Introduction
Drug abuse has been a public health and social
problem. According to the survey, nearly 80% of drug users are
abused of new synthetic drugs (1).
Compared with the traditional drugs, the new synthetic drug
produced long-lasting and severe damage to the central nervous
system (CNS) and more easily leaded to addiction (2). Methamphetamine and ketamine are the
most popular new synthetic drugs in recent years. With a chemical
structure of CNS stimulants, methamphetamine has a strong central
excitatory effect and will produce a mental dependence response
after long-term use (3). Ketamine,
commonly known as ‘K powder’, is a new synthetic drug which is
abused worldwide (4). However, the
mechanism of ketamine addiction is poorly understood.
At present, CPP test is one of the most effective
methods for the determination of animal reward (5). It is a simple but effective means of
evaluating drug dependence potential and looking for anti-addiction
drugs (6). The essence of drug
addiction is a pathological memory based on drug-induced gene
expression and changes in synaptic plasticity (7). In recent years, the important
biological functions of post-transcriptional regulation have caught
widespread attention and extensive research, such as the role of
miRNA in drug addiction. Many studies have reported that drug
addiction, such as cocaine addiction, nicotine addiction, opioid
addiction and alcohol addiction are associated with miRNAs
(8–11). Research into the regulatory role
played by miRNAs in methamphetamine and ketamine addiction is in a
preliminary stage.
Studies have shown that some miRNAs are encapsulated
in exosomes which can be picked up by target cells. In this
situation, these miRNAs act as the type of endogenous miRNA that
silences target genes (12). As a
molecular medium, exosomes can transmit information between cells
to cause disease, and can carry a variety of substances including
protein, mRNA, miRNA, DNA, and lipid (13). Moreover, exosomes are involved in
processes of cell migration, cell communication, angiogenesis,
tumor-cell growth and tumor immunity (14–16).
Therefore, our study replicated a CPP test of methamphetamine and
ketamine dependence in rats. Furthermore, by comparing the
expression of miRNAs in serum exosomes of methamphetamine and
ketamine-dependence rats, this study aims to provide insights into
the miRNA-mediated relationship between the two model groups. A
total of 10 differentially expressed (DE) co-miRNAs both in
methamphetamine and ketamine group were further analyzed by
bioinformatics.
Materials and methods
Animals
Sprague-Dawley rats (weight, 170–210 g; age, 2
months) were provided by the Southern Medical University's
Experimental Animal Center (no. SCXK GD 2011–0015; Guangzhou,
China). All animal experimental protocols and operating procedures
were in accordance with the National Institutes of Health (NIH;
Bethesda, MD, USA) guidelines for the experimental use of
laboratory animals. The experiments were approved by the
Experimental Animal Ethics Committee of Southern Medical
University.
Reagents and drugs
Ketamine Hydrochloride Injection (no. 1311-7603) was
purchased from Fujian Gutian Pharmaceutical Co., Ltd. (Fujian,
China). Methamphetamine (no. 1212-9802) was obtained from the
National Narcotics Laboratory (Beijing, China). The
TruSeq® Small RNA Sample Prep kit (no. 4472908) was
purchased from Illumina Corporation. The Qubit® dsDNA HS
Assay kit (no. 4367809) was purchased from Thermo Fisher
Scientific, Inc. (Waltham, MA, USA). U6 was used as the calibration
gene to provide an internal control. Primers were synthesized by
the Guangzhou Ruibo Biotechnology Co., Ltd. (Guangzhou, China). All
other chemicals used in these experiments were of analytical
grade.
Apparatus
The CPP apparatus consisted of two equal-sized
compartments (30 cm long × 30 cm wide × 30 cm high), including one
having a white interior and the other having a black interior that
separated by a wall with a sliding door. Bio-Rad Gradient PCR
(Bio-Rad Laboratories, Inc., Hercules, CA, USA); Agilent 2200
Bioanalyzer; Stratagene Mx3005P Real-time polymerase chain reaction
(PCR) instrument (Agilent Technologies, Inc., Santa Clara, CA,
USA); Agilent 2200 TapeStation (Agilent Technologies, Inc.); DU800
Nucleic Acid/Protein Analyzer (Beckman Coulter, Inc., Brea, CA,
USA); IX53 Fluorescence Inverted Microscope (Olympus Corporation,
Tokyo, Japan); Qubit 2.0 (Thermo Fisher Scientific, Inc.); Hiseq
2500 (Illumina, Inc., San Diego, CA, USA) and ND-1000 Nanodrop
(Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Conditioned place preference (CPP)
paradigm
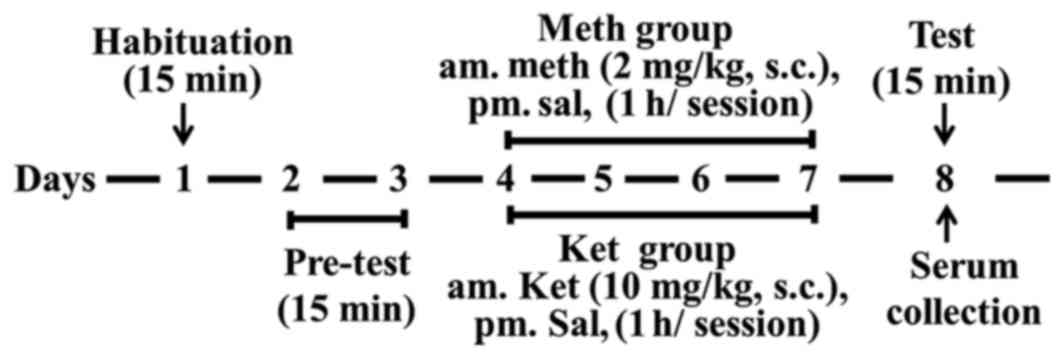
Rats were tested in a CPP apparatus as described
previously. Suitable rats were randomly divided into three groups
of ten: Control group, methamphetamine group and ketamine group.
The method of the CPP paradigm was copied from our previous reports
(17,18) and training process for CPP test is
shown in Fig. 1.
RNA extraction and gene-chip
sequencing
The extraction of RNA from the exosome was performed
in accordance with QIAGEN's miRNeasy Mini lit manual. The TRIzol
method was used for sample extraction and samples were tested with
the Agilent 2200 Tape station system and the Nano drop ND-1000
Spectrophotometer. Total RNA (initial volume 1 µl) from each
acceptable sample was used to construct a miRNA library using the
TruSeq® Small RNA Sample Prep kit (Illumina, Inc.). Then
the library of acceptable miRNAs was submitted to sample
preparation according to the method described in the lllumina HiSeq
2500 User Guide, the final concentration of sample being 10 pM.
Validation of selected DE miRNAs by
reverse transcription-quantitative PCR (RT-qPCR)
Then, 1 µl total RNA from each sample was taken and
synthesized into cDNA by reverse transcription according to the
manual of the reverse transcription kit. One microliter of
reverse-transcribed cDNA was taken and evaluated according to the
instructions for SYBR® Select Master Mix dye by
Real-Time PCR. The 7900HT Fast Real-Time PCR System was used to
amplify the different genes. The program used for amplification was
as follows: 2 min at 50°C, 2 min at 95°C, followed by 40 cycles of
15 sec at 95°C for denaturation, and 1 min at 60°C for annealing
and extension. U6 was used as a reference gene. Three independent
experiments were performed.
miRNA sequence analysis and functional
analysis
The expression of miRNAs in each library was
estimated using the free web-server tool sRNAbench (http://seqanswers.com), where the normalized read
count of each miRNA was shifted by the following formula:
RPM=(miRNAs read number/total map reads) × 1,000,000. Evaluation of
DE of miRNAs was employed to focus our sugar-edge study (http://pubs.acs.org/doi/abs/10.1021/jp053379q). miRNAs
were characterized by having log2FC >1 or <-1,
P<0.01 (19). Target Scan
(http://targetscan.org/), DIANA Tools (http://diana.imis.athena-innovation.gr/DianaTools/),
and miRDB (http://www.mirdb.org/cgi-bin/search.cgi) were used to
predict the target genes of selected DE miRNAs. GO (Gene Ontology)
and KEGG (Kyoto Encyclopedia of Genomes and Genes), accessed on the
David site (http://david.abcc.ncifcrf.gov). The top 10 GO terms
and KEGG pathways were identified. A threshold of P<0.05 was
used to enrich each miRNA's significant biological functions. The
net map of protein-protein interaction (PPI) was analyzed by STRING
online analysis software (https://string-db.org/).
Statistical analysis
Data were expressed as mean ± SD and analyzed by
using one-way analysis of variance, followed by the two-tailed,
least-significant-difference post hoc test. Statistical analyses
were performed using SPSS software (version 17.0; SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
The behavioral alteration induced by
methamphetamine and ketamine
The effects of methamphetamine and ketamine on CPP
rats were shown in Table I. Both
methamphetamine and ketamine greatly increased the activity time
and activity distance in the non-preferred (white) compartment
compared with that of the control rats (P<0.01). Rat activity
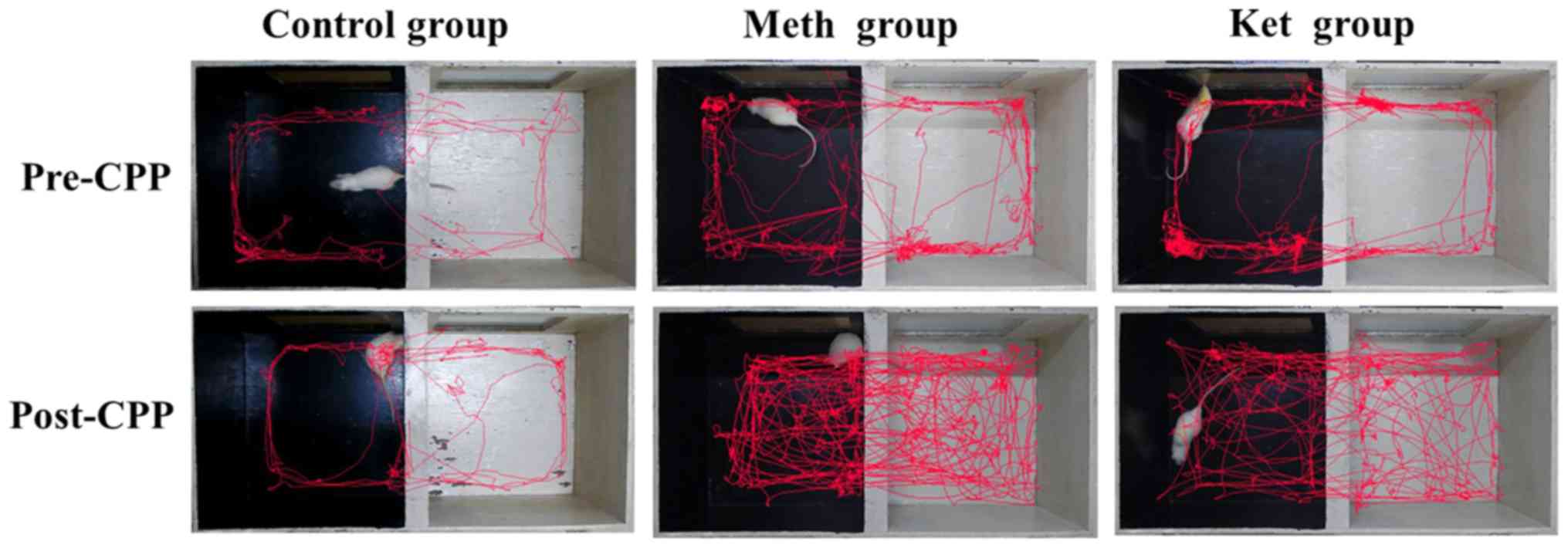
routes in CPP are shown in Fig. 2.
In comparison with those of the control rats, the activity routes
in the non-preferred compartment were increased greatly by
methamphetamine and ketamine.
 | Table I.Time span and exercise distance of
rats in the white compartment following injections of
methamphetamine and ketamine. |
Table I.
Time span and exercise distance of
rats in the white compartment following injections of
methamphetamine and ketamine.
|
| Activity time in
white side (sec) | Activity distance in
white side (cm) |
|---|
|
|
|
|
|---|
| Group | Pre-CPP | Post-CPP | Pre-CPP | Post-CPP |
|---|
| Control | 192.93±22.89 | 202.67±28.24 | 3,182.89±323.22 | 3,349.26±412.28 |
| Methamphetamine | 201.47±25.98 |
460.47±66.71a | 3,242.86±308.59 |
4,475.97±585.74a |
| Ketamine | 195.11±19.32 |
435.09±50.34a | 3,123.37±317.30 |
4,501.02±577.21a |
| F-value | 0.910 | 63.462 | 0.243 | 58.732 |
| P-value | 0.532 |
1.715×10−5 | 0.668 |
2.303×10−4 |
The alteration of miRNAs expression
induced by methamphetamine and ketamine
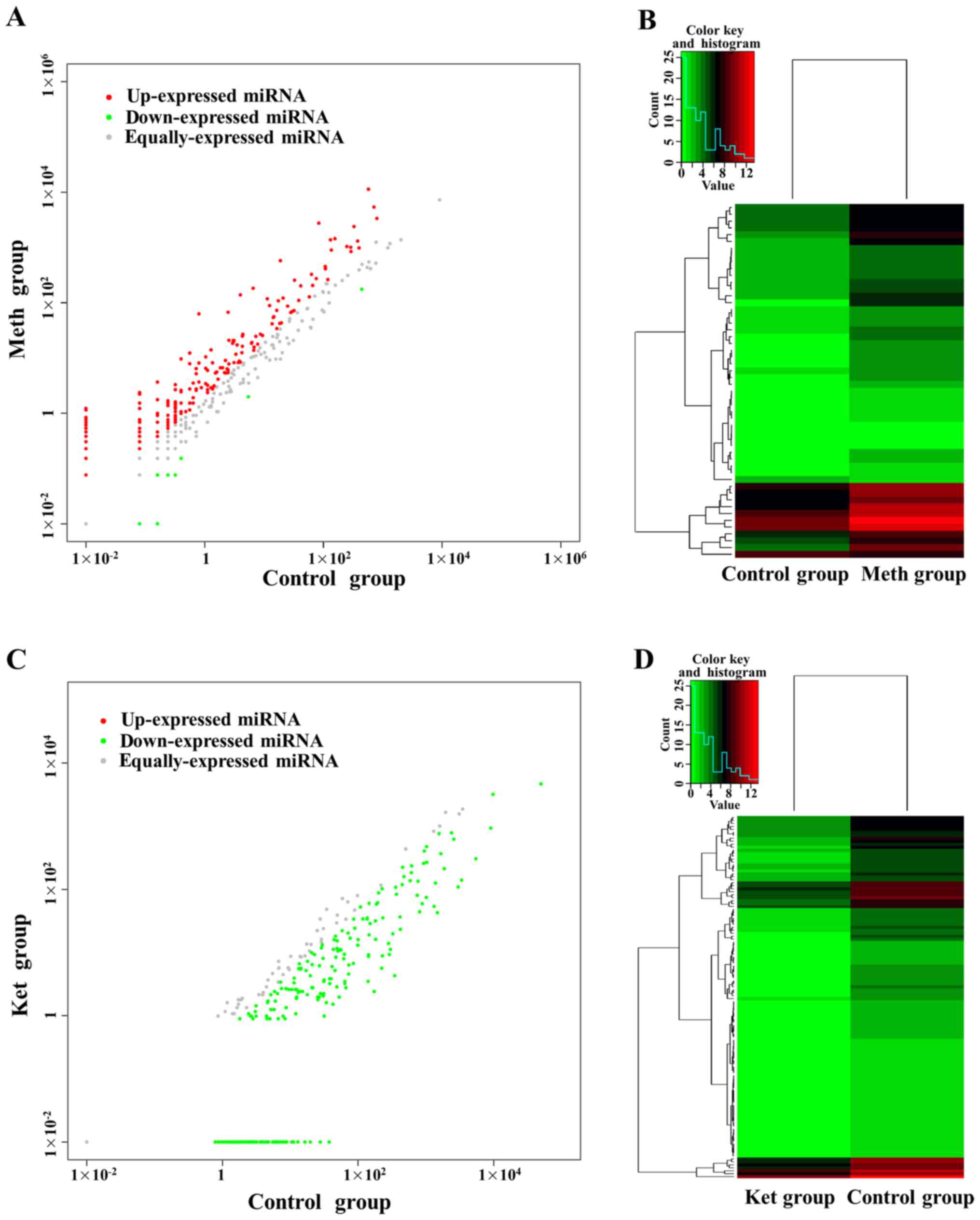
Compared with control group, there were 301 miRNAs
DE in methamphetamine group. Among them, there were 276 miRNAs high
expressed after methamphetamine induction, and 25 were low
expressed (Fig. 3A). Among 276 high
expressed miRNAs, there are 50 miRNAs were significantly DE (log2FC
>1, P<0.01), accounting for 96.15% of the number of
significant DE miRNAs. Among 25 low expressed miRNAs, there are two
miRNAs were significantly DE (log2FC <-1, P<0.01), accounting
for 3.85% of the number of significant DE miRNAs. A heat map of 52
(50 high expressed and two low expressed) significantly DE miRNAs
in serum exosomes of methamphetamine rats showed in Fig. 3B.
Compared with control group, there were 267 miRNAs
DE in ketamine group (Fig. 3C).
Among them, there were all low expressed after ketamine induction,
and 122 miRNAs were DE (log2FC <-1, P<0.01). A heat map of
122 DE miRNAs in serum exosomes of ketamine rats showed in Fig. 3D. Further analyzed of 52 DE miRNAs in
methamphetamine group and 122 DE miRNAs in ketamine group by venn
diagram, we found that ten miRNAs were DE co-miRNAs (Table II). Most of these miRNAs were
up-regulated by methamphetamine, but down-regulated by
ketamine.
 | Table II.Ten differentially expressed co-miRNAs
in the methamphetamine and ketamine groups. |
Table II.
Ten differentially expressed co-miRNAs
in the methamphetamine and ketamine groups.
|
| Control vs. Meth | Control vs.
Ket |
|---|
|
|
|
|
|---|
| miRNA_ID | log2
(fold-change) | P-value | log2
(fold-change) | P-value |
|---|
| rno-miR-128-3p | −1.3445 |
5.83×10−4 | −4.6258 |
5.79×10−5 |
|
rno-miR-133a-3p | 4.7943 |
3.51×10−7 | −7.4342 |
1.98×10−2 |
| rno-miR-152-3p | 2.5283 |
2.52×10−2 | −4.3439 |
2.88×10−4 |
|
rno-miR-181a-5p | 2.6260 |
1.77×10−2 | −4.7517 |
3.51×10−5 |
| rno-miR-192-3p | 6.9336 |
6.15×10−3 | −7.4342 |
1.98×10−2 |
| rno-miR-194-5p | 4.7535 |
9.42×10−7 | −5.3395 |
2.23×10−6 |
| rno-miR-218b | 2.6077 |
2.35×10−2 | −3.6964 |
1.07×10−2 |
| rno-miR-22-5p | 2.7264 |
1.72×10−2 | −3.5319 |
1.31×10−2 |
| rno-miR-362-3p | 2.8471 |
3.30×10−2 | −3.9071 |
4.44×10−3 |
| rno-miR-674-3p | 3.1519 |
1.13×10−2 | −10.9521 |
9.89×10−9 |
Functional analysis
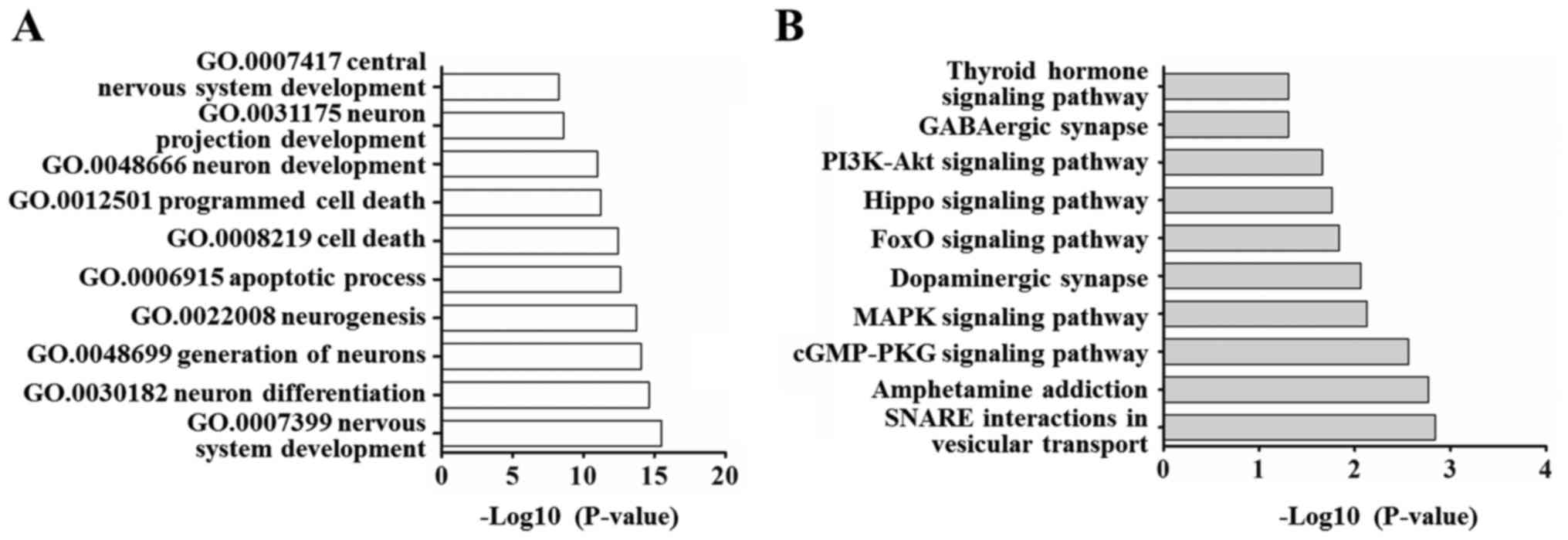
Based on the above analysis, we selected these ten
DE co-miRNAs for target gene prediction. A total of 1151 targets of
these miRNAs were predicted by TargetScan, miRDB, and DIANA tools.
Table III gives more information
on these miRNAs. These predicted targets were classed into 633
kinds according to the biological processes of the GO categories
(P<0.05), specially including nervous system development, neuron
generation and differentiation, apoptotic process as well as cell
death (Fig. 4A). For KEGG pathway
analysis, there were 26 KEGG pathways have statistically
significant (P<0.05), specially including SNARE interactions in
vesicular transport, amphetamine addiction, cGMP-PKG signaling
pathway, dopaminergic synapse and GABAergic synapse. The above
results suggested that the DE miRNAs and their target genes are
related to vesicle transport, amphetamine addiction and
neurotransmitter delivery (Fig.
4B).
 | Table III.Information on selected microRNAs and
the number of candidate genes. |
Table III.
Information on selected microRNAs and
the number of candidate genes.
| miRNA ID | Accession
number | miRNA sequence | No. candidate
genes |
|---|
| rno-miR-128-3p | MIMAT0000834 |
UCACAGUGAACCGGUCUCUUU | 106 |
|
rno-miR-133a-3p | MIMAT0000839 |
UUUGGUCCCCUUCAACCAGCUG | 57 |
| rno-miR-152-3p | MIMAT0000854 |
UCAGUGCAUGACAGAACUUGG | 177 |
| rno-miR-192-3p | MIMAT0017147 |
CUGCCAGUUCCAUAGGUCACAG | 133 |
| rno-miR-218b | MIMAT0017838 |
CAUGGUUAGAUCAAGCACAA | 108 |
| rno-miR-22-5p | MIMAT0003152 |
AGUUCUUCAGUGGCAAGCUUUA | 66 |
| rno-miR-362-3p | MIMAT0017357 |
AACACACCUGUUCAAGGAUUCA | 280 |
| rno-miR-674-3p | MIMAT0005330 |
CACAGCUCCCAUCUCAGAACAA | 63 |
| rno-miR-194-5p | MIMAT0000869 |
UGUAACAGCAACUCCAUGUGGA | 53 |
|
rno-miR-181a-5p | MIMAT0000858 |
AACAUUCAACGCUGUCGGUGAGU | 108 |
Proteins-proteins interactions (PPI)
analysis
A total of 1,151 targets of these ten DE miRNAs were
analyzed by STRING software, and the PPI net map was showed in
Fig. 5. The PPI shows a complex
interaction of target genes encoding proteins. STX5, BET1, CLTB,
CLTC, ACLY, ENO2, TNFRSF1B, SNAP23, VAMP2, STX6 and
VAMP4 have core positions in PPI network.
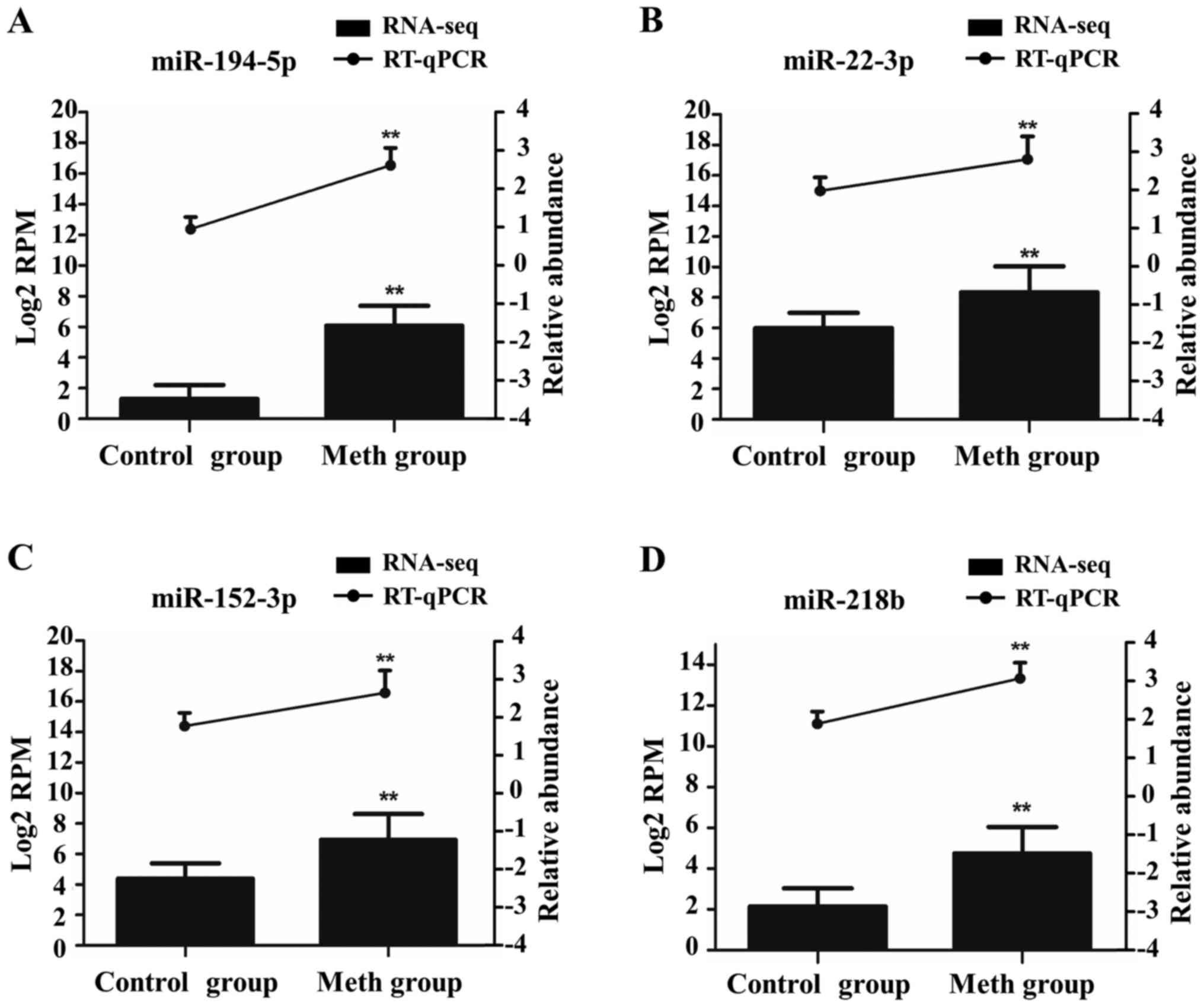
Validation of miRNAs by RT-qPCR
To further verify the accuracy of our gene-chip
analysis, we selected four DE miRNAs from these ten DE co-miRNAs of
methamphetamine group for verification by RT-qPCR. The
dissolution-curve analysis showed no heterozygotes, indicating that
the amplification products were single and free from nonspecific
amplification. The amplification curve indicated that all samples
had entered the expansion platform period, indicating that the
reaction conditions had been set accurately. From Fig. 6, we see that the RT-qPCR results are
consistent with those of gene-chip sequencing.
Discussion
The CPP test is the classic experiment to detect
psychological drug dependence (20).
Our results showed that methamphetamine and ketamine significantly
increased the activity time and distance of rats in the
non-preferred compartment after CPP, which suggest that we have
replicated the CPP test successfully. From the difference analysis
of miRNA expression, methamphetamine could up regulate the
expression of most miRNA, and ketamine down regulate the expression
of miRNA. These suggest that different addictive drugs have
different effects on the expression of miRNA in rats.
There are ten DE miRNAs both in the two model
groups, such as miR-128-3p, miR-133a-3p, miR-152-3p and
miR-181a-5p. By GO analysis, we found that the biological processes
of these ten miRNAs target genes mainly enriched in nervous system
development, neuron generation and differentiation, apoptotic
process as well as cell death. The essence of drug addiction is a
pathological memory based on drug-induced gene expression and
changes in synaptic plasticity (21).
From KEGG pathway analysis, we also found that these
target genes mainly located in SNARE interactions in vesicular
transport, amphetamine addiction, cGMP-PKG signaling pathway,
dopaminergic synapse and GABAergic synapse. Many of these pathways
involved in drug addiction. In our study, these ten DE miRNAs were
all from serum exosome. Exosome is a nano-sized vesicular body
(22). It can transport miRNA to
target organs or cells, so pathway of SNARE interactions in
vesicular transport may related to DE co-miRNAs transportion and
then involved in drug addiction. The mesolimbic dopamine system is
involved in the rewarding effects of almost all addictive drugs
(23). Central nervous stimulants
increased the release of dopamine and the concentration of dopamine
in synaptic gap, and then caused addictive behavior (24). GABA energy system is the most
important inhibitory system in the CNS (25). It has been found that the withdrawal
of morphine addiction leads to an increase in the inhibitory effect
of GABA on systemic (26),
suggesting that morphine addiction is closely related to the GABA
system.
The PPI shows a complex interaction of target genes
encoding proteins. Many target genes are related to vesicle
transport, neurons protection and apoptosis. As the main structural
component of the lattice-type cytoplasmic face of coated pits and
vesicles, CLTB entrap specific macromolecules during
receptor-mediated endocytosis (27).
SNAP23 is an essential component of the high affinity
receptor for the general membrane fusion machinery and is an
important regulator of transport vesicle docking and fusion
(28). In nervous tissue,
ACLY may be involved in the biosynthesis of acetylcholine
(29). The activity of cholinergic
transmitters in the brain is closely related to cognitive
processes. Knockout studies in mice also suggest a role of
TNFRSF1B in protecting neurons from apoptosis by stimulating
antioxidative pathways (30).
RAD21 plays a role in apoptosis, via its cleavage by
caspase-3/CASP3 or caspase-7/CASP7 during early steps of apoptosis
(31).
In this study, methamphetamine significantly
increased the activity time and distance of rats in the
non-preferred compartment after CPP and up regulated most miRNAs
expression. Ketamine could also increase the activity time and
distance of rats in the non-preferred compartment after CPP, but
down regulated all miRNAs expression. It suggests that the
mechanism of methamphetamine and ketamine addiction is different.
As to the biological processes and pathways are involved in the
development of addiction at multiple stages, we speculate these ten
DE miRNAs both in methamphetamine group and ketamine group are
closely related to drug addiction. These biological processes of
target genes may be common or partly involved in the process of
methamphetamine and ketamine addiction, but further studies are
necessary. Because of their role in regulating important molecules,
our results are important for theory.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (no. 81673628 and 81229003), Guangdong Science
and Technology Project (no. 2016A020226003), Natural Science
Foundation of Guangdong Province (no. 2014A030310251) and State of
traditional Chinese medicine of Guangdong province (no. 20162085
and 20171165).
References
|
1
|
Tomás-Rosselló J, Rawson RA, Zarza MJ,
Bellows A, Busse A, Saenz E, Freese T, Shawkey M, Carise D, Ali R
and Ling W: United nations office on drugs and crime international
network of drug dependence treatment and rehabilitation resource
centres: Treatnet. Subst Abus. 31:251–263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vinogradova MV, Kryukova EM, Kulyamina OS,
Vapnyarskaya OI and Sokolova AP: Approaches to the study of the
status and trends of drug abuse, rehabilitation and reintegration
of drug users. Biosci Biotechnol Res Asia. 11:1505–1514. 2014.
View Article : Google Scholar
|
|
3
|
Baptista S, Lourenço J, Milhazes N, Borges
F, Silva AP and Bacci A: Long-term treatment with low doses of
methamphetamine promotes neuronal differentiation and strengthens
long-term potentiation of glutamatergic synapses onto dentate
granule neurons. eNeuro. 3:pii: ENEURO2016. View Article : Google Scholar
|
|
4
|
Stoicea N, Versteeg G, Florescu D, Joseph
N, Fiorda-Diaz J, Navarrete V and Bergese SD: Ketamine-based
anesthetic protocols and evoked potential monitoring: A
risk/benefit overview. Front Neurosci. 10:372016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tzschentke TM: Measuring reward with the
conditioned place preference (CPP) paradigm: Update of the last
decade. Addict Biol. 12:227–462. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tzschentke TM: Measuring reward with the
conditioned place preference paradigm: A comprehensive review of
drug effects, recent progress and new issues. Prog Neurobiol.
56:613–672. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun W: Dopamine neurons in the ventral
tegmental area: Drug-induced synaptic plasticity and its role in
relapse to drug-seeking behavior. Curr Drug Abuse Rev. 4:270–285.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hollander JA, Im HI, Amelio AL, Kocerha J,
Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD and Kenny
PJ: Striatal microRNA controls cocaine intake through CREB
signalling. Nature. 466:197–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maccani MA, Avissar-Whiting M, Banister
CE, McGonnigal B, Padbury JF and Marsit CJ: Maternal cigarette
smoking during pregnancy is associated with downregulation of
mir-16, mir-21, and mir-146a in the placenta. Epigenetics.
5:583–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng H, Zeng Y, Zhang X, Chu J, Loh HH
and Law PY: mu-Opioid receptor agonists differentially regulate the
expression of miR-190 and NeuroD. Mol Pharmacol. 77:102–109. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baek MN, Jung KH, Halder D, Choi MR, Lee
BH, Lee BC, Jung MH, Choi IG, Chung MK, Oh DY and Chai YG:
Artificial microRNA-based neurokinin-1 receptor gene silencing
reduces alcohol consumption in mice. Neurosci Lett. 475:124–128.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Emmanouilidou E, Melachroinou K,
Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L and
Vekrellis K: Cell-produced alpha-synuclein is secreted in a
calcium-dependent manner by exosomes and impacts neuronal survival.
J Neurosci. 30:6838–6851. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Simons M and Raposo G: Exosomes-vesicular
carriers for intercellular communication. Curr Opin Cell Biol.
21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai R, Yeo R, Tan S, Zhang B, Yin Y, Sze
N, Choo A and Lim S: Biochemical potential of MSC exosome.
Cytotherap. 16:S432014. View Article : Google Scholar
|
|
15
|
Sahoo S, Klychko E, Thorne T, Misener S,
Schultz KM, Millay M, Ito A, Liu T, Kamide C, Agrawal H, et al:
Exosomes from human CD34(+) stem cells mediate their proangiogenic
paracrine activity. Circ Res. 109:724–728. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang HW, Wang J, Ding F, Callahan K,
Bratkowski MA, Butler JS, Nogales E and Ke A: Architecture of the
yeast Rrp44 exosome complex suggests routes of RNA recruitment for
3′ end processing. Proc Natl Acad Sci USA. 104:pp. 16844–16849.
2007; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu W, Peng QX, Lin XL, Luo CH, Jiang MJ,
Mo ZX and Yung KK: Effect of rhynchophylline on the expression of
p-CREB and sc-Fos in triatum and hippocampal CA1 area of
methamphetamine-induced conditioned place preference rats.
Fitoterapia. 92:16–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Liu W, Peng Q, Jiang M, Luo C, Guo
Y, Liu Y, Fang M and Mo Z: Effect of rhynchophylline on conditioned
place preference on expression of NR2B in methamphetamine-dependent
mice. Biochem Biophys Res Commun. 452:695–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Manzanedo C, Aguilar MA, Rodriguez-Arias M
and Miñarro J: Conditioned place preference paradigm can be a mouse
model of relapse to opiates. Neurosci Res Commun. 28:23–29. 2001.
View Article : Google Scholar
|
|
21
|
Van den Oever MC, Spijker S and Smit AB:
The synaptic pathology of drug addiction. Adv Exp Med Biol.
970:469–491. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gonda DD, Akers JC, Kim R, Kalkanis SN,
Hochberg FH, Chen CC and Carter BS: Neuro-oncologic applications of
exosomes, microvesicles, and other nano-sized extracellular
particles. Neurosurgery. 72:501–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang MH, Jung MS, Lee MJ, Yoo KH, Yook YJ,
Park EY, Choi SH, Suh YJ, Kim KW and Park JH: Gene expression
profiling of the rewarding effect caused by methamphetamine in the
mesolimbic dopamine system. Mol Cells. 26:121–130. 2008.PubMed/NCBI
|
|
24
|
During MJ, Bean AJ and Roth RH: Effects of
CNS stimulants on the in vivo release of the colocalized
transmitters, dopamine and neurotensin, from rat prefrontal cortex.
Neurosci Lett. 140:129–133. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vaz GC, Bahia AP, de Figueiredo
Müller-Ribeiro FC, Xavier CH, Patel KP, Santos RA, Moreira FA,
Frézard F and Fontes MA: Cardiovascular and behavioral effects
produced by administration of liposome-entrapped GABA into the rat
central nervous system. Neuroscience. 285:60–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nugent FS: VTA GABAergic plasticity: An
inhibitory synaptic model of drug addiction. Springer; New York,
NY: 2011
|
|
27
|
Borner GH, Antrobus R, Hirst J, Bhumbra
GS, Kozik P, Jackson LP, Sahlender DA and Robinson MS: Multivariate
proteomic profiling identifies novel accessory proteins of coated
vesicles. J Cell Biol. 197:141–160. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bauer MC, O'Connell DJ, Maj M, Wagner L,
Cahill DJ and Linse S: Identification of a high-affinity network of
secretagogin-binding proteins involved in vesicle secretion. Mol
Biosyst. 7:2196–2204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Byrne FL, Poon IK, Modesitt SC, Tomsig JL,
Chow JD, Healy ME, Baker WD, Atkins KA, Lancaster JM, Marchion DC,
et al: Metabolic vulnerabilities in endometrial cancer. Cancer Res.
74:5832–5845. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Surhone LM, Tennoe MT and Henssonow SF:
Tnfrsf1b. Betascript Publishing; 2011
|
|
31
|
Pati D, Zhang N and Plon SE: Linking
sister chromatid cohesion and apoptosis: Role of Rad21. Mol Cell
Biol. 22:8267–8277. 2002. View Article : Google Scholar : PubMed/NCBI
|