Introduction
Endometriosis is a benign gynecological disorder
that occurs in 10% of women of reproductive age (1). The main symptoms include infertility
and chronic pelvic pain (2).
Although there are a number of studies on endometriosis, the
majority of the mechanisms are not well understood (3–6).
Identifying disease biomarkers and their interaction networks is
important to improve the understanding of the causes of
endometriosis, as well as to improve medical care.
Several databases have been developed that store
associations between genes and diseases, such as the Online
Mendelian Inheritance in Man (7),
the Human Gene Mutation Database (8)
and the Genetic Association Database (9). Due to the nature of the database
curation process, the data are incomplete. Some gene-disease
databases that combine gene-associated diseases from several
expert, public and curated data sources also exist (10,11).
With the rapid accumulation of gene-disease data, increasing
research has been utilizing the gene-disease database as a
start-point to mine disease biomarkers (12–14).
Protein-protein interaction (PPI) networks include
information on the biological processes and molecular functions of
cells and have been widely used to characterize the underlying
mechanisms of genes associated with complex diseases (15,16). The
majority of human diseases are caused by a group of correlated
molecules or a network, rather than a single gene (17). Thus, identification and validation of
biomarker networks is critical to disease diagnosis, prognosis and
treatment.
In the present study, a disease network of
endometriosis that integrated human PPIs and known disease-causing
genes was constructed. Endometriosis-causing genes were identified
from gene-disease databases. Subsequently, bioinformatics
approaches, including PPI network construction, module analysis,
functional enrichment analysis and text mining, were utilized in
the research. The results of the present study may provide new
targets for endometriosis therapy and identify the potential
mechanisms of the disease.
Materials and methods
Seed gene selection
Endometriosis-related genes were obtained from
Genotator (http://genotator.hms.harvard.edu/) (10) and DisGeNET (http://www.disgenet.org) (11). For each tool, gene lists were
extracted using the query term, endometriosis. Genotator provides
high quality gene-disease associations based upon data from 11
trustworthy resources. DisGeNET is a discovery platform that
integrates information on gene-disease associations from several
public data sources and literature (11). Thus, a list of genes that had been
experimentally validated to be associated with endometriosis were
obtained.
Disease-gene network construction
Endometriosis-associated genes were submitted to
atBioNet (https://www.fda.gov/ScienceResearch/BioinformaticsTools/ucm285284.htm)
and PPIs were obtained. atBioNet is a network analysis tool that
provides a systematic insight into gene interactions by examining
significant functional modules (18). The default option is ‘Human Database’
that combines data from a variety of public PPI sources, including
BioGRID (19), the Database of
Interacting Proteins (20), the
Human Protein Reference Database (21), IntAct (22), the Molecular INTeraction database
(23), REACTOME (24) and the Signaling Pathways Integrated
Knowledge Engine (25). The protein
interaction network included 12,043 human proteins and 132,605
interactions. SCAN algorithm was used to identify functional
modules and perform assessment of generated gene networks for
biomarker discovery (26).
Pathway enrichment analysis
To identify potential roles of genes in
endometriosis, the Kyoto Encyclopedia of Genes and Genomes (KEGG)
(27) pathway analysis component in
atBioNet was used. Overrepresented KEGG pathways for each module
were ranked according to the P-value obtained from Fisher's exact
tests.
Literature mining
To identify the genes associated with endometriosis,
mining from the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed) with keywords
‘gene symbol’ and ‘endometriosis’ was conducted. Subsequently, the
articles associated with endometriosis were screened manually. A
high number of papers indicated that the relationship between
potential biomarker genes and endometriosis is well studied and
documented.
Results
Screening of seed genes related to
endometriosis
A total of 271 and 229 genes were extracted from
Genotator and DisGeNET, respectively. The common genes, of which
there were 100, were used as seed genes to generate functional
modules.
Construction of biomarker
networks
Of 100 input genes, 96 were found in GenBank
(https://www.ncbi.nlm.nih.gov/genbank/), and network
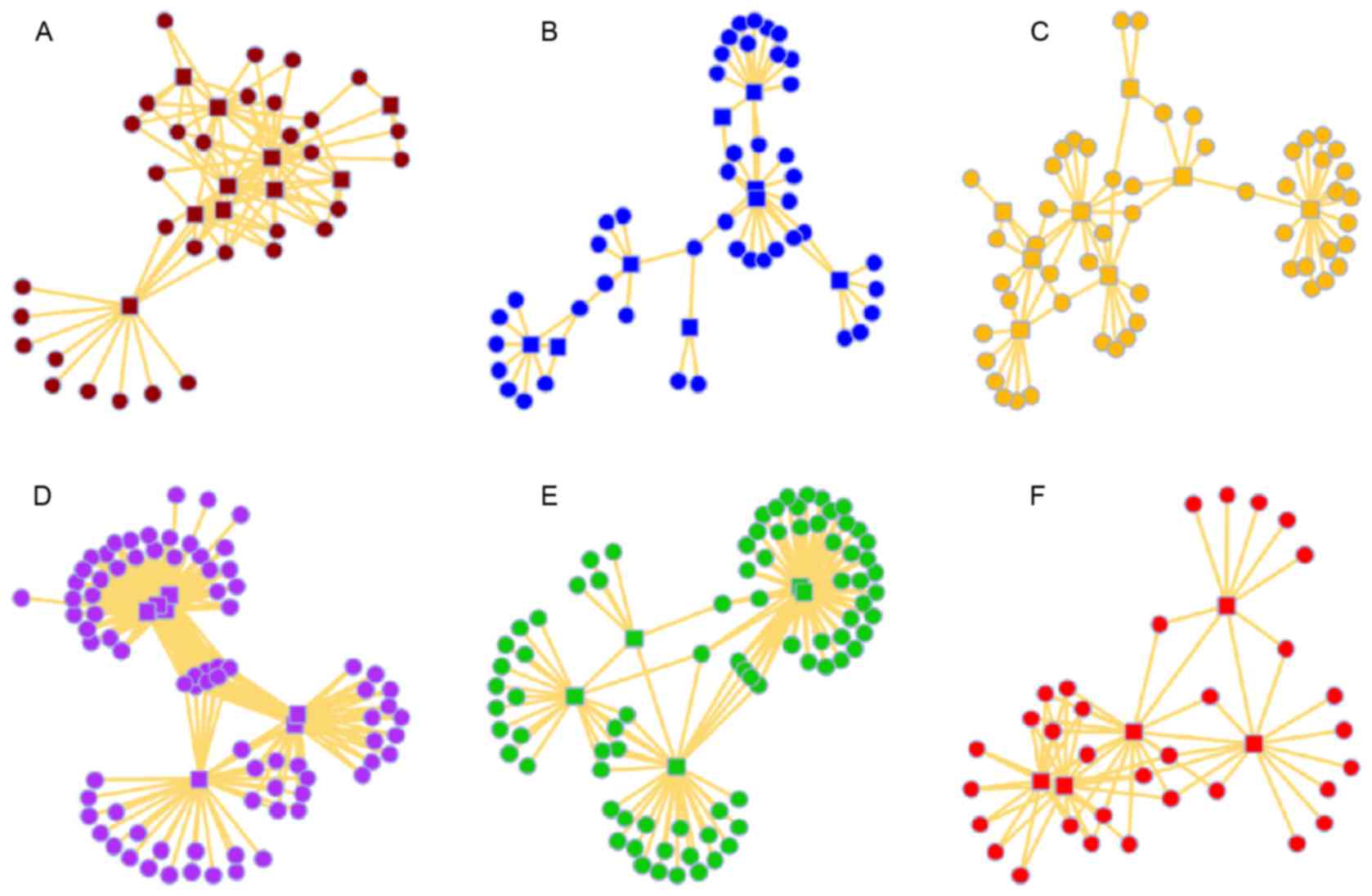
clustering identified six major sub network modules from the
original PPI network (Fig. 1). Hub
genes in each module were identified (Table I).
 | Table I.Hub genes in each module. |
Table I.
Hub genes in each module.
| Module | Gene ID | Gene symbol |
|---|
| A |
196 | AHR |
| A |
367 | AR |
| A |
405 | ARNT |
| A | 2099 | ESR1 |
| A | 8204 | NRIP1 |
| A | 2100 | ESR2 |
| A | 7157 | TP53 |
| A | 2516 | NR5A1 |
| A | 2908 | NR3C1 |
| A | 5241 | PGR |
| B | 3557 | IL1RN |
| B | 3552 | IL1A |
| B | 3554 | IL1R1 |
| B | 3553 | IL1B |
| B | 3560 | IL2RB |
| B | 3600 | IL15 |
| B | 3565 | IL4 |
| B | 3586 | IL10 |
| B | 3606 | IL18 |
| C | 4316 | MMP7 |
| C | 3479 | IGF1 |
| C | 3484 | IGFBP1 |
| C | 4312 | MMP1 |
| C | 5069 | PAPPA |
| C | 7077 | TIMP2 |
| C | 4322 | MMP13 |
| C | 4321 | MMP12 |
| D | 3106 | HLA-B |
| D | 3105 | HLA-A |
| D | 3107 | HLA-C |
| D | 3115 | HLA-DPB1 |
| D | 3117 | HLA-DQA1 |
| D | 3119 | HLA-DQB1 |
| D | 3123 | HLA-DRB1 |
| E |
328 | APEX1 |
| E | 4968 | OGG1 |
| E | 7515 | XRCC1 |
| E | 2068 | ERCC2 |
| E | 2073 | ERCC5 |
| F |
355 | FAS |
| F |
356 | FASLG |
| F | 7132 | TNFRSF1A |
| F | 7124 | TNF |
| F | 4049 | LTA |
KEGG pathway analysis
A total of 2,429 genes from the KEGG human database
were added to the PPI network and genes in each module were
selected for pathway enrichment analysis. The top 10 significantly
enriched KEGG pathways for the six modules in endometriosis are
demonstrated in Table II. Module A
was a cancer cell proliferation module. The majority of the
pathways in the first module were related to the proliferation of
cancer cells and were associated with pathways in cancer, the cell
cycle, oocyte meiosis, adherens junctions and the Wnt signaling
pathway. The enriched pathways in module B were associated with the
immune system and infectious diseases, including cytokine-cytokine
receptor interaction, the mitogen-activated protein kinase
signaling pathway, the Janus kinase-signal transducer and activator
of transcription (JAK-STAT) signaling pathway, the intestinal
immune network for immunoglobulin (Ig) A production and Toll-like
receptor signaling pathways. Module C was associated with
complement and coagulation cascades, extracellular matrix-receptor
interaction, focal adhesion, and proteasome and hematopoietic cell
lineages associated with immune and metastasis. The enriched
pathways in module D were associated with inflammatory responses,
including phagosome, cell adhesion molecules, antigen processing
and presentation, natural killer cell mediated cytotoxicity, T cell
receptor signaling pathways and the intestinal immune network for
IgA production. The majority of the pathways in module E were
related to processes of replication and repair, including DNA
replication, base excision repair, nucleotide excision repair,
mismatch repair and homologous recombination.
 | Table II.Top 10 KEGG pathways ranked by
P-value for the top six modules in endometriosis. |
Table II.
Top 10 KEGG pathways ranked by
P-value for the top six modules in endometriosis.
| Functional modules
(no. of genes) | Map title in
KEGG | No. of genes mapped
in the pathway |
P-valuea |
|---|
| Module A
(n=42) | Pathways in cancer
(hsa05200) | 7 | <0.0001 |
|
| Thyroid cancer
(hsa05216) | 3 | <0.0001 |
|
| Prostate cancer
(hsa05215) | 4 | 0.0001 |
|
| Oocyte meiosis
(hsa04114) | 4 | 0.0003 |
|
| Neurotrophin
signaling pathway (hsa04722) | 4 | 0.0005 |
|
| Cell cycle
(hsa04110) | 4 | 0.0005 |
|
| Basal transcription
factors (hsa03022) | 3 | 0.0005 |
|
| Colorectal cancer
(hsa05210) | 3 | 0.0008 |
|
| Wnt signaling
pathway (hsa04310) | 4 | 0.0009 |
|
| Renal cell
carcinoma (hsa05211) | 3 | 0.0011 |
| Module B
(n=54) | Cytokine-cytokine
receptor interaction (hsa04060) | 19 | <0.0001 |
|
| Apoptosis
(hsa04210) | 11 | <0.0001 |
|
| JAK-STAT signaling
pathway (hsa04630) | 15 | <0.0001 |
|
| Pertussis
(hsa05133) | 9 | <0.0001 |
|
| Measles
(hsa05162) | 11 | <0.0001 |
|
| Tuberculosis
(hsa05152) | 11 | <0.0001 |
|
| Toxoplasmosis
(hsa05145) | 9 | <0.0001 |
|
| Leishmaniasis
(hsa05140) | 7 | <0.0001 |
|
| Intestinal immune
network for IgA production (hsa04672) | 6 | <0.0001 |
|
| Toll-like receptor
signaling pathway (hsa04620) | 7 | <0.0001 |
| Module C
(n=60) | Complement and
coagulation cascades (hsa04610) | 6 | <0.0001 |
|
| Hypertrophic
cardiomyopathy (hsa05410) | 4 | 0.0009 |
|
| ECM-receptor
interaction (hsa04512) | 4 | 0.0010 |
|
| Dilated
cardiomyopathy (hsa05414) | 4 | 0.0012 |
|
| Hematopoietic cell
lineage (hsa04640) | 3 | 0.0111 |
|
| Focal adhesion
(hsa04510) | 4 | 0.0208 |
|
| Proteasome
(hsa03050) | 2 | 0.0237 |
|
| Pathways in cancer
(hsa05200) | 5 | 0.0280 |
|
| Vitamin B6
metabolism (hsa00750) | 1 | 0.0316 |
|
| Staphylococcus
aureus infection (hsa05150) | 2 | 0.0355 |
| Module D
(n=87) | Phagosome
(hsa04145) | 17 | <0.0001 |
|
| Cell adhesion
molecules (hsa04514) | 24 | <0.0001 |
|
| Antigen processing
and presentation (hsa04612) | 24 | <0.0001 |
|
| Natural killer cell
mediated cytotoxicity (hsa04650) | 13 | <0.0001 |
|
| T cell receptor
signaling pathway (hsa04660) | 18 | <0.0001 |
|
| Intestinal immune
network for IgA production (hsa04672) | 13 | <0.0001 |
|
| Type I diabetes
mellitus (hsa04940) | 17 | <0.0001 |
|
| Leishmaniasis
(hsa05140) | 14 | <0.0001 |
|
| Toxoplasmosis
(hsa05145) | 13 | <0.0001 |
|
| Staphylococcus
aureus infection (hsa05150) | 13 | <0.0001 |
| Module E
(n=87) | Purine metabolism
(hsa00230) | 19 | <0.0001 |
|
| Pyrimidine
metabolism (hsa00240) | 19 | <0.0001 |
|
| RNA polymerase
(hsa03020) | 12 | <0.0001 |
|
| DNA replication
(hsa03030) | 17 | <0.0001 |
|
| Base excision
repair (hsa03410) | 22 | <0.0001 |
|
| Nucleotide excision
repair (hsa03420) | 31 | <0.0001 |
|
| Mismatch repair
(hsa03430) | 14 | <0.0001 |
|
| Homologous
recombination (hsa03440) | 7 | <0.0001 |
|
| Huntington's
disease (hsa05016) | 13 | <0.0001 |
|
| Basal transcription
factors (hsa03022) | 6 | <0.0001 |
| Module F
(n=38) | Cytokine-cytokine
receptor interaction (hsa04060) | 12 | <0.0001 |
|
| Apoptosis
(hsa04210) | 14 | <0.0001 |
|
| RIG-I-like receptor
signaling pathway (hsa04622) | 7 | <0.0001 |
|
| Tuberculosis
(hsa05152) | 8 | <0.0001 |
|
| Pathways in cancer
(hsa05200) | 10 | <0.0001 |
|
| Natural killer cell
mediated cytotoxicity (hsa04650) | 7 | <0.0001 |
|
| Chagas disease
(American trypanosomiasis; hsa05142) | 6 | <0.0001 |
|
| Alzheimer's disease
(hsa05010) | 7 | <0.0001 |
|
| Osteoclast
differentiation (hsa04380) | 6 | <0.0001 |
|
| Type I diabetes
mellitus (hsa04940) | 4 | <0.0001 |
Endometriosis-associated genes
identified in literature
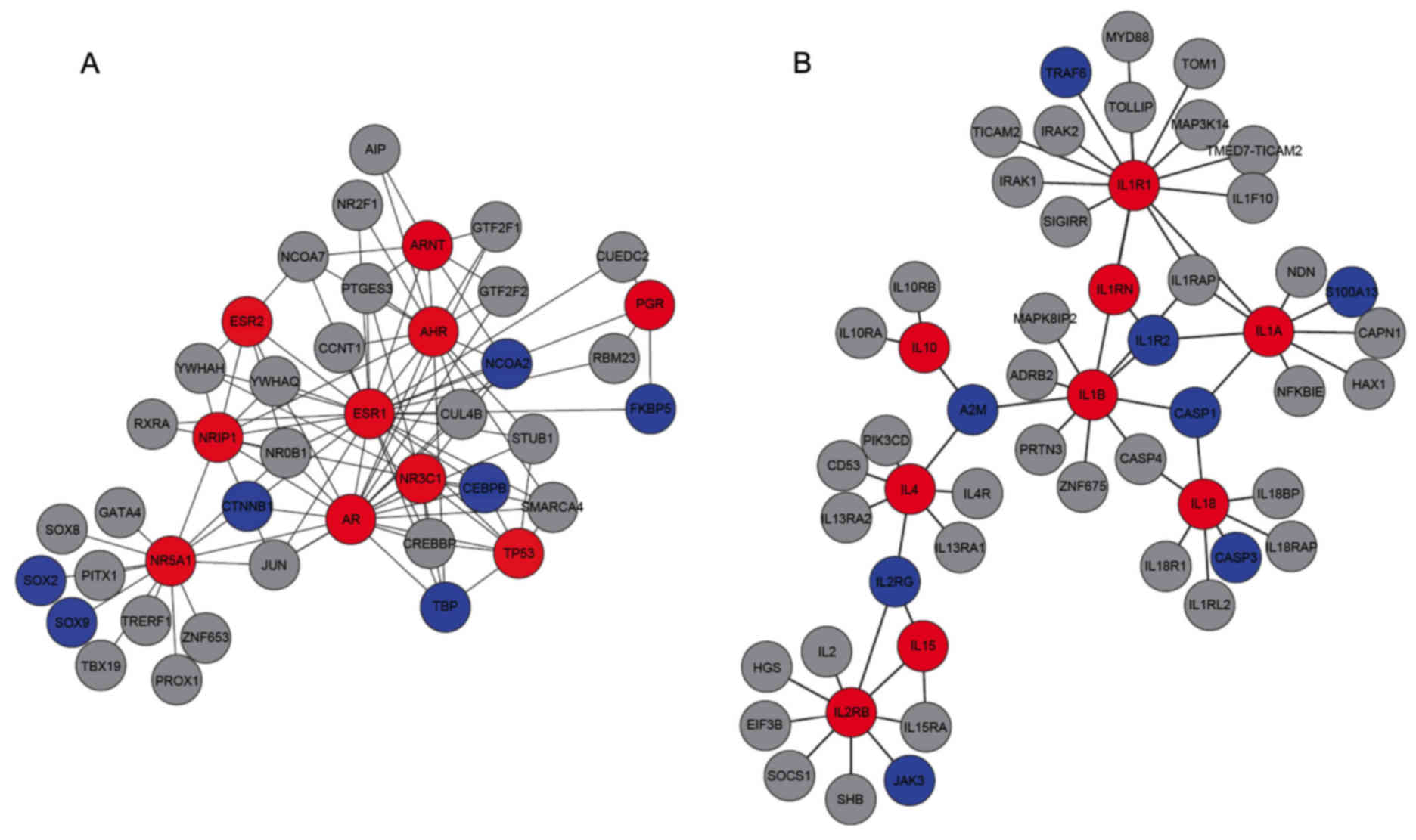
A total of 15 genes, seven in the first module and
eight in the second module, have previously been reported in
literature to be candidate biomarkers for endometriosis (Fig. 2). For example, women with
endometriosis had significantly higher SOX2 expression levels
compared to controls (Fig. 2A)
(28). Also, various genes
identified in the second module (Fig.
2B), including CASP3, S100A13 and IL1R2, have been reported to
be associated with endometriosis (29–31).
Details for the 15 literature-confirmed potential endometriosis
biomarkers are listed in Table
III.
 | Table III.Details of the 15 potential
endometriosis biomarkers in modules A and B demonstrated in
literature. |
Table III.
Details of the 15 potential
endometriosis biomarkers in modules A and B demonstrated in
literature.
| Gene ID | Gene symbol | Module | PMID | Description |
|---|
| 1051 | CEBPB | A | 23097472 | A novel functional
link between C/EBPβ and STAT3 that is a critical regulator of
endometrial differentiation in women. |
| 1499 | CTNNB1 | A | 23765252 | CTNNB1 mutations
are significantly different in low-grade ovarian endometrioid
carcinomas (53%) compared with low-grade endometrial endometrioid
carcinomas (28%; P<0.0057). |
| 2289 | FKBP5 | A | 22279148 | No significant
endometriosis-related change was observed for FKBP5. |
| 6908 | TBP | A | 18252806 | TBP inhibits the
TNF-α-induced expression of endome triotic genes in 12Z
endometriotic epithelial cells. |
| 10499 | NCOA2 | A | 12050280 | Abnormal increases
in endometrial TIF2 and SRC-3 levels are also associated with
infertility in women with polycystic ovary syndrome. |
| 6657 | SOX2 | A | 23670619 | Samples from
endometriosis patients had higher mRNA expression levels of Oct-4,
CXCR4, SOX2 and MET compared with that of the normal controls. |
| 6662 | SOX9 | A | 23847113 | Cells in ectopic
endometriosis lesions also expressed SSEA-1 and nuclear SOX9. |
| 2 | A2M | B | 2454848 | Women with
endometriosis had significantly lower amounts of functional α-2M
than did women without endometriosis. |
| 836 | CASP3 | B | 24246915 | Significantly lower
expression of caspase-3 protein was found in ectopic (3.20±1.24)
and eutopic endometrium (3.88±1.93) as compared with the control
group (6.49±1.85; P<0.01). |
| 7189 | TRAF6 | B | 20130413 | TRAF2, TRAF6 and
TAK1 were constitutively activated and were unaffected by TSA
treatment in endometriotic cells. |
| 834 | CASP1 | B | 17094974 | Eutopic and ectopic
ECs from women with endome triosis expressed decreased transcript
abundance of p53 and Caspase-1 compared to ECs from women without
endometriosis. |
| 6284 | S100A13 | B | 15821778 | Expression of
S100A13 corresponds to the activation of the endothelial cells in
the process of endometriotic angiogenesis. |
| 7850 | IL1R2 | B | 17482186 | IL-1RII can
neutralize IL-1β and counteract its effect on endometrial stromal
cells, and may provide a new clinical strategy for the treatment of
endometriosis. |
| 3561 | IL2RG | B | 16759924 | IL2RG was
demonstrated to be significantly differen tially expressed in blood
lymphocytes between endome triosis patients and controls. |
| 3718 | JAK3 | B | 17631002 | JAK3 inhibitors,
especially JANEX-1, may prove useful to prevent or alleviate the
symptoms of endometriosis. |
Discussion
The cause of endometriosis is not entirely
understood. No single theory is able to explain all cases of
endometriosis. The present study implemented PPI for endometriosis
biomarker network analysis and identified biologically relevant
functional modules. A number of genes and pathways identified in
the modules have already been reported to participate in the
pathogenesis of endometriosis (32–36).
Although endometriosis is a benign disorder, several
common characteristics of this disease are shared with invasive
cancer (37). Previous epidemiologic
studies have demonstrated that women with endometriosis have an
increased risk of ovarian and breast cancer (38,39).
Coincidentally, the three chromosomal regions (9p, 11q and 22q)
that have demonstrated loss of heterozygosity in ovarian
endometriosis were the same that were observed in ovarian tumors
(40). These studies have
demonstrated that the inactivation of tumor suppressor genes has an
important role in the development of endometriosis. The results of
the present study demonstrated that expression of cancer-related
pathways are significantly imbalanced in endometriosis in module A.
The hub genes identified were AHR, AR, ARNT, ESR1, NRIP1, ESR2,
TP53, NR5A1, NR3C1 and PGR.
The enriched pathways in module B were associated
with the immune system and infectious diseases. The presence of
proinflammatory cytokines in the peritoneal fluid of patients with
endometriosis has been reported in previous studies (41–43).
Cytokines may regulate the actions of leukocytes in the peritoneal
fluid or may act directly on the ectopic endometrium (44). Dysregulation of the JAK-STAT pathway
is associated with various immune disorders (45), which was also demonstrated in the
results of the present study. IL10RA, IL15, IL10 and JAK3 from the
Toll-like receptor signaling pathway and CASP1, IL18, IL1B and
TRAF6 from the NOD-like receptor signaling pathway, which are
important for generating mature proinflammatory cytokines, were
also identified in this module and are confirmed by previous
studies (35,46). Module B also included the
osteoclastogenesis pathway, which is predominantly regulated by
signaling pathways activated by immune receptors (47).
Matrix metalloproteinases (MMPs) are a family of
proteolytic enzymes that share a conserved domain structure. MMPs
are capable of degrading various types of extracellular matrix
(ECM) and serve an important function in tissue remodeling
associated with various physiological and pathological processes
(48). The expression of several
MMPs is maximal during the menstrual phase in the human endometrium
(49). MMPs also have a vital role
in the pathogenesis of endometriosis and cancer, particularly in
the processes of metastasis and invasion (33,50).
MMP1, MMP7, MMP12, MMP13, IGF1, IGFBP1, PAPPA and TIMP2 were
identified as the hub genes in module C. ECM-receptor interaction,
focal adhesion and proteasomes were also identified in this module,
as in previous studies (32,51,52).
The immune response is one of the major factors
influencing pathogenesis of endometriosis. Numerous genes in the
fourth module are involved in the function of the immune system.
Hub genes in this module are members of the HLA gene family,
including HLA-A, -B, -C, -DPB1, -DQA1, -DQB1 and -DRB1, which have
key roles in the immune response, and it appears that endometriosis
shares many similarities with autoimmune diseases (34,53). It
has been demonstrated that patients with endometriosis display a
significantly higher expression of HLA I and II molecules compared
with individuals without endometriosis (54).
Oxidative stress has been proposed as a potential
factor involved in the pathophysiology of endometriosis (55). Accumulation of reactive oxygen
species may induce cellular injury, such as DNA damage. The present
study demonstrated that the majority of the pathways in module E
were related to replication and repair. APEX1, OGG1, XRCC1, ERCC2
and ERCC5 were the seed genes identified in this module. APEX1 and
XRCC1 are key genes involved in the base excision repair pathway,
which removes DNA adducts induced predominantly by oxidation and
alkylation (56). APEX1 is an
essential enzyme and has a central role in the DNA repair system;
however, a study by Hsu et al (57) demonstrated that APEX1 Asp148Glu was
not associated with endometriosis in patients in Taiwan. Future
studies may confirm the association between APEX1 and the risk of
endometriosis. XRCC1 has been demonstrated to physically interact
with several enzymes known to be involved in the repair of
single-strand breaks in DNA (58). A
study by Hsieh et al (36)
indicated that XRCC1 Arg399Gln polymorphism is correlated with a
higher susceptibility to endometriosis.
In conclusion, the pathogenesis of endometriosis is
likely multifactorial. The present study constructed a disease
network of endometriosis that integrated human protein-protein
interactions and known disease-causing genes. The present study has
identified a number of biological mechanisms that may be associated
with endometriosis. Further studies on the specific function and
interactions of the genes in related modules are required to
improve the understanding of endometriosis.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81360336)
and the Joint Special Funds for the Department of Science and
Technology of Yunnan Province-Kunming Medical University (grant no.
2015FB017).
References
|
1
|
Podgaec S, Abrao MS, Dias JA Jr, Rizzo LV,
de Oliveira RM and Baracat EC: Endometriosis: An inflammatory
disease with a Th2 immune response component. Hum Reprod.
22:1373–1379. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Braun DP and Dmowski WP: Endometriosis:
Abnormal endometrium and dysfunctional immune response. Curr Opin
Obstet Gynecol. 10:365–369. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sha G, Wu D, Zhang L, Chen X, Lei M, Sun
H, Lin S and Lang J: Differentially expressed genes in human
endometrial endothelial cells derived from eutopic endometrium of
patients with endometriosis compared with those from patients
without endometriosis. Hum Reprod. 22:3159–3169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kato N, Sasou S and Motoyama T: Expression
of hepatocyte nuclear factor-1beta (HNF-1beta) in clear cell tumors
and endometriosis of the ovary. Mod Pathol. 19:83–89. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kvaskoff M, Mu F, Terry KL, Harris HR,
Poole EM, Farland L and Missmer SA: Endometriosis: A high-risk
population for major chronic diseases? Hum Reprod Update.
21:500–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahn SH, Monsanto SP, Miller C, Singh SS,
Thomas R and Tayade C: Pathophysiology and immune dysfunction in
endometriosis. Biomed Res Int. 2015:7959762015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamosh A, Scott AF, Amberger JS, Bocchini
CA and McKusick VA: Online mendelian inheritance in man (OMIM), a
knowledgebase of human genes and genetic disorders. Nucleic Acids
Res. 33(Database issue): D514–D517. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stenson PD, Mort M, Ball EV, Howells K,
Phillips AD, Thomas NS and Cooper DN: The human gene mutation
database: 2008 pdate. Genome Med. 1:132009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Becker KG, Barnes KC, Bright TJ and Wang
SA: The genetic association database. Nature Genet. 36:431–432.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wall DP, Pivovarov R, Tong M, Jung JY,
Fusaro VA, DeLuca TF and Tonellato PJ: Genotator: A
disease-agnostic tool for genetic annotation of disease. BMC Med
Genomics. 3:502010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bauer-Mehren A, Rautschka M, Sanz F and
Furlong LI: DisGeNET: A Cytoscape plugin to visualize, integrate,
search and analyze gene-disease networks. Bioinformatics.
26:2924–2926. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lim J, Hao T, Shaw C, Patel AJ, Szabó G,
Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al: A
protein-protein interaction network for human inherited ataxias and
disorders of Purkinje cell degeneration. Cell. 125:801–814. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pujana MA, Han JD, Starita LM, Stevens KN,
Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al:
Network modeling links breast cancer susceptibility and centrosome
dysfunction. Nat Genet. 39:1338–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jia P, Kao CF, Kuo PH and Zhao Z: A
comprehensive network and pathway analysis of candidate genes in
major depressive disorder. BMC Syst Biol. 5 Suppl 3:S122011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vidal M, Cusick ME and Barabási AL:
Interactome networks and human disease. Cell. 144:986–998. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu G, Feng X and Stein L: A human
functional protein interaction network and its application to
cancer data analysis. Genome Biol. 11:R532010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schadt EE: Molecular networks as sensors
and drivers of common human diseases. Nature. 461:218–223. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding Y, Chen M, Liu Z, Ding D, Ye Y, Zhang
M, Kelly R, Guo L, Su Z, Harris SC, et al: atBioNet-an integrated
network analysis tool for genomics and biomarker discovery. BMC
Genomics. 13:3252012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stark C, Breitkreutz BJ, Chatr-Aryamontri
A and Tyers M: The BioGRID Interaction Database: 2011 pdate.
Nucleic Acids Res. 39(Database issue): D698–D704. 2010.PubMed/NCBI
|
|
20
|
Xenarios I, Rice DW, Salwinski L, Baron
MK, Marcotte EM and Eisenberg D: DIP: The database of interacting
proteins. Nucleic Acids Res. 28:289–291. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prasad TS Keshava, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37(Database issue):
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aranda B, Achuthan P, Alam-Faruque Y,
Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S,
Khadake J, et al: The IntAct molecular interaction database in
2010. Nucleic Acids Res. 38(Database issue): D525–D531. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Licata L, Briganti L, Peluso D, Perfetto
L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP,
Santonico E, et al: MINT, the molecular interaction database: 2012
pdate. Nucleic Acids Res. 40(Database issue): D857–D861. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Matthews L, Gopinath G, Gillespie M, Caudy
M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B,
et al: Reactome knowledgebase of human biological pathways and
processes. Nucleic Acids Res. 37(Database issue): D619–D622. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elkon R, Vesterman R, Amit N, Ulitsky I,
Zohar I, Weisz M, Mass G, Orlev N, Sternberg G, Blekhman R, et al:
SPIKE-a database, visualization and analysis tool of cellular
signaling pathways. BMC bioinformatics. 9:1102008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu X, Yuruk N, Feng Z and Schweiger T:
SCAN: A structural clustering algorithm for networks. Proceedings
of the 13th ACM SIGKDD international conference on Knowledge
Discovery and Data Mining. ACM. San Jose, CA. pp. 824–833.
2007;
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hwang JH, Oh JJ, Wang T, Jin YC, Lee JS,
Choi JR, Lee KS, Joo JK and Lee HG: Identification of biomarkers
for endometriosis in eutopic endometrial cells from patients with
endometriosis using a proteomics approach. Mol Med Rep. 8:183–188.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wei WD, Ruan F, Tu FX, Zhou CY and Lin J:
Expression of suppressor of cytokine signaling-3 and caspase-3 in
endometriosis and their correlation. Zhonghua Bing Li Xue Za Zhi.
42:515–518. 2013.(In Chinese). PubMed/NCBI
|
|
30
|
Hayrabedyan S, Kyurkchiev S and Kehayov I:
Endoglin (cd105) and S100A13 as markers of active angiogenesis in
endometriosis. Reprod Biol. 5:51–67. 2005.PubMed/NCBI
|
|
31
|
Hou Z, Zhou J, Ma X, Fan L, Liao L and Liu
J: Role of interleukin-1 receptor type II in the pathogenesis of
endometriosis. Fertil Steril. 89:42–51. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Selam B, Kayisli UA, Garcia-Velasco JA and
Arici A: Extracellular matrix-dependent regulation of Fas ligand
expression in human endometrial stromal cells. Biol Reprod. 66:1–5.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Osteen KG, Yeaman GR and Bruner-Tran KL:
Matrix metalloproteinases and endometriosis. Semin Reprod Med.
21:155–164. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Bakker PI, McVean G, Sabeti PC, Miretti
MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M,
et al: A high-resolution HLA and SNP haplotype map for disease
association studies in the extended human MHC. Nat Genet.
38:1166–1172. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kumar H, Kawai T and Akira S: Toll-like
receptors and innate immunity. Biochem Biophys Res Commun.
388:621–625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hsieh YY, Chang CC, Chen SY, Chen CP, Lin
WH and Tsai FJ: XRCC1 399 Arg-related genotype and allele, but not
XRCC1 His107Arg, XRCC1 Trp194Arg, KCNQ2, AT1R and hOGG1
polymorphisms, are associated with higher susceptibility of
endometriosis. Gynecol Endocrinol. 28:305–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang QY and Wu RJ: Growth mechanisms of
endometriotic cells in implanted places: A review. Gynecol
Endocrinol. 28:562–567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vlahos NF, Economopoulos KP and Fotiou S:
Endometriosis, in vitro fertilisation and the risk of
gynaecological malignancies, including ovarian and breast cancer.
Best Pract Res Clin Obstet Gynaecol. 24:39–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pollacco J, Sacco K, Portelli M,
Schembri-Wismayer P and Calleja-Agius J: Molecular links between
endometriosis and cancer. Gynecol Endocrinol. 28:577–581. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang X, Hitchcock A, Bryan EJ, Watson RH,
Englefield P, Thomas EJ and Campbell IG: Microsatellite analysis of
endometriosis reveals loss of heterozygosity at candidate ovarian
tumor suppressor gene loci. Cancer Res. 56:3534–3539.
1996.PubMed/NCBI
|
|
41
|
Hsieh YY, Chang CC, Tsai FJ, Hsu CM, Lin
CC and Tsai CH: Interleukin-2 receptor beta (IL-2R beta)-627*C
homozygote but not IL-12R beta 1 codon 378 or IL-18 105
polymorphism is associated with higher susceptibility to
endometriosis. Fertil Steril. 84:510–512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ayaz L, Celik SK, Cayan F, Aras-Ates N and
Tamer L: Functional association of interleukin-18 gene −607 C/A
promoter polymorphisms with endometriosis. Fertil Steril.
95:298–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Monsanto SP, Edwards AK, Zhou J,
Nagarkatti P, Nagarkatti M, Young SL, Lessey BA and Tayade C:
Surgical removal of endometriotic lesions alters local and systemic
proinflammatory cytokines in endometriosis patients. Fertil Steril.
105:968–977. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Harada T, Iwabe T and Terakawa N: Role of
cytokines in endometriosis. Fertil Steril. 76:1–10. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shuai K and Liu B: Regulation of JAK-STAT
signalling in the immune system. Nat Rev Immunol. 3:900–911. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Petrilli V, Dostert C, Muruve DA and
Tschopp J: The inflammasome: A danger sensing complex triggering
innate immunity. Curr Opin Immunol. 19:615–622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Harada M, Osuga Y, Hirata T, Hirota Y,
Koga K, Yoshino O, Morimoto C, Fujiwara T, Momoeda M, Yano T, et
al: Concentration of osteoprotegerin (OPG) in peritoneal fluid is
increased in women with endometriosis. Hum Reprod. 19:2188–2191.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nissinen L and Kähäri VM: Matrix
metalloproteinases in inflammation. Biochim Biophys Acta.
1840:2571–2580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cominelli A, Chevronnay HP Gaide, Lemoine
P, Courtoy PJ, Marbaix E and Henriet P: Matrix metalloproteinase-27
is expressed in CD163+/CD206+ M2 macrophages in the cycling human
endometrium and in superficial endometriotic lesions. Mol Hum
Reprod. 20:767–775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nagase H, Visse R and Murphy G: Structure
and function of matrix metalloproteinases and TIMPs. Cardiovasc
Res. 69:562–573. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mu L, Zheng W, Wang L, Chen XJ, Zhang X
and Yang JH: Alteration of focal adhesion kinase expression in
eutopic endometrium of women with endometriosis. Fertil Steril.
89:529–537. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Celik O, Hascalik S, Elter K, Tagluk ME,
Gurates B and Aydin NE: Combating endometriosis by blocking
proteasome and nuclear factor-kappaB pathways. Hum Reprod.
23:2458–2465. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nothnick WB: Treating endometriosis as an
autoimmune disease. Fertil Steril. 76:223–231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kitawaki J, Obayashi H, Kado N, Ishihara
H, Koshiba H, Maruya E, Saji H, Ohta M, Hasegawa G, Nakamura N, et
al: Association of HLA class I and class II alleles with
susceptibility to endometriosis. Hum Immunol. 63:D1033–D1038. 2002.
View Article : Google Scholar
|
|
55
|
Zhang X, Sharma RK, Agarwal A and Falcone
T: Effect of pentoxifylline in reducing oxidative stress-induced
embryotoxicity. J Assist Reprod Genet. 22:415–417. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wood RD, Mitchell M, Sgouros J and Lindahl
T: Human DNA repair genes. Science. 291:1284–1289. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hsu CM, Chang WS, Hwang JJ, Wang JY, Hsiao
YL, Tsai CW, Liu JC, Ying TH and Bau DT: The role of
apurinic/apyrimidinic endonuclease DNA repair gene in
endometriosis. Cancer Genomics Proteomics. 11:295–301.
2014.PubMed/NCBI
|
|
58
|
Brem R and Hall J: XRCC1 is required for
DNA single-strand break repair in human cells. Nucleic Acids Res.
33:2512–2520. 2005. View Article : Google Scholar : PubMed/NCBI
|