CP, Bloom O,
Quraishi S, Overby MC, Levine M and Chahine NO: Serum levels of the
proinflammatory cytokine interleukin-6 vary based on diagnoses in
individuals with lumbar intervertebral disc diseases. Arthritis Res
Ther. 18:32016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li W, Liu T, Wu L, Chen C, Jia Z, Bai X
and Ruan D: Blocking the function of inflammatory cytokines and
mediators by using IL-10 and TGF-β: A potential biological
immunotherapy for intervertebral disc degeneration in a beagle
model. Int J Mol Sci. 15:17270–17283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang W, Zhang X, Hao J, Shen J, Fang J,
Dong W, Wang D, Zhang X, Shui W, Luo Y, et al: SIRT1 protects
against apoptosis by promoting autophagy in degenerative human disc
nucleus pulposus cells. Sci Rep. 4:74562014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen D, Xia D, Pan Z, Xu D, Zhou Y, Wu Y,
Cai N, Tang Q, Wang C, Yan M, et al: Metformin protects against
apoptosis and senescence in nucleus pulposus cells and ameliorates
disc degeneration in vivo. Cell Death Dis. 7:e24412016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang XH, Zhu L, Hong X, Wang YT, Wang F,
Bao JP, Xie XH, Liu L and Wu XT: Resveratrol attenuated
TNF-α-induced MMP-3 expression in human nucleus pulposus cells by
activating autophagy via AMPK/SIRT1 signaling pathway. Exp Biol Med
(Maywood). 241:848–853. 2016. View Article : Google Scholar : PubMed/NCBI
|