Introduction
Down's syndrome (DS), or 21-trisomy syndrome, is a
kind of chromosomal abnormal genetic disease caused by a local or
total copy of chromosome 21, whereby patients suffer from delayed
body growth, special facies, mild to moderate mental retardation
and other symptoms, seriously affecting the life of patients
(1). According to statistics,
approximately 1 of 1,000 infants is inflicted by this disease
annually, and the mortality rate of DS patients reached 0.5% in
2015 (2). The fundamental way to
reduce DS risk is to avoid the birth of newborn patients; thus,
prenatal screening for pregnant women is imperative. At present,
the conventional method for the prenatal examination of DS is the
amniotic cell chromosome analysis, in which fetal cells are
obtained via B ultrasound-guided puncture of the amniotic cavity
for karyotype analysis. However, this method is invasive and the
fetal loss rate is high. Consequently, the majority of high-risk
pregnant women refuse this method (3).
In previous years, non-invasive DS prenatal
diagnosis techniques have been developed, including ultrasound
diagnosis, karyotype analysis of peripheral blood cell chromosome
in pregnant women, and peripheral-free fetal nucleic acid analysis,
of which free fetal nucleic acid has rapidly become developed as a
method in the prenatal diagnosis of DS (4). Free fetal nucleic acid is characterized
by rich content and stable existence in the peripheral blood of
pregnant women (5), and is more
suitable for the DS prenatal diagnosis. However, the maternal
nucleic acid also exists in peripheral blood, and is essential to
distinguish maternal from fetal nucleic acid. Findings have shown
that the maternal nucleic acid in peripheral blood is derived from
hematopoietic cells, while the free fetal nucleic acid is derived
from placental trophocytes. Owing to the different tissues, the
level of DNA methylation is also different between them (6,7), which
can provide new ideas for the study on DS prenatal diagnosis and
maternal-fetal epigenetic differences.
Down's syndrome critical region 4 (DSCR4)
gene is a kind of highly expressed gene in placenta and is
expressed in human choriocarcinoma cell lines JEG3 and BeWo.
DSCR4 is located in the DSCR region on chromosome 21q22.2
(8). Previous findings showed that
obstacles are formed in the syncytiotrophoblast in the placenta of
DS patients. Thus, the process of secretion of hormones in the
maternal circulation is affected (9). DSCR4 is located in DSCR and expressed
in placental tissues and plays a unique role in placental
development and participates in the pathological process of DS. In
addition, the expression level of DSCR4 is related to the
methylation level in its promoter region, and there is also a
difference in methylation between maternal and fetal DNA (10). Therefore, DSCR4 gene
methylation is a characteristic marker with maternal-fetal
epigenetic differences.
The aim of the study was to examine the association
between the DSCR4 gene methylation level in plasma in
high-risk pregnant women with DS in early pregnancy (hereinafter
referred to as pregnant women in early pregnancy) and DS in order
to screen the biomarkers with maternal-fetal epigenetic differences
for the non-invasive prenatal diagnosis of DS and provide new
perspectives for the prognosis and treatment of DS.
Materials and methods
General data
Twenty pregnant women in early pregnancy, admitted
to Outpatient Department in The Third Xiangya Hospital of Central
South University (Hunan, China) from January 2016 to May 2016 were
included in the study. All 20 were high-risk pregnant women with DS
with a gestation period of 8–12 weeks, age of 28.45±3.77 years and
weight of 108.43±17.83 kg. Subjects were spontaneously pregnant
with single birth for the first time without pregnancy
complications, medical and surgical diseases, tumors or other
diseases.
Study participation was agreed to by the subjects
and the informed consent form was signed. The present study was
approved by the Ethics Committee of The Third Xiangya Hospital of
Central South University.
Main reagents
Human choriocarcinoma cell lines JEG3 and BeWo were
purchased from Cobioer Biosciences (Nanjing, China). An EZ DNA
methylation-gold kit (Zymo Research Corp., Irvine, CA, USA); QIAamp
DNA mini and DNA blood mini kit (both from Qiagen, Inc., Valencia,
CA, USA); TaqαI, TRIzol reagent, Prime Script® RT
reagent kit with gDNA Eraser and SYBR® Premix Ex
Taq™ II (all from Takara Biotechnology Co., Ltd.,
Dalian, China); F12, Dulbecco's modified Eagle's medium (DMEM) and
Lipofectamine 2000 (both from Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA); Cell Counting kit-8 (CCK-8) (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan); and primers
(Shanghai Shenggong Biology Engineering Technology Service, Ltd.,
Shanghai, China) were used in the present study.
Extraction of DNA in peripheral blood
cells and plasma
Fresh peripheral blood (4 ml) was drawn from
pregnant women in early pregnancy, and the plasma and blood cells
were completely isolated via centrifugation after ethylene diamine
tetraacetic acid was added for anticoagulation. Isopyknic
phosphate-buffered saline was added into the blood cells to
resuspend the cells, while a sodium dodecyl sulfonate lysis buffer
was added into the plasma. After incubation at 37°C for 2 h and
ultrafiltration centrifugation, poly d(T)18 was added and mixed
evenly. DNA in blood cells and plasma was extracted according to
the instructions of QIAamp DNA mini and DNA blood mini kit.
Hydrosulphite treatment
DNA (400 ng) in blood cells and plasma was taken
quantitatively and the water was complemented until 20 µl. The
solution was treated with hydrosulphite according to the
instructions of the EZ DNA methylation-gold kit. The products were
eluted for subsequent experiments.
Methylation-specific polymerase chain
reaction (MSP)
DNA (2 µl) in blood cells, and 2 µl DNA in plasma
after hydrosulphite treatment were taken, and the PCR amplification
system and reaction process were optimized routinely. The gene
coding regions of undermethylated DSCR4 (U-DSCR4) and
hypermethylated DSCR4 (M-DSCR4) were amplified using the hot-start
method. The amplification products were 148 and 143 bp, and they
were observed via electrophoresis and photographed. The primer
sequences are shown in Table I.
 | Table I.Primers and siRNA sequences. |
Table I.
Primers and siRNA sequences.
| Primer and oligo | Sequence (5′-3′) |
|---|
| U-DSCR4-F |
AGTGAAATTTTGATTTAAAAAGTGA |
| U-DSCR4-R |
AACAAAAACCATCTTATCCACTCAT |
| M-DSCR4-F |
GCGAAATTTCGATTTAAAAAGC |
| M-DSCR4-R |
AAAACCGTCTTATCCACTCGTT |
| ACTB-F |
TGGTGATGGAGGAGGTTTAGTAAGT |
| ACTB-R |
AACCAATAAAACCTACTCCTCCCTTAAGGCUGAGUAGGUCCACAAATTUU |
| DSCR4 siRNA |
UGUGGACCUACUCAGCCTT |
Restriction endonuclease analysis
The amplification product gel was recycled and 500
ng gel was taken and added with 10 U TaqαI endonuclease. After
incubation via water bath at 65°C for 2 h, the enzyme-digested
products were observed via 2% agarose gel electrophoresis and
photographed. At the same time, the non-enzyme digestion group was
set as a control.
Reverse transcriptase-quantitative PCR
(RT-qPCR) analysis
The reverse transcriptase-quantitative
methylation-specific PCR (RT-qMSP) reaction system was prepared
using hydrosulphite-treated DNA in blood cells and plasma as the
template according to the protocol of the kit. Additionally, this
DNA was detected in CFX-96 real-time PCR detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Data were collected and
treated in the relative expression form using the comparative cycle
threshold method. β-actin was taken as the internal reference. The
primer sequences are shown in Table
I.
Cell transfection
The DNA-Lipofectamine 2000 complex was prepared as
per the instructions of Lipofectamine 2000, and the diluted plasmid
DNA was mixed with Lipofectamine 2000 at a ratio of 1/2-1/3
(µg/µl). The mixture was placed at room temperature for 20 min, and
for another 10 min at room temperature after small interfering RNA
(siRNA) oligo was added. The mixture was added into the 96-well
plates and incubated with CO2 at 37°C for 24–48 h. The
transfected cells were used for subsequent experiments, and the
siRNA sequences of DSCR4 are shown in Table I.
Cell proliferation analysis
The transfected cells (5×103) were
inoculated onto the 96-well plate according to the instructions of
the CCK-8 kit. CCK-8 solution (10 µl) was added into each well at
0, 1, 2, 3 and 4 days and incubated at 37°C for 2 h. Absorbance was
measured at 450 nm.
Cell invasion and migration
analysis
The artificial basement membrane was added into the
culture wells. DMEM containing 3×104 cells after
transfection for 48 h was inoculated into the upper chamber of the
basement membrane, while the lower chamber was filled with DMEM
containing 20% fetal bovine serum. After incubation for 24 h, the
membrane was fixed with 4% methanol and stained with 0.1 % crystal
violet three times, followed by cell migration analysis.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
(SPSS, Chicago, IL, USA). The experiments were repeated three times
and the data were presented as mean ± standard deviation.
Kruskal-Wallis one-way analysis of variance and a two-tailed
t-value test were performed between groups. P<0.05 was
considered to indicate a statistically significant diference.
Results
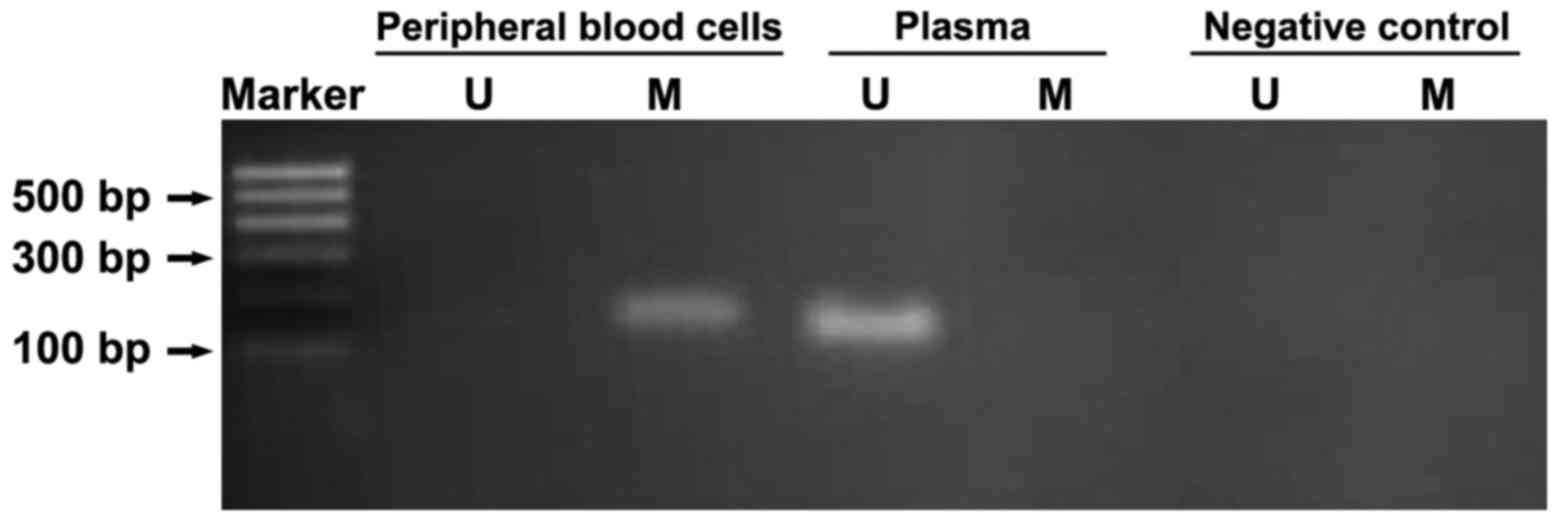
Methylation difference of DSCR4 gene
in peripheral blood DNA in high-risk pregnant women with DS
MSP was performed for the hydrosulphite-treated DNA
in peripheral blood cells and plasma of pregnant women in early
pregnancy to amplify U-DSCR4 and M-DSCR4. U-DSCR4 was detected in
plasma, but not in blood cells, whereas M-DSCR4 was detected in
peripheral blood cells in pregnant women, but not in plasma,
indicating that there is a difference in the methylation level
between DSCR4 genes in peripheral blood and plasma in
high-risk pregnant women with DS (Fig.
1).
Verification of the methylation
difference of DSCR4 in peripheral blood DNA via restriction
endonuclease
The methylation difference of DSCR4 between
peripheral blood cells and plasma in high-risk pregnant women with
DS was verified via restriction endonuclease analysis. Since the
TaqαI recognition site was TCGA, if C in CpG was unmethylated, the
amplification product would mutate into T following hydrosulphite
treatment, changing the digestion site and disabling TaqαI
digestion. Fig. 2 shows that the
DSCR4 amplification products in peripheral blood cells in pregnant
women were digested under the action of TaqαI, and the
enzyme-digested products were 102 and 130 bp, indicating that DSCR4
is in a hypermethylated status in peripheral blood cells. However,
the products in plasma were not digested, suggesting that DSCR4 is
in an unmethylated status in plasma.
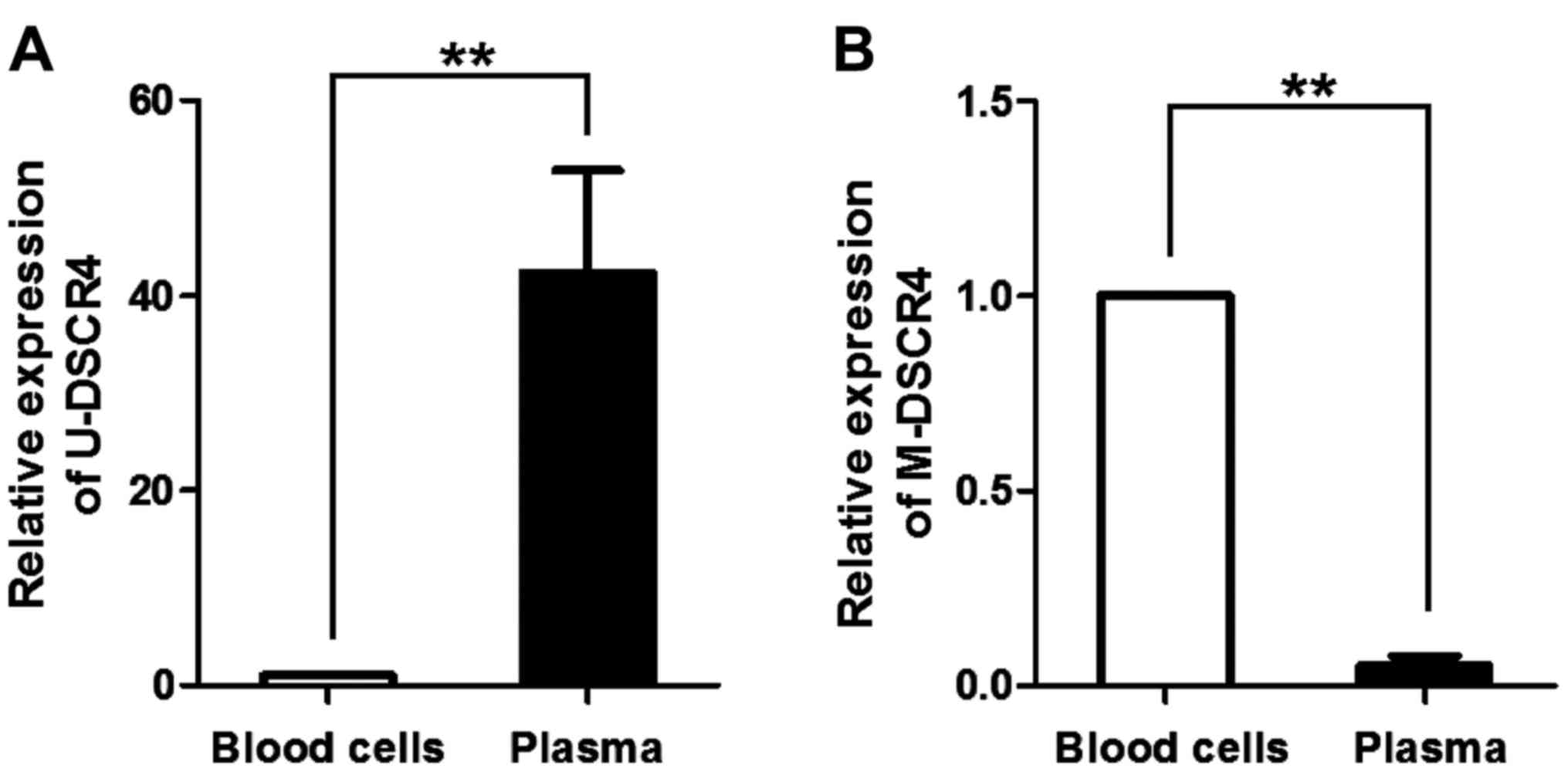
Expression of DSCR4 with different
methylation levels in the peripheral blood of pregnant women
The expression levels of U-DSCR4 and M-DSCR4 in
peripheral blood cells and plasma of pregnant women in early
pregnancy were detected via RT-qPCR. The expression level of
U-DSCR4 in plasma was significantly higher than that in blood cells
(Fig. 2A), U-DSCR4 mainly existed in
plasma DNA. However, the expression level of M-DSCR4 in plasma was
obviously lower than that in blood cells, M-DSCR4 mainly existed in
blood cell DNA (Fig. 2B).
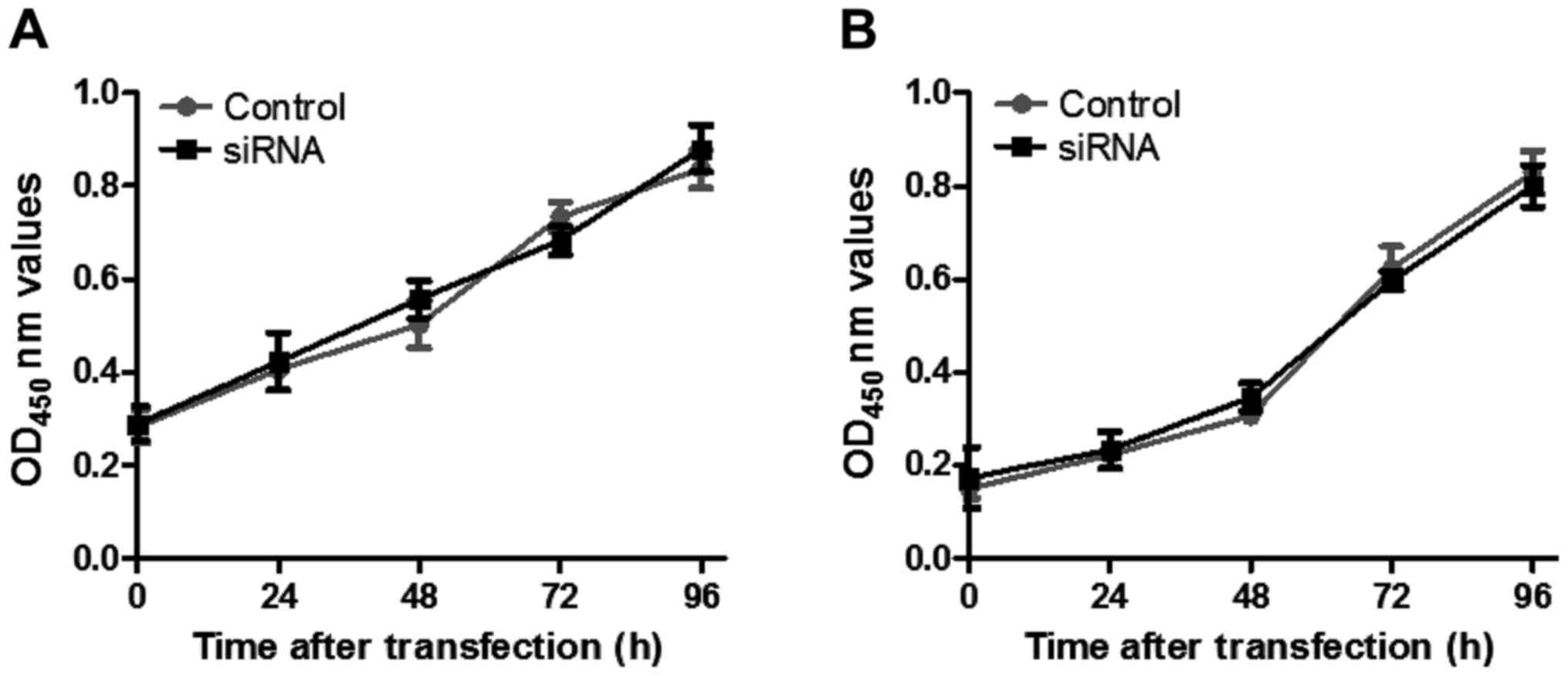
Effect of DSCR4 on cell
proliferation
The proliferation of JEG3 and BeWo cells transfected
with DSCR4 siRNA was detected via CCK-8. There was no
significant effect on the proliferation of JEG3 and BeWo cells in
the DSCR4 intervention group compared with that in the control
group (Fig. 3).
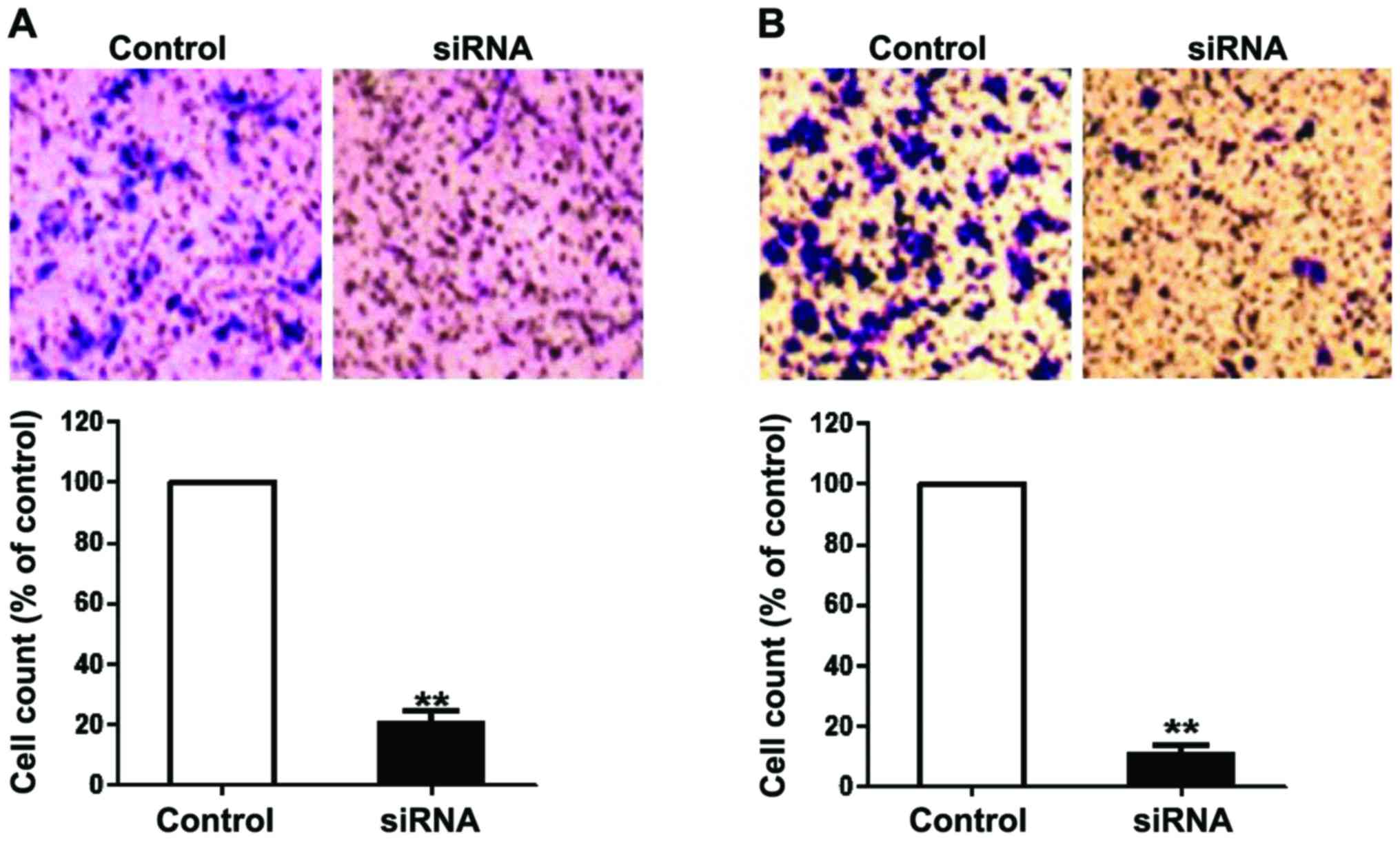
Effect of DSCR4 on cell invasion and
migration
The invasion and migration of JEG3 and BeWo cells
transfected with DSCR4 siRNA for 48 h were analyzed. The
results showed that the intervention in DSCR4 expression
significantly decreased the invasion and migration capacities of
JEG3 and BeWo cells compared with the control group, indicating
that DSCR4 can promote the invasion and migration of JEG3 and BeWo
cells (Fig. 4).
Discussion
DS is a kind of chromosomal abnormal disease, and
patients often suffer from dysplasia, immune system defects and
mental retardation accompanied with congenital heart disease
(11). The diagnosis and treatment
of DS poses a significant financial burden on the patient's family.
At present, the commonly used diagnostic method is invasive
examination of pregnant women based on serological screening.
However, this method has varying shortcomings and the establishment
of non-invasive prenatal diagnosis is imperative to resolved this
issue. Lo et al used the heterozygous SNP on the PLAC4 mRNA
specifically expressed in placenta to calculate the ratio of PLAC4
allele for DS diagnosis, but the detection rate was low (12). This method provides a new idea for
the free fetal nucleic acid in peripheral blood of pregnant women
as a target for a non-invasive DS diagnosis.
Therefore, it is crucial to differentiate fetal from
maternal DNA in peripheral blood. Previous findings have shown that
the methylation level of the SERPINB5 gene promoter region
on chromosome 18q21.3 has a difference between peripheral blood
cells and placental tissues of pregnant women, the former of which
exists in a hypermethylated state, and the latter of which exists
in an undermethylated state and can be considered as coming from
the fetus (13). Consequently,
maternal DNA methylation differences can be used as a key factor in
distinguishing maternal-fetal DNA. DNA methylation is a common form
of epigenetic modification, which adds methyl selectively on C of
the CG dinucleotide through the effect of methyltransferase. As a
result, the gene sequence does not change, albeit the heritable
changes occur to the gene expression (14). At present, placental-derived
non-methylated genes, such as U-SERPINB5 and U-RASSF1A, can be
successfully detected in the free nucleic acid in plasma of
pregnant women and used in clinical diagnosis (15,16).
Since the pathogenesis basis of DS is the local or
total copy of chromosome 21, screening the genes with
maternal-fetal methylation differences on chromosome 21 is
beneficial to reveal the pathological process and molecular
mechanisms of DS and provide new targets for the prenatal diagnosis
of DS. Chim et al systematically studied the marker
sequences with maternal-fetal methylation differences on chromosome
21 (17). The findings showed that
the fetal HLCS gene on chromosome 21 is hypermethylated and
can be used to detect DS in male fetuses (18). In addition, studies have shown that
there is a region of 5.4 Mb in length on chromosome 21q22.2 known
as DS critical region (19). This
region includes a variety of DS-related genes, of which
DSCR4 is specifically expressed in placenta and may be
associated with the abnormal placental trophoblast of DS, and
involved in the regulation of the pathological process of DS
(20).
Findings of this study indicated that there was a
difference in methylation of DSCR4 gene in peripheral blood
DNA in high-risk pregnant women with DS in early pregnancy.
DSCR4 was in an undermethylated status in plasma and in a
hypermethylated status in blood cells. The peripheral blood DNA
contains both maternal and free fetal DNA, of which the maternal
nucleic acid comes from the hematopoietic cells and mainly exists
in the blood cells, and the free fetal nucleic acid is derived from
the placental trophoblast cells and mainly exists in plasma
(6,7). Thus, there is a difference in the
methylation level of DSCR4 in maternal DNA, and DSCR4 can be used
as a biomarker for DS prenatal diagnosis and distinguishing
maternal-fetal epigenetic differences. In general, the degree of
DNA methylation is negatively correlated with gene expression, and
our findings also revealed that U-DSCR4 was highly expressed in the
plasma of pregnant women in early pregnancy, indicating it was
highly expressed in fetal DNA. M-DSCR4 was highly expressed in
blood cells, indicating it was highly expressed in maternal DNA.
Thus, U-DSCR4 is an epigenetic specific marker of fetal DS. In
addition, the intervention in DSCR4 expression suppressed
the invasion and migration of trophoblast cells, but had no effect
on cell proliferation, suggesting that DSCR4 regulates the
abnormality of DS placental trophoblast by promoting cell migration
and invasion and plays a role in early placental development. In
conclusion, this study provided a theoretical basis for the
non-invasive prenatal diagnosis of DS and screened new biomarkers
for maternal-fetal epigenetic differences. Additionally, it
provided a new perspective for studying the role of DSCR4 in
pathological process of DS and placental development.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Weijerman ME and de Winter JP: Clinical
practice. The care of children with Down syndrome. Eur J Pediatr.
169:1445–1452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vos T, Allen C, Arora M, Barber RM, Bhutta
ZA, Brown A, Carter A, Casey DC, Charlson FJ, Chen AZ, et al: GBD
2015 disease and injury incidence and prevalence collaborators:
Global, regional, and national incidence, prevalence, and years
lived with disability for 310 diseases and injuries, 1990–2015: A
systematic analysis for the Global Burden of Disease Study 2015.
Lancet. 388:1545–1602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ekelund CK, Jørgensen FS, Petersen OB,
Sundberg K and Tabor A: Danish fetal medicine research group:
Impact of a new national screening policy for Down's syndrome in
Denmark: Population based cohort study. BMJ. 337:a25472008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chiu RW and Lo YM: Non-invasive prenatal
diagnosis by fetal nucleic acid analysis in maternal plasma: The
coming of age. Semin Fetal Neonatal Med. 16:88–93. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Angert RM, LeShane ES, Lo YM, Chan LY,
Delli-Bovi LC and Bianchi DW: Fetal cell-free plasma DNA
concentrations in maternal blood are stable 24 hours after
collection: Analysis of first- and third-trimester samples. Clin
Chem. 49:195–198. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW
and Lo YM: Predominant hematopoietic origin of cell-free DNA in
plasma and serum after sex-mismatched bone marrow transplantation.
Clin Chem. 48:421–427. 2002.PubMed/NCBI
|
|
7
|
Li B, Feng ZH, Sun H, Zhao ZH, Yang SB and
Yang P: The blood genome-wide DNA methylation analysis reveals
novel epigenetic changes in human heart failure. Eur Rev Med
Pharmacol Sci. 21:1828–1836. 2017.PubMed/NCBI
|
|
8
|
Nakamura A, Hattori M and Sakaki Y: A
novel gene isolated from human placenta located in Down syndrome
critical region on chromosome 21. DNA Res. 4:321–324. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wright A, Zhou Y, Weier JF, Caceres E,
Kapidzic M, Tabata T, Kahn M, Nash C and Fisher SJ: Trisomy 21 is
associated with variable defects in cytotrophoblast differentiation
along the invasive pathway. Am J Med Genet A. 130A:1–364. 2004.
View Article : Google Scholar
|
|
10
|
Alldred SK, Deeks JJ, Guo B, Neilson JP
and Alfirevic Z: Second trimester serum tests for Down's syndrome
screening. Cochrane Database Syst Rev. CD0099252012.PubMed/NCBI
|
|
11
|
Roper RJ and Reeves RH: Understanding the
basis for Down syndrome phenotypes. PLoS Genet. 2:e502006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lo YM, Tsui NB, Chiu RW, Lau TK, Leung TN,
Heung MM, Gerovassili A, Jin Y, Nicolaides KH, Cantor CR, et al:
Plasma placental RNA allelic ratio permits noninvasive prenatal
chromosomal aneuploidy detection. Nat Med. 13:218–223. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong YK, Ding C, Chiu RW, Gerovassili A,
Chim SS, Leung TY, Leung TN, Lau TK, Nicolaides KH and Lo YM:
Noninvasive prenatal detection of fetal trisomy 18 by epigenetic
allelic ratio analysis in maternal plasma: Theoretical and
empirical considerations. Clin Chem. 52:2194–2202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zemach A, McDaniel IE, Silva P and
Zilberman D: Genome-wide evolutionary analysis of eukaryotic DNA
methylation. Science. 328:916–919. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chim SS, Tong YK, Chiu RW, Lau TK, Leung
TN, Chan LY, Oudejans CB, Ding C and Lo YM: Detection of the
placental epigenetic signature of the maspin gene in maternal
plasma. Proc Natl Acad Sci USA. 102:pp. 14753–14758. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu LJ and Zhang SL: Expression of RASSF1A
in epithelial ovarian cancers. Eur Rev Med Pharmacol Sci.
19:813–817. 2015.PubMed/NCBI
|
|
17
|
Chim SS, Jin S, Lee TY, Lun FM, Lee WS,
Chan LY, Jin Y, Yang N, Tong YK, Leung TY, et al: Systematic search
for placental DNA-methylation markers on chromosome 21: Toward a
maternal plasma-based epigenetic test for fetal trisomy 21. Clin
Chem. 54:500–511. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tong YK, Jin S, Chiu RW, Ding C, Chan KC,
Leung TY, Yu L, Lau TK and Lo YM: Noninvasive prenatal detection of
trisomy 21 by an epigenetic-genetic chromosome-dosage approach.
Clin Chem. 56:90–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Korenberg JR: Down syndrome phenotypic
mapping. Prog Clin Biol Res. 373:43–52. 1991.PubMed/NCBI
|
|
20
|
Massin N, Frendo JL, Guibourdenche J,
Luton D, Giovangrandi Y, Muller F, Vidaud M and Evain-Brion D:
Defect of syncytiotrophoblast formation and human chorionic
gonadotropin expression in Down's syndrome. Placenta. 22:93–97.
2001. View Article : Google Scholar
|