Introduction
The development of high-throughput RNA sequencing
technology has led to the discovery of thousands of non-coding RNA
(ncRNA) genes (1,2). Until now, the number of newly
identified ncRNA genes has been increasing and outnumbers the
number of coding transcripts (3,4).
Additionally, ncRNAs have been demonstrated to participate in a
plethora of physiological and pathological processes (5–7).
Previously, ncRNAs were believed to be the product of random
transcription without any intrinsic function (8,9). With
the development of ncRNA research, accumulating evidence has
unveiled that ncRNAs are functionally important molecules in the
cell (5). In general, ncRNAs are a
heterogeneous group of molecules and may be categorized into
microRNA (miR), long ncRNA (lncRNA), circular RNA (circRNA),
endogenous small interfering RNA, Piwi-associated small RNA, small
nucleolar RNA (snoRNA), sno-derived RNA, transcription initiation
RNA and miR-offset RNA (10). Among
them, the function of miR, lncRNA and circRNA have attracted
attention from academia. To date, miRs are the most studied ncRNAs,
with hundreds of potential target genes, most of which have been
bioinformatically predicted (11–13).
miR-449a, located in the first intron of cell
division cycle (CDC)20B (chromosome 5q11), belongs to the
miR-34/miR-449 families and has been reported to be downregulated
in tumors (14). Previous studies
also revealed that miR-449a functions as a tumor suppressor by
regulating cell proliferation, cycle procession, apoptosis,
migration and invasion in multiple human malignances (15–17). As
studies focusing on miR-449a and hepatocellular carcinoma (HCC) are
currently rather scarce, the role of miR-449a in HCC tumorigenesis
remains unclarified.
The incidence of liver cancer is high throughout the
world, particularly in East and South-East Asia and Northern and
Western Africa (18). Every year,
cases in China alone account for ~50% of the total number of cases
and mortalities (19). It has been
reported that HCC is the most common type (~80%) of primary liver
cancer in the world (18).
Currently, with the improvement of hygiene and sanitation,
hepatitis B virus (HBV) and hepatitis C virus (HCV) infection are
decreasing (20). The understanding
of the molecular pathogenesis of HCC has also significantly
improved. The incidence of liver cancer is decreasing in some
high-risk areas, including China and Japan (18,20).
However, HCC remains to have a high incidence rate; therefore, it
is urgent that more effective diagnostic techniques or replacement
therapies are identified for patients with HCC. Collective data
from high-throughput analyses of a large number of samples have
offered a precise landscape of HCC genetic alterations (21). Indeed, it has enabled the delineation
of some key events that may dominate tumor development and
progression. It is hoped that translation of this knowledge into
targets and biomarkers may impact HCC decision-making and
ultimately improve patient outcomes.
The aim of the present work was to systematically
review the current literature and to utilize the public Gene
Expression Omnibus (GEO) databases to determine the role of
miR-449a and its significance in HCC, thereby helping to understand
and explore the pathogenesis of miR-449a in HCC. In addition, a
preliminary bioinformatics analysis was conducted for miR-449a to
investigate the novel potential pathways that miR-449a may
participate in.
Materials and methods
Literature retrieval
To identify the role of miR-449a in HCC, a
systematic literature search was conducted in the electronic
databases PubMed (https://www.ncbi.nlm.nih.gov/pubmed), Embase
(https://www.embase.com/), Web of Science
(https://apps.webofknowledge.com/), China
Biology Medicine disc (CBMdisc) (http://www.sinomed.ac.cn/zh/), China National
Knowledge Infrastructure (CNKI) (http://www.cnki.net/), WANFANG DATA (http://g.wanfangdata.com.cn/), Chongqing VIP
(http://qikan.cqvip.com/) and Google Scholar
(https://scholar.google.com) until
October 2017. The key words were as follows: ‘Malignan* OR cancer
OR tumor OR tumor OR neoplas* OR carcinoma,’ ‘hepatocellular OR
liver OR hepatic OR HCC’ and ‘miR-449a OR miRNA-449a OR
microRNA-449a OR miR 449a OR miRNA 449a OR microRNA 449a.’ The
articles were selected for the study if they met the following
criteria: i) Studies published in Chinese or English; ii) studies
assessing the dysregulation of miR-449a in HCC; iii) the expression
profiles of miR-449a in patients with HCC and healthy controls were
available; and iv) studies conducted on human tissues or body
fluids, such as serum and urine. A total of 9 papers from PubMed, 5
from Google Scholar and 19 from the Web of Science were identified
in the initial search, while only one study was found in the four
Chinese databases (CBMdisc, CNKI, WANFANG DATA and Chongqing VIP).
Following the elimination of duplicate studies, eight studies met
the inclusion criteria. Review articles were not included in the
present study. In the second step, the references and related
citations of the eight articles were checked for additional
information.
GEO microarray chip screening
To guarantee more comprehensive data collection, the
GEO database of the National Center for Biotechnology Information
of the National Institute of Health of USA (http://ncbi.nlm.nih.gov/geo/) was searched, which is a
public functional genomics data repository. The following terms
were used: ‘Malignan* OR cancer OR tumor OR tumor OR neoplas* OR
carcinoma’ AND ‘hepatocellular OR liver OR hepatic OR HCC.’
Eligible microarrays were downloaded to further extract the
miR-449a expression values, which contained two sides of the
miR-449a expression level (HCC and normal control). In each
dataset, the mean value and standard deviation were calculated.
Following this, all datasets were gathered for the assessment of
miR-449a expression in HCC with STATA 12.0 (StataCorp LLC, College
Station, TX, USA). The standardized mean difference (SMD) and 95%
confidence interval (CI) were determined for the pooled values. As
the SMD is the mean difference (MD) divided by the standard
deviation (SD) and MD is the mean difference of the tumor group and
the control group (22), SMD=0 meant
that there was no significant difference of miR-449a expression in
HCC tissues and normal control, SMD<0 means that miR-449a was
downregulated in HCC and SMD>0 means that miR-449a was
upregulated in HCC. A sensitivity analysis was also conducted with
STATA 12.0 ‘metainf’ command line (23).
Screening differentially expressed
genes and predicting target genes
GEO Profiles (https://www.ncbi.nlm.nih.gov/geoprofiles/) was
searched to explore the differentially expressed genes associated
with miR-449a in HCC using the terms ‘HCC AND miR-449a.’ Finally,
one dataset (GSE74710) (24)
demonstrated that the transient transfection of miR-449a mimics
into an HCC cell line, HLE, identified putative target genes of
miR-449a. The dataset was downloaded to calculate the fold change
(FC) between the group transfected with the miR-449a mimics and the
negative control by R/Bioconductor (version 3.4.2, https://www.R-project.org/) FC<0.5 and P<0.05
were selected.
On the other hand, target genes of miR-449a were
predicted using miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
(25), which combined 12 existing
miR-target prediction programs (miRWalk, Microt4, miRanda,
mirbridge, miRDB, miRMap, miRNAMap, Pictar2, PITA, RNA22, RNAhybrid
and TargetScan) to provide comprehensive potential targets for
miR-449a. The genes predicted in more than five prediction software
programs were regarded as reliable targets of miR-449a.
In addition, a novel computerized approach, Natural
Language Processing (NLP), was performed to obtain all the genes
related to HCC (details have been described previously) (26). Finally, three components that were
overlapped were identified for further bioinformatics analysis.
Functional and signaling pathway
analysis
The functional and signaling pathway analyses of the
selected genes were performed on a public database platform, the
Database for Annotation, Visualization and Integrated Discovery
(DAVID) (http://david.ncifcrf.gov/). The
analyses included Gene Ontology (GO) function analysis (http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) analysis. The GO function
analysis included three categories, namely, the biological
processes (BP), cellular components (CC) and molecular functions
(MF). The results of the GO analysis were visualized as
co-expression networks via Cytoscape v3.4.0 (http://cytoscape.org/) with the BiNGO plug-in, and the
result of the KEGG analysis was visualized as a bar chart.
Protein-protein interaction (PPI)
networks analysis
The potential targets were also input into the
STRING database (http://string-db.org/) to explore the hub genes
involved in HCC. Network nodes represented the proteins, and edges
represented the protein-protein associations. All the parameter
settings were selected as defaults.
Statistical analysis
The scatterplots that exhibited miR-449a expression
from all the GEO datasets were generated using GraphPad Prism 5.0
(GraphPad Software, Inc., La Jolla, CA, USA). The significance of
the difference between the HCC and non-cancerous liver tissues was
analyzed using Student's t-tests. P<0.05 was considered to
indicate a statistically significant difference. For meta-analysis,
fixed-effects models or random-effects models were used for pooling
SMD depending on inconsistency statistics (I2). When
I2 >50%, a random-effects model was selected; when
I2 <50%, a fixed-effects model was preferentially
used (27). Furthermore, Begg's
funnel plot was applied for calculating publication bias (28).
Results
Systematic review of original
literature
With the search and screen criteria, eight studies
exploring the role of miR-449a in HCC were obtained. The data from
the selected studies included HCC cell lines, animal models and
human biopsy specimens. However, the studies on miR-449a are
currently limited and effective data could not be extracted from
the articles. Thus, it was not possible to perform a meta-analysis.
The characteristics of the eight included studies are presented in
Table I.
 | Table I.Characteristics of the eight included
studies. |
Table I.
Characteristics of the eight included
studies.
| Author, year | Country | Tissue | Sample, n
(HCC/control) | miR | Targets | (Refs.) |
|---|
| Wang et al,
2017 | China | Liver cancer | 18/18 | miR-449a | CXCL5 | (29) |
| Liu et al,
2016 | China | HCC | 40/40 | miR-449a | ADAM10 | (30) |
| Chen et al,
2015 | China | HCC | 66/18 | miR-449a | FOS, MET | (31) |
| Liu et al,
2016 | China | Liver cancer | 48/48 | miR-449a | POU2F1, CAPN6 | (32) |
| Zhang et al,
2016 | China | HCC cell lines | – | miR-449a | CREB5 | (33) |
| Buurman et
al, 2012 | Germany | HCC | 23/0 | miR-449 family | C-MET, SOX4 | (36) |
| Sandbothe et
al, 2017 | Germany | Liver cancer | 61/4 | miR-449 family | SOX4 | (24) |
| Sarma et al,
2012 | America | HCV, AH, | 10 HCV, 10 AH | miR-449a | NOTCH1 | (34) |
|
|
| NASH | 10 NASH, 10
control |
|
|
|
The first study conducted by Wang et al
(29) collected 18 liver cancer
tissues along with their adjacent normal controls to explore the
effect of miR-449a on liver cancer migration and invasion. This
study provided preliminary evidence that miR-449a could serve as a
tumor suppressor to influence the migration and invasion of liver
cancer through targeting C-X-C motif chemokine 5 (CXCL5).
Consistent results were also observed in four HCC cell lines.
Another study conducted by Liu et al
(30) measured miR-449a expression
in 40 pairs of HCC tissues and adjacent normal tissues, as well as
four HCC cell lines, and demonstrated that miR-449a expression was
decreased in the HCC tissues and four cell lines. They further
explored the mechanism underlying the inhibitory effects of
miR-449a on the growth and metastasis of HCC cells and revealed
that miR-449a functioned as a tumor suppressor miR by inhibiting
the cell proliferation, colony formation, migration and invasion of
HCC by partially repressing a disintegrin and metalloproteinase
domain-containing protein 10 (ADAM10) expression.
Chen et al (31) investigated the miR-449a expression
level in 77 HCCs and 18 normal controls. They found that the level
of miR-449a in HCC was notably lower when compared to that in the
normal controls. Furthermore, the portal vein tumor thrombus
tissues displayed a more significant reduction of miR-449a
expression, which indicated that the reduction of miR-449a in HCCs
was notably associated with a more aggressive tumor phenotype. They
also conducted a log-rank test, which revealed that the reduction
of miR-449a was associated with short disease-free survival in
patients with HCC. The study revealed that miR-449a may suppress
the epithelial-mesenchymal transition (EMT) and the metastasis of
HCC by inhibiting FOS and Met expression and subsequently
suppressing the downstream signaling.
Another study by Liu et al (32) explored the cellular function of
miR-449a in 48 cases of liver cancer. By comparison with adjacent
tissues, it was revealed that miR-449a was decreased in the liver
cancer tissues, and the loss of miR-449a in the liver cancer
tissues was associated with tumor progression, which suggested that
miR-449a may be a suppressor of cancer metastasis. The results
obtained from four human liver cancer cell lines (HepG2, 7404, 7721
and 7405) were consistent with the liver tissues. Additionally,
they performed a series of bioinformatics analyses and validated
experiments, which revealed that miR-449a could induce
G1 arrest of the liver cancer cells, suppress cell
proliferation and promote cell apoptosis. It was also identified
that miR-449a affected the biological behavior of liver cancer
cells by the downregulation of calpain 6 (CAPN6) and POU class 2
homeobox 1 (POU2F1).
The study conducted by Zhang et al (33) explored the molecular mechanisms of
the interactions between miR-449a and HBV infection and identified
that miR-449a expression was downregulated in the HCC cells.
Ectopic expression of miR-449a in HCC cells, to a large extent,
boosted HBV replication, transcription, progeny virion secretion
and antigen expression in a dose-dependent manner. They reported
that miR-449a directly targeted the cyclic adenosine monophosphate
(cAMP)-responsive factor and bound to cAMP responsive element
binding protein 5 (CREB5), which influenced Farnesol-X-Receptor
(FXR)α expression and facilitated HBV replication. Similarly, Sarma
et al (34) analyzed the
genome-wide miR in liver biopsies obtained from patients with
chronic HCV infection and observed a downregulation of miR-449a
compared with the level in the normal liver. It is well known that
patients with HCV infection have a high risk of developing HCC
(35).
Buurman et al (36) investigated 23 primary HCCs of various
Gleason grades, as well as HCC cell lines and tumor xenografts, all
of which demonstrated a reduced expression of miR-449a and an
increased expression of c-MET in the samples. This indicated that
miR-449a may function through targeting c-MET in
hepatocarcinogenesis. Furthermore, analysis of the tissue samples
revealed that the lowest concentrations of miR-449 were associated
with high levels of c-MET, which appeared in Gleason grade 1,
suggesting that the deregulation of miR-449a was mainly effective
in the early stages of HCC tumorigenesis.
Recently, Sandbothe et al (24) further investigated the function of
the miR-449 family using microarray analysis and public databases
to identify their binding specificities, putative target genes and
regulated pathways. They demonstrated that miR-449 family members
significantly regulated cell cycle control, transforming growth
factor (TGF)-β signaling, hepatocyte growth factor signaling and
the Wnt/β-catenin signaling pathways that were frequently altered
in HCC. Following comprehensive analysis, they focused their study
on the signaling of TGF-β by targeting SOX4, which served a dual
role in HCC, acting as a tumor suppressor during the early stages
of liver damage, but promoting tumor progression and metastasis in
advanced HCC.
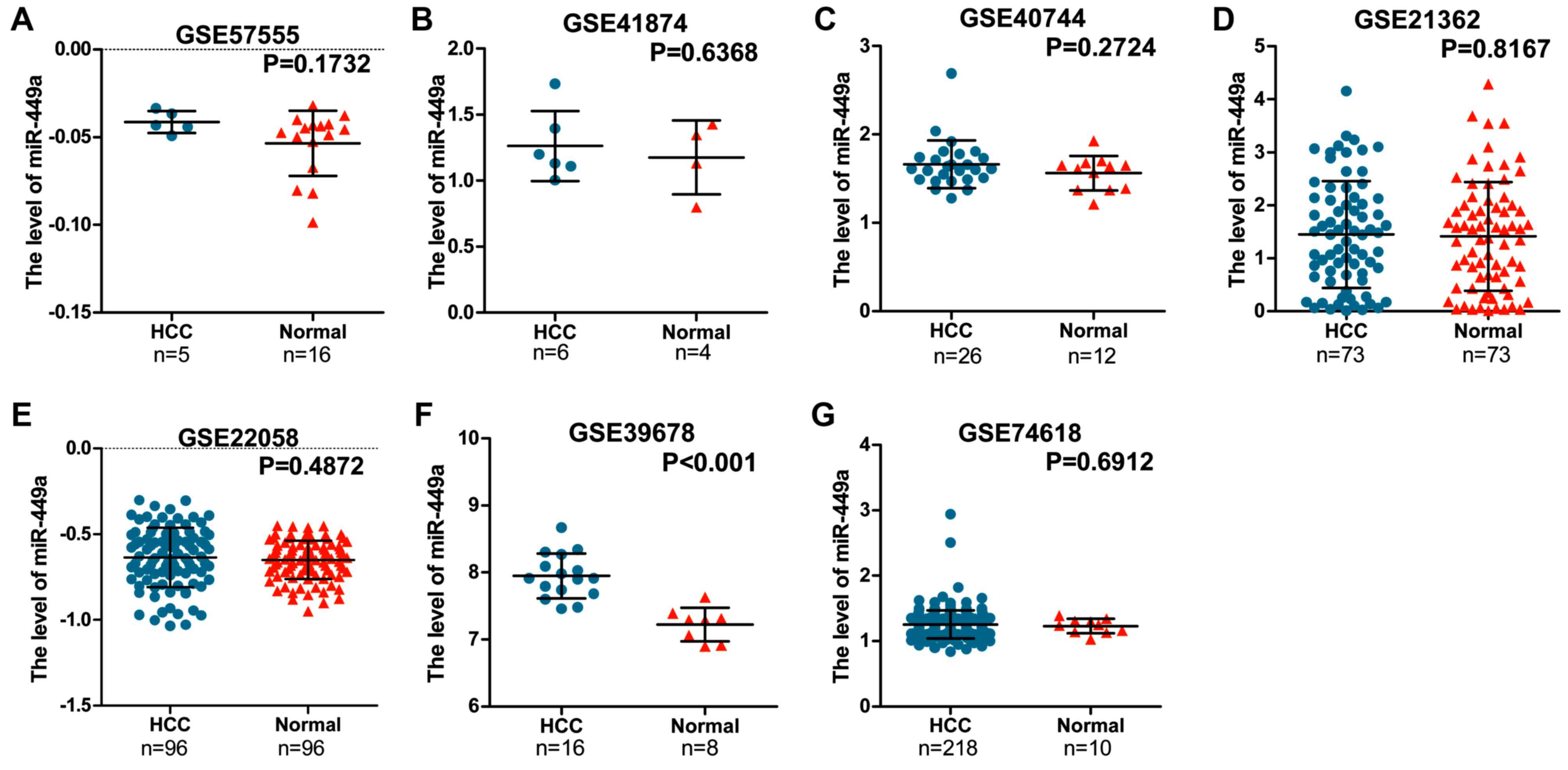
Determining the expression of miR-449a
in HCC by gathering seven GEO datasets
GEO datasets were searched to assess the expression
of miR-449a between HCC and non-cancer tissues. miR-449a levels in
all seven microarray chips [GSE57555 (37), GSE41874 (38), GSE40744 (39), GSE21362 (40), GSE22058 (41), GSE39678 (42) and GSE74618 (43)] were presented in Fig. 1. Meta-analysis was conducted with
seven datasets to assess the miR-449a expression level in patients
with HCC in another manner, on account of the fact that the GEO
microarrays could not present the role of miR-449a in HCC directly.
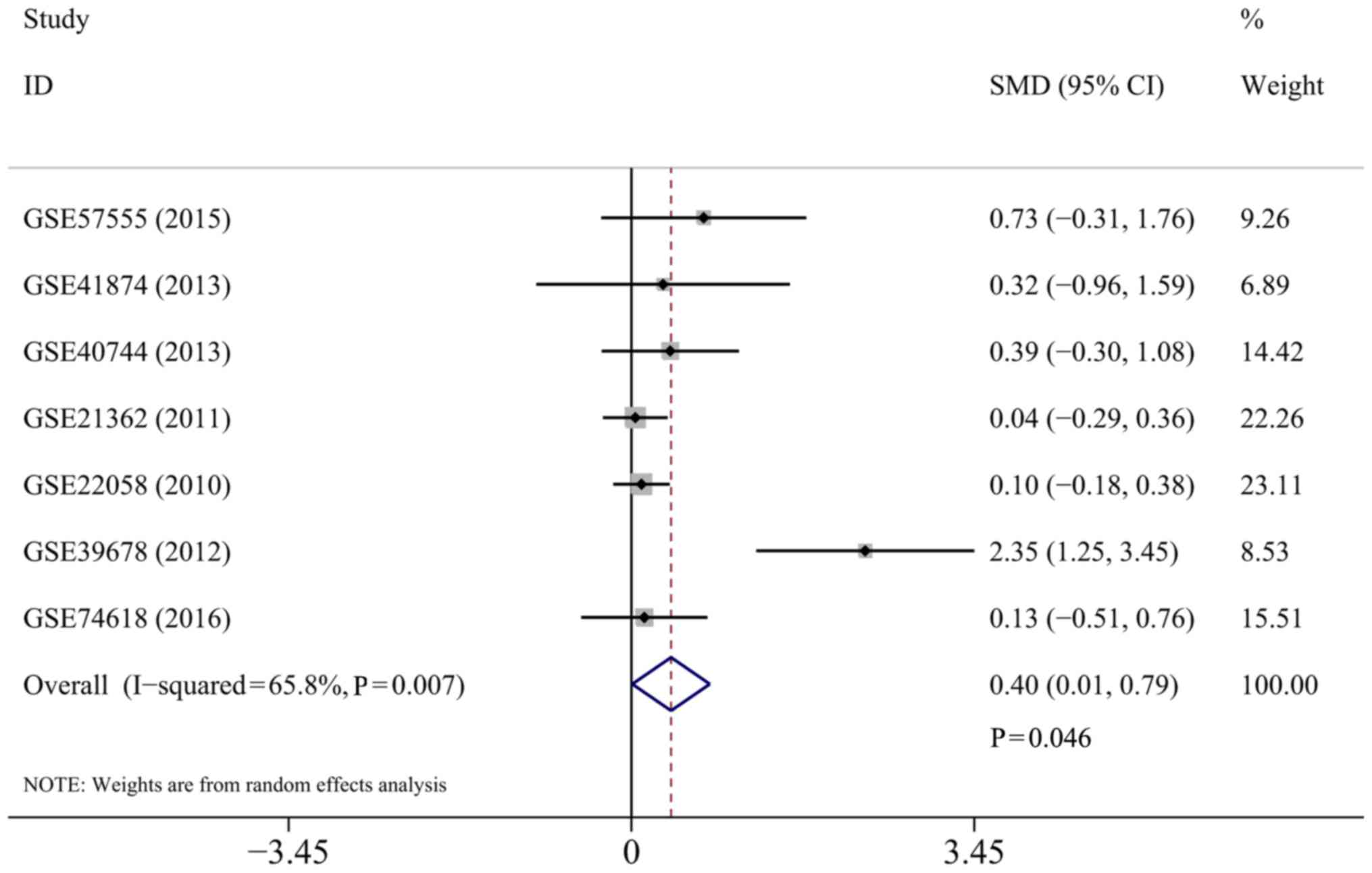
In total, 440 patients with HCC and 219 normal controls were
selected for the meta-analysis. A random-effects model was selected
to calculate the pooled SMD and 95% CI, and significant
heterogeneity was identified among individual datasets
(I2=65.8%; P=0.007). The results demonstrated that a
significant difference was observed between the HCC groups and the
normal controls (SMD=0.40; 95% CI, 0.01–0.79; P=0.046), suggesting
that the miR-449a expression levels were increased in patients with
HCC rather than in the normal controls (Fig. 2).
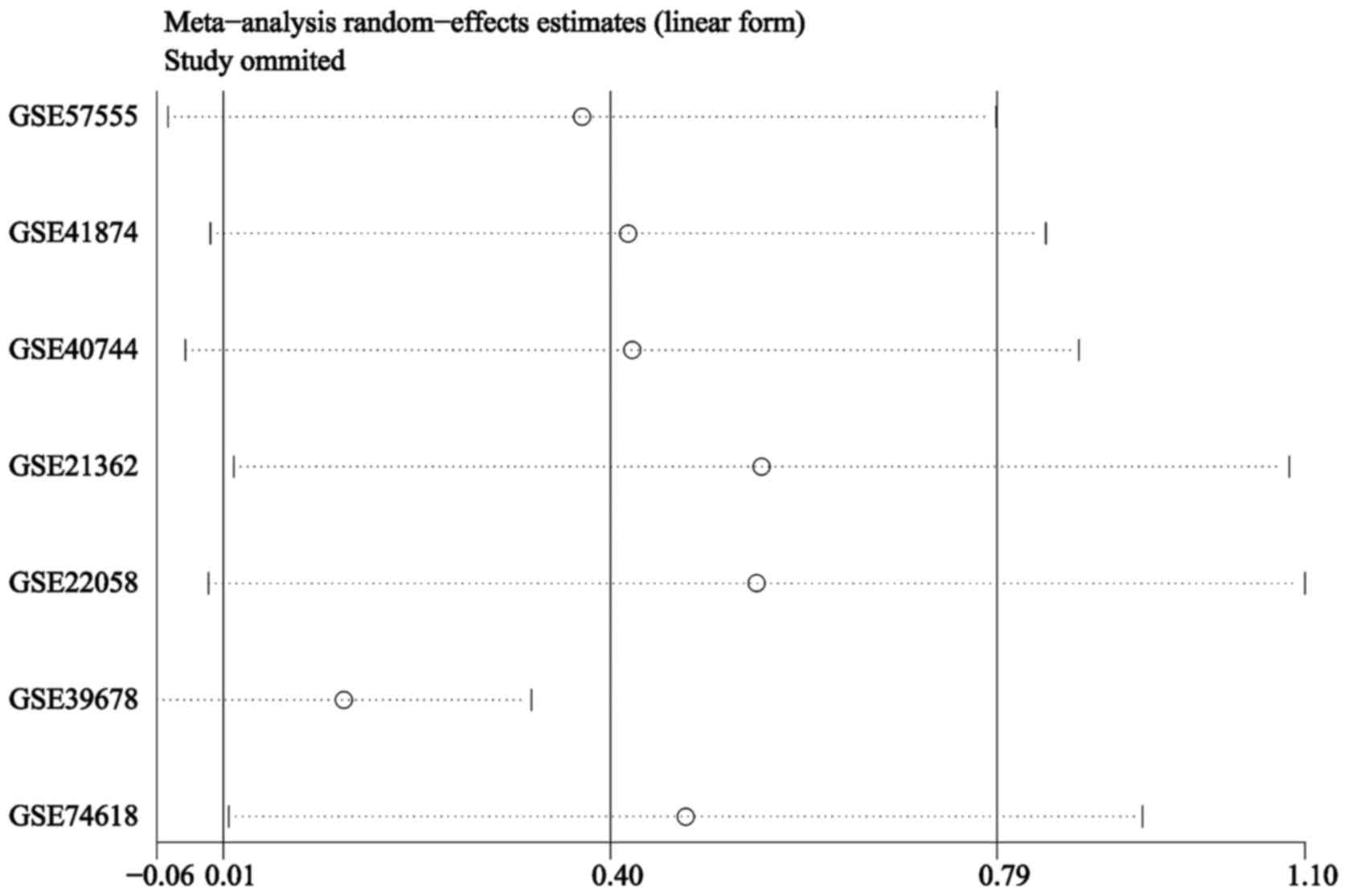
The results of the sensitivity analysis revealed
that heterogeneity may be confounded by the GSE39678 dataset
(Fig. 3). The heterogeneity
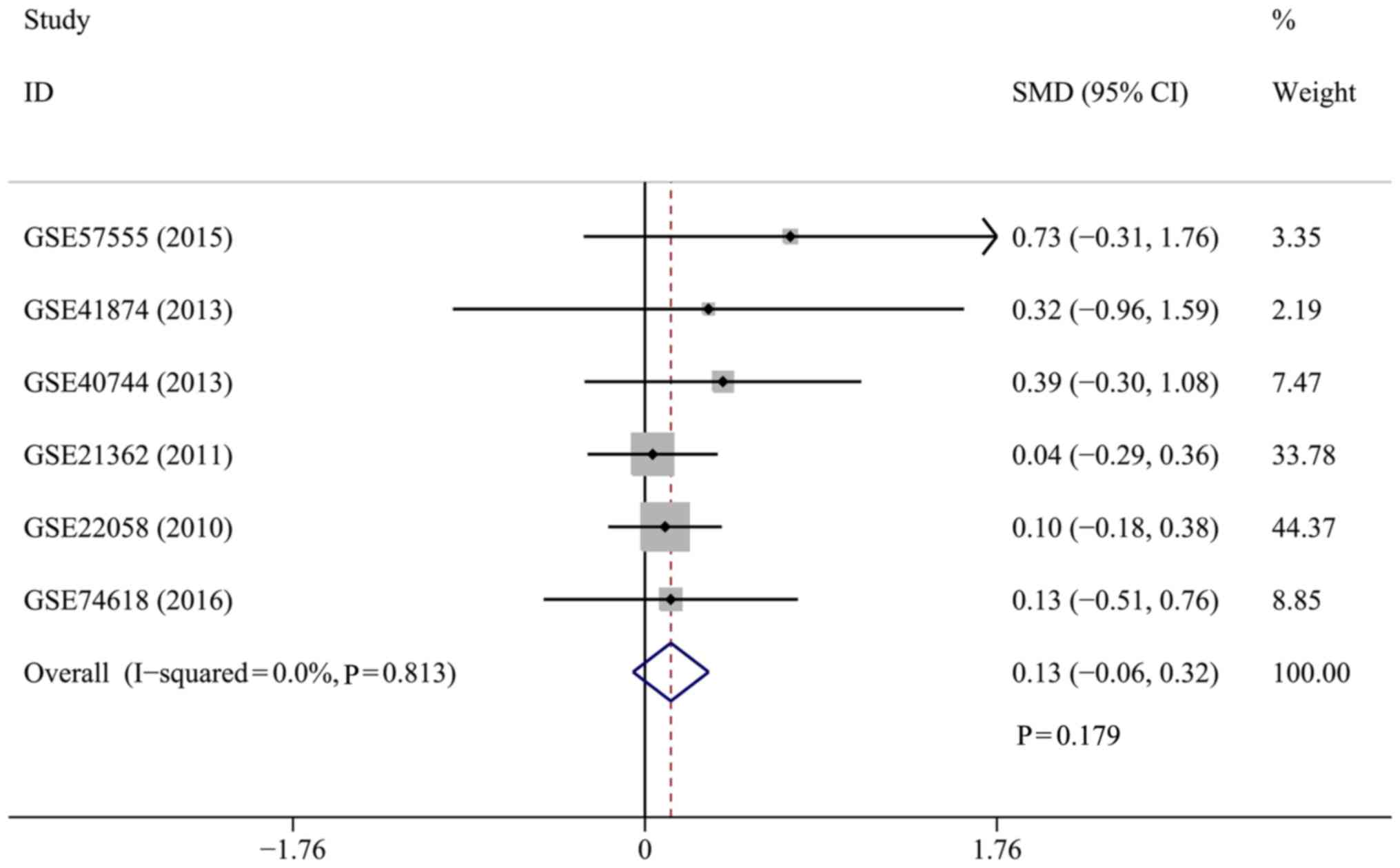
disappeared when the GSE39678 dataset was removed
(I2=0.0%; P=0.813). Hence, the SMD and 95% CI were
pooled again as a fixed-effects model. However, no significant
difference was observed in the miR-449a expression level between
the patients with HCC and healthy controls (SMD=0.13; 95% CI,
−0.06–0.32; P=0.179) (Fig. 4). The
results suggested that the conclusion for the present meta-analysis
was unreliable according to the inconsistent values when pooled by
the separate random- and fixed-effects models.
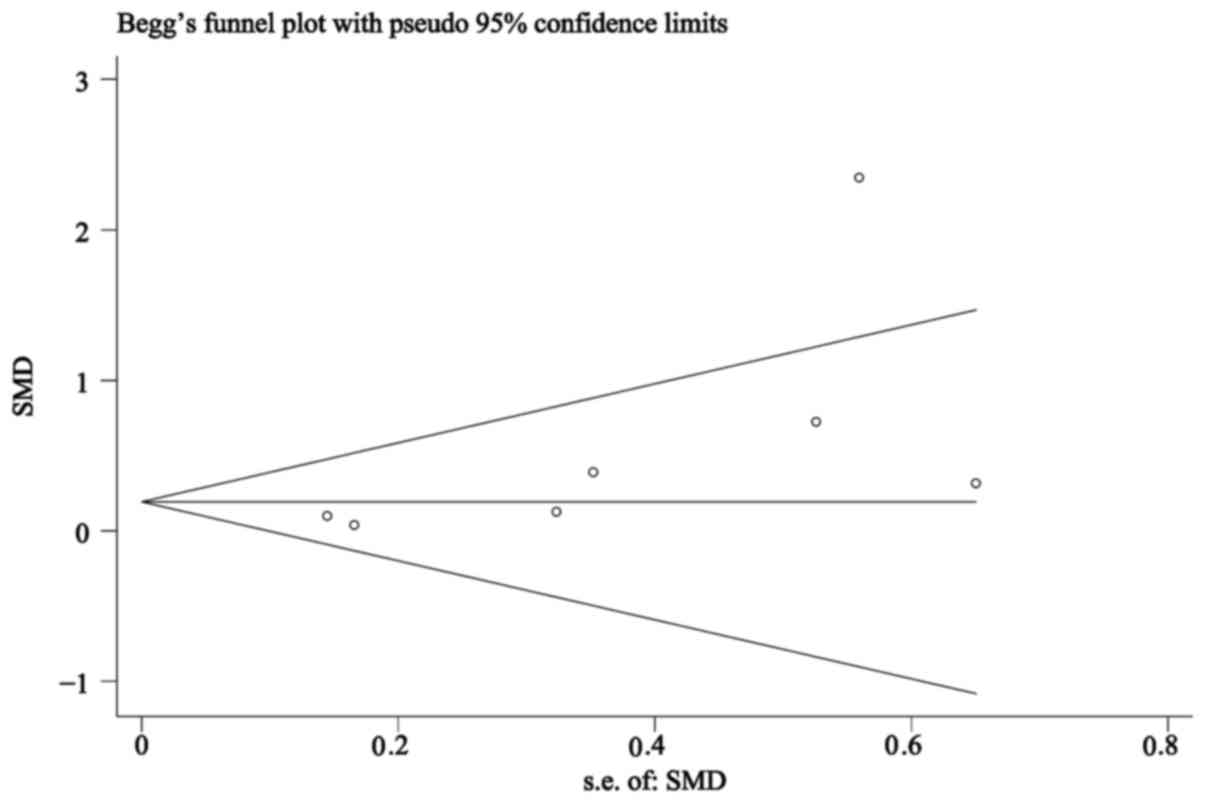
Publication bias analysis revealed that the Begg's
funnel plots (Fig. 5) were almost
symmetrical, with the obtained Pr>|z|=0.072>0.05 for miR-449a
(where Pr represented the Begg's P-value and z represented the
standard normal variate for the confidence level specified by
option level), suggesting that publication bias from the included
studies was absent in the present study.
Potential target genes of miR-449a in
HCC
In the GSE74710 dataset, since the HCC cell line,
HLE, was transfected with miR-449a mimics, the downregulated genes
(FC<0.5 and P<0.05) were screened as potential miR-449a
target genes, and 617 eligible genes were ultimately extracted.
Additionally, potential targets of miR-449a in silico were
also predicted. A total of 3,662 genes that appeared in more than
five databases were selected. Furthermore, a total of 1,800
HCC-related genes were recognized from NLP.
To narrow down the area and obtain more reliable
miR-449a targets, three kinds of genes were overlapped. Finally, 23
genes were left for further GO and KEGG pathway analysis and PPI
network analysis, which were largely representative of the
prospective molecular mechanism of miR-449a in HCC (data not
shown).
Potential pathways of miR-449a in
HCC
The GO and KEGG enrichment analyses were performed
on the potential target genes of miR-449a that were associated with
HCC by the functional annotation tool of the DAVID database. The
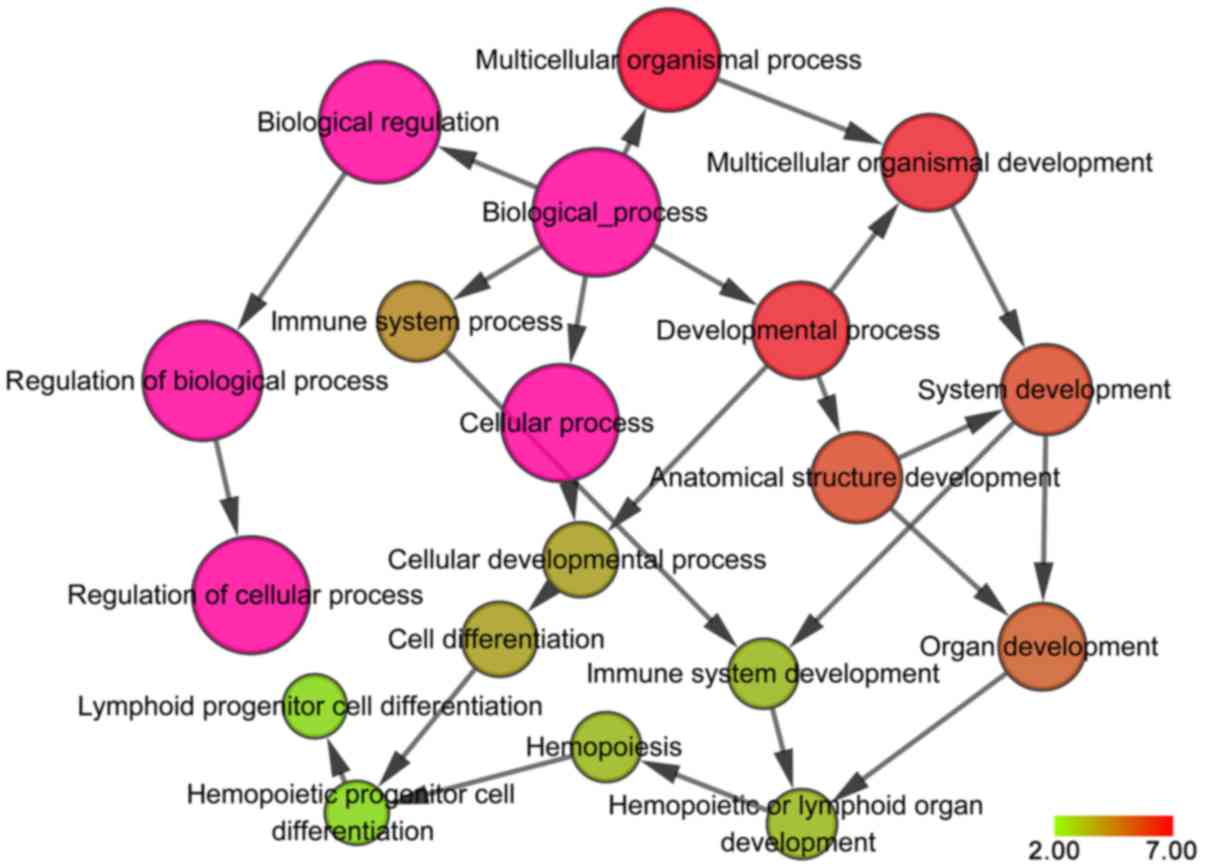
most enriched GO and KEGG terms were obtained. It was revealed that
in the GO_BP category, these genes were predominantly involved in
‘positive regulation of transcription from RNA polymerase II
promoter’ (GO: 0045944; P=1.778×10−4), ‘positive
regulation of apoptotic process’ (GO: 0043065;
P=5.67×10−4) and ‘negative regulation of cell
proliferation’ (GO: 0008285; P=0.0016). In the GO_CC category, the
genes tended to affect the ‘cell surface’ (GO: 0009986; P=0.0265),
‘cytoplasm’ (GO: 0005737; P=0.0279) and ‘nucleus’ (GO: 0005634;
P=0.0361), while in the GO_MF category, they mainly participated in
‘transmembrane receptor protein tyrosine kinase activity’ (GO:
0004714; P=0.0011), ‘protein binding’ (GO:0005515; P=0.0038) and
‘transcription factor activity, and sequence-specific DNA binding’
(GO: 0003700; P=0.0069). The results of the GO analysis were
presented in Table II and
visualized as co-expression networks using Cytoscape v3.4.0 with
the BiNGO plug-in, as demonstrated in Fig. 6.
 | Table II.Top ten GO functional annotation for
most significantly related targets of microRNA-449a. |
Table II.
Top ten GO functional annotation for
most significantly related targets of microRNA-449a.
| GO ID | GO term | Count (%) | P-value | Gene symbol |
|---|
| Biological
process |
|
|
|
|
|
GO:0009615 | Response to
virus | 6 (13.3) |
9.75×10−6 | DUOX2, CDK6, HMGA2,
FOXP3, CXCL12, FOSL1 |
|
GO:0043065 | Positive regulation
of apoptotic process | 8 (17.8) |
1.18×10−5 | TXNIP, DAB2IP,
NOTCH1, DIABLO, SOX4, ARHGEF9, HMGA2, FOSL1 |
|
GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | 11 (24.4) |
1.78×10−4 | DAB2IP, NOTCH1,
E2F5, MET, SOX4, CTCFL, HMGA2, MYB, FOXP3, FOSL1, FOXP1 |
|
GO:0051591 | Response to
cAMP | 4 (8.9) |
2.36×10−4 | DUOX2, COL1A1,
AREG, FOSL1 |
|
GO:0042542 | Response to
hydrogen peroxide | 4 (8.9) |
3.20×10−4 | TXNIP, COL1A1,
AREG, FOSL1 |
|
GO:0008285 | Negative regulation
of cell proliferation | 7 (15.6) |
5.48×10−4 | SPRY1, DAB2IP,
NOTCH1, SOX4, CDK6, FOXP3, FOSL1 |
|
GO:0035019 | Somatic stem cell
population maintenance | 4 (8.9) |
6.55×10−4 | ZHX2, SOX4, KIT,
HMGA2 |
|
GO:0045892 | Negative regulation
of transcription, DNA-templated | 7 (15.6) |
1.81×10−3 | DAB2IP, NOTCH1,
ZHX2, HMGA2, MYB, FOXP3, FOXP1 |
|
GO:0045893 | Positive regulation
of transcription, DNA-templated DNA | 7 (15.6) |
2.12×10−3 | NOTCH1, SOX4,
CTCFL, COL1A1, HMGA2, MYB, FOXP3 |
|
GO:0000122 | Negative regulation
of transcription from RNA polymerase II promoter | 8 (17.8) |
2.48×10−3 | TXNIP, DAB2IP,
NOTCH1, ZHX2, HMGA2, MYB, FOXP3, FOXP1 |
| Cellular
component |
|
|
|
|
|
GO:0009986 | Cell surface | 7 (15.6) |
1.82×10−3 | CLCN3, NOTCH1, MET,
AXL, TGFA, CFTR, AREG |
|
GO:0000139 | Golgi membrane | 6 (13.3) |
1.35×10−2 | CLCN3, NOTCH1,
ST6GAL1, FUT8, TGFA, AREG |
|
GO:0030141 | Secretory
granule | 3 (6.7) |
1.38×10−2 | CLCN3, COL1A1,
RAB27B |
|
GO:0005615 | Extracellular
space | 9 (20.0) |
1.42×10−2 | LPO, AXL, TGFA,
IGFBP1, COL1A1, KIT, AREG, CXCL12, TIMP3 |
|
GO:0016324 | Apical plasma
membrane | 4 (8.9) |
3.30×10−2 | NOTCH1, DUOX2,
CFTR, RAB27B |
|
GO:0005634 | Nucleus | 20 (44.4) |
3.97×10−2 | TXNIP, GLRX5, E2F5,
ZHX2, SOX4, CTCFL, CDK6, HMGA2, FOXP3, TIMP3, CDC25A, FOXP1, MBP,
NOTCH1, HPSE, MSI1, TGFA, AREG, MYB, FOSL1 |
|
GO:0070062 | Extracellular
exosome | 12 (26.7) |
6.662×10−2 | GSTM2, LPO, DAB2IP,
ST6GAL1, FUT8, DUOX2, ARHGAP1, AXL, CFTR, RAB27B, CXCL12,
TIMP3 |
|
GO:0016323 | Basolateral plasma
membrane | 3 (6.7) |
7.01×10−2 | LPO, TGFA,
CFTR |
|
GO:0005829 | Cytosol | 13 (28.9) |
9.0×10−2 | TXNIP, DAB2IP,
CDK6, CFTR, ARHGEF9, CDC25A, GSTM2, SPRY1, NOTCH1, ATG4B, ARHGAP1,
DIABLO, FOSL1 |
|
GO:0009897 | External side of
plasma membrane | 3 (6.7) |
9.35×10−2 | CLCN3, KIT,
CXCL12 |
| Molecular
function |
|
|
|
|
|
GO:0005515 | Protein
binding | 35 (77.8) |
5.07×10−4 | CLCN3, E2F5, SOX4,
CTCFL, KIT, TIMP3, MBP, GSTM2, SPRY1, HPSE, ARHGAP1, TGFA, DIABLO,
RAB27B, MYB, etc. |
|
GO:0001077 | Transcriptional
activator activity, RNA polymerase II core promoter proximal region
sequence-specific binding | 5 (11.1) |
3.26×10−3 | SOX4, CTCFL, HMGA2,
MYB, FOSL1 |
|
GO:0004714 | Transmembrane
receptor protein tyrosine kinase activity | 3 (6.7) |
4.40×10−3 | MET, AXL, KIT |
|
GO:0001047 | Core promoter
binding | 3 (6.7) |
1.21×10−2 | NOTCH1, HMGA2,
FOXP3 |
|
GO:0002020 | Protease
binding | 3 (6.7) |
2.85×10−2 | KIT, TIMP3,
MBP |
|
GO:0046982 | Protein
heterodimerization activity | 5 (11.1) |
3.24×10−2 | CLCN3, NOTCH1, AXL,
ZHX2, SOX4 |
|
GO:0003700 | Transcription
factor activity, sequence-specific DNA binding | 7 (15.6) |
3.73×10−2 | NOTCH1, E2F5, ZHX2,
SOX4, FOXP3, FOSL1, FOXP1 |
|
GO:0017124 | SH3 domain
binding | 3 (6.7) |
3.85×10−2 | DAB2IP, FUT8,
ARHGAP1 |
|
GO:0042803 | Protein
homodimerization activity | 6 (13.3) |
4.03×10−2 | GSTM2, CLCN3,
DAB2IP, ZHX2, KIT, FOXP3 |
|
GO:0008191 |
Metalloendopeptidase inhibitor
activity | 2 (4.4) |
4.09×10−2 | RECK, TIMP3 |
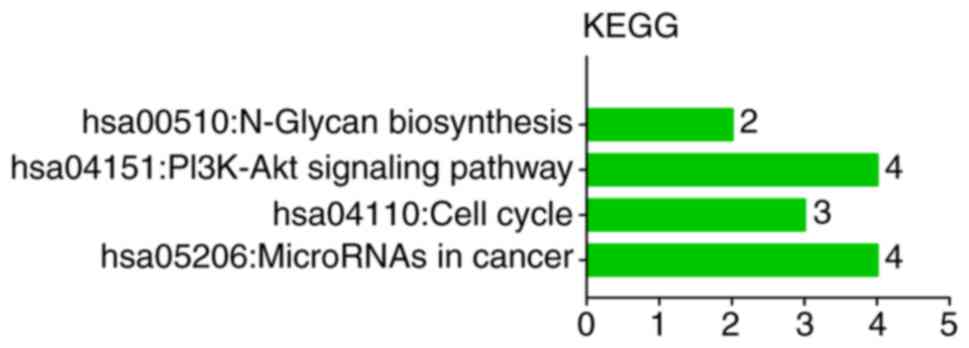
KEGG pathway investigation indicated that these
target genes participated in the following categories: ‘MicroRNAs
in cancer’ (hsa05206; P=0.0146), ‘Cell cycle’ (hsa04110; P=0.0219),
‘PI3K-Akt signaling pathway’ (hsa04151; P=0.0243) and ‘N-Glycan
biosynthesis’ (hsa00510; P=0.0884) (Table III and Fig. 7).
 | Table III.KEGG functional annotation for most
significantly related targets of microRNA-449a. |
Table III.
KEGG functional annotation for most
significantly related targets of microRNA-449a.
| KEGG ID | KEGG term | Count (%) | P-value | Gene symbol |
|---|
| hsa05206 | MicroRNAs in
cancer | 7 (15.6) |
5.35×10−4 | RECK, NOTCH1, MET,
CDK6, HMGA2, TIMP3, CDC25A |
| hsa02151 | PI3K-Akt signaling
pathway | 5 (11.1) |
3.83×10−2 | MET, CDK6, COL1A1,
KIT, MYB |
| hsa05200 | Pathways in
cancer | 5 (11.1) |
5.71×10−2 | MET, TGFA, CDK6,
KIT, CXCL12 |
| hsa04110 | Cell cycle | 3 (6.7) |
7.85×10−2 | E2F5, CDK6,
CDC25A |
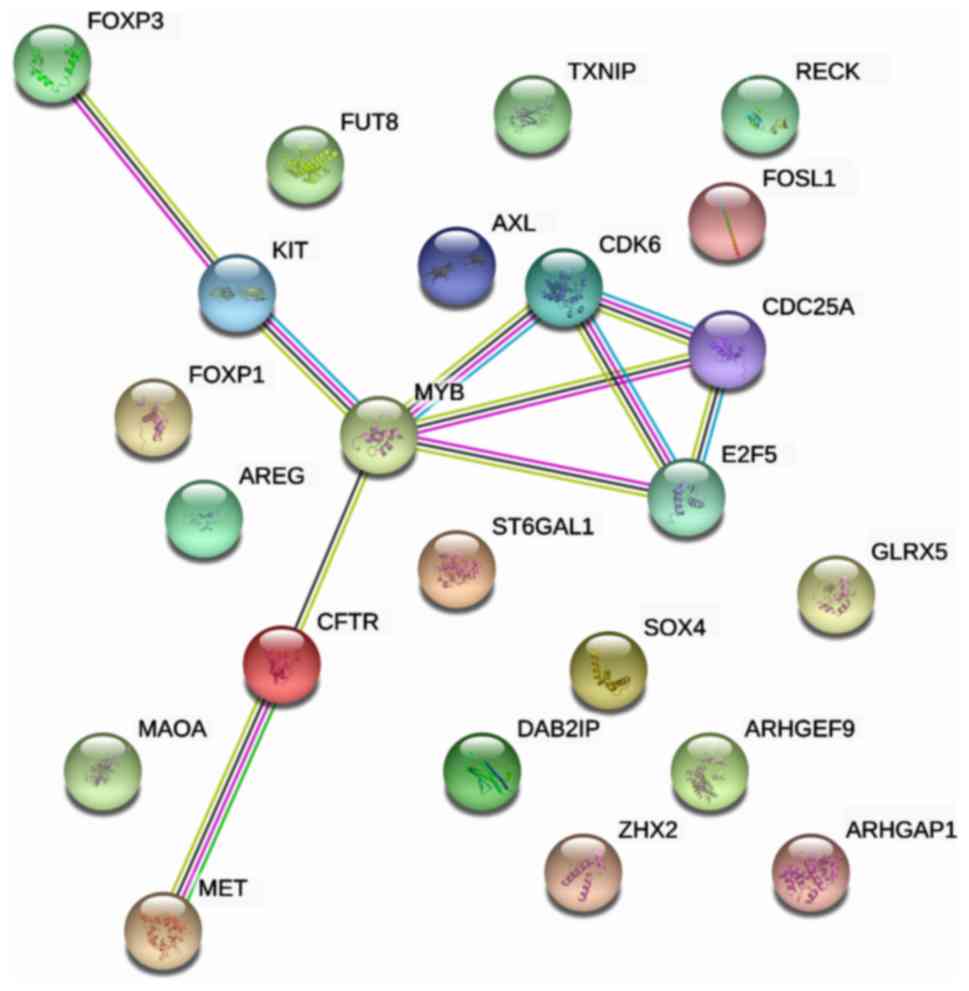
In the PPI network analysis, 23 genes were input to
construct an interaction network, and the PPI enrichment P-value
was 0.0096, with 23 interaction edges (Fig. 8). Meanwhile, the local clustering
coefficient was 0.23. With comprehensive analysis of the network,
the hub nodes, MYB, E2F5, CDK6 and CDC25A, were identified as the
most essential genes in HCC, which may also function as novel
targets for medical treatment.
Discussion
Accumulating evidence (44–47) has
revealed that the altered expression of miR contribute to the
initiation and progression of various diseases, including cancer.
miR are post-transcriptional regulators of gene expression and are
involved in the silencing of mRNA translation (48). Investigating the altered expression
of miR in malignancies may provide a novel and effective
therapeutic target for HCC in the future.
The present systematic review and meta-analysis
summarized eight original studies and seven microarray chips to
investigate the function of miR-449a in HCC. In the original
studies, all the researchers held the same viewpoint, that the
miR-449a expression level was reduced in HCC. However, there were
still some differences in their research. Wang et al
(29) explored the effect of
miR-449a on liver cancer and found that miR-449a predominantly
regulated CXCL5 expression, which encodes a chemokine that was
suggested to bind the G-protein coupled receptor chemokine (C-X-C
motif) receptor 2 to recruit neutrophils, promote angiogenesis and
remodel connective tissues (49).
The increased expression of this gene would promote cancer cell
proliferation, migration and invasion. Liu et al (30) also found that miR-449a functioned as
a tumor suppressor by obstructing cell proliferation, colony
formation, migration and invasion of HCC by partially repressing
ADAM10 expression. However, different from these two findings, Chen
et al (31) documented the
involvement of EMT in HCC progression. They revealed that miR-449a
likely suppressed EMT and metastasis of HCC by inhibiting FOS and
Met expression and suppressing the downstream signaling. The
mechanism of EMT in HCC metastasis has been presented by Yang et
al (50). In addition, Liu et
al (32) reported that miR-449a
may function by downregulating CAPN6, which is a potential oncogene
in tumors and may serve a role in tumor formation by inhibiting
apoptosis and promoting angiogenesis.
Another notable discovery was documented by Zhang
et al (33) and Sarma et
al (34). Zhang et al
dissected the relationship between miR-449a and HBV infection and
revealed that miR-449a acted as a linker between histone
deacetylation and HBV replication, directly targeting CREB5, and
CREB5 knockdown induced FXRα expression. FXRα also referred to as
nuclear receptor subfamily 1 group H member 4, which is a
transcription factor that binds two motifs within the HBV enhancer
II and the core promoter regions to promote HBV transcription and
replication (51). Sarma et
al (34) demonstrated that HCV
downregulated miR-449a expression in human livers, which could
elevate the levels of transcription factors, leading to an increase
in the inflammatory response and promoting cell proliferation,
which resulted in HCC.
Buurman et al (36) investigated the miR-449 family for
several years, and identified at least one member (miR-449a,
miR-449b or miR-449c) of the miR-449 family-targeted pathways,
including Wnt/β-catenin, p53/cell cycle control, mitogen-activated
protein kinase and TGF-β. These are promising targets for HCC
therapy (52). Buurman et al
(36) focused on the signaling of
TGF-β and discovered that this pathway served a dual role in HCC,
acting as a tumor suppressor during early stages of liver damage,
but promoting tumor progression and metastasis in advanced HCC.
Furthermore, their first clinical trial for miR replacement therapy
was initiated in 2013 to evaluate the safety of miR-449a in
patients with unresectable primary liver cancer or other selected
solid or hematologic malignancies (53). miR replacement therapy may provide
hope for patients with liver cancer in the future.
In summary, previous original studies have shared
the same views, that miR-449a is downregulated in HCC tissues and
involved in various pathways by targeting multiple mRNA. However,
to our surprise, the pooled results of seven microarrays revealed
that miR-449a was overexpressed in HCC, which was completely
contrary to the original studies. The causes of this discrepancy
were analyzed, and it was speculated that microarray analysis was
not sensitive enough to detect the RNA expression level compared to
the method of reverse transcription-quantitative polymerase chain
reaction. The majority of the microarrays gathered in the present
study demonstrated no significant difference in the miR-449a
expression between HCC and normal controls.
Since the original studies indicated that miR-449a
may be involved in multiple signaling pathways and target multiple
mRNAs, the potential targets of miR-449a in HCC were overlapped to
explore whether miR-449a may exert its function in other pathways.
The potential targets, including GEO downregulated genes, in
silico predicted genes, as well as HCC-related genes recognized
from NLP, were merged with three different resources to obtain more
reliable targets. However, limitations remained in the present
study. The GEO downregulated genes were required to have an FC
value of <0.5 and a P-value of <0.05 for inclusion, and the
adjusted P-value was also taken into consideration, which provides
a more accurate indication of the level of false positives for a
given cut-off value. However, the adjusted P-values of all the
eligible genes were distributed from 0.1–0.3. The results revealed
that more false positives may exist in these included genes.
Therefore, the miRWalk database, which contains 12 prediction
algorithms, and NLP were used to identify more reliable targets.
The present results revealed that, in addition to ‘cell cycle
pathway’ (hsa04110), miR-449a may participate in ‘MicroRNAs in
cancer’ (hsa05206), ‘PI3K-Akt signaling pathway’ (hsa04151) and
‘N-Glycan biosynthesis pathways’ (hsa00510) to regulate HCC
tumorigenesis.
In addition, PPI network analysis was conducted, and
hub nodes, including MYB, E2F5, CDK6 and CDC25A, were identified as
the most essential genes in HCC. Among them, MYB encoded a protein
with three helix-turn-helix DNA-binding domains, which functioned
as transcription regulators (54).
It was reported that MYB expression was increased at the mRNA level
in HCC (55). E2F5 is a member of
the E2F transcription factor family and binds to the promoters of
target genes involved in cell cycle control, and consequently
modulates the expression of these targets (56). The members of the E2F family have
been divided into the activator (E2F1-E2F3) and repressor
(E2F4-E2F8) subclasses (57). The
overexpression or amplification of the E2F5 gene has been reported
in various solid tumors, such as HCC. For example, Jiang et
al (58) provided evidence that
E2F5 was commonly upregulated in HCC and E2F5 knockdown notably
inhibited the growth of HCC cells. Whether MYB or E2F5 deregulation
in HCC is targeted by miR-449a requires further investigation. The
phosphorylation of CDKs is required for timely progression through
the cell division cycle (59). CDK6
has been demonstrated to phosphorylate and regulate the activity of
the tumor suppressor protein, retinoblastoma (60,61).
Altered expression of this gene has been observed in multiple human
cancer types (62–64). Likewise, the dysregulation of CDK6 in
HCC has also been previously reported (65,66).
CDC25A, a member of the CDC25 family of phosphatases, is a cell
cycle and apoptotic regulator (67).
Previous research has reported that dephosphorylation of CDC25 is
the rate-limiting step for CDK activation. Their phosphorylation
removes the inhibitory phosphorylation on the CDK and regulates the
G1-S phase progression in the cell cycle (68).
In conclusion, as the number of studies in the
present systematic review was small and these studies provided
limited data to conduct a meta-analysis, it is difficult to deduce
any definite conclusion. Based on the evidence presented here,
miR-449a was revealed to be reduced in HCC and is likely to be
involved in various kinds of signaling pathways that target
multiple mRNAs to exert its function in HCC. Larger samples and
further investigation are required to validate this conclusion.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao
Y, Zhang H and Lin J: Identification and characterization of small
non-coding RNAs from Chinese fir by high throughput sequencing. BMC
Plant Biol. 12:1462012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bussotti G, Notredame C and Enright AJ:
Detecting and comparing non-coding RNAs in the high-throughput era.
Int J Mol Sci. 14:15423–15458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Ke C, Ma X, Zhao Q, Yang M, Zhang
W and Wang J: MicroRNA-92 promotes invasion and chemoresistance by
targeting GSK3β and activating Wnt signaling in bladder cancer
cells. Tumour Biol. 37:16295–16304. 2016. View Article : Google Scholar
|
|
5
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dhamija S and Diederichs S: From junk to
master regulators of invasion: lncRNA functions in migration, EMT
and metastasis. Int J Cancer. 139:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu W, Liu S, Liang Y, Zhou Z and Liu X:
MiR-7 inhibits progression of hepatocarcinoma by targeting KLF-4
and promises a novel diagnostic biomarker. Cancer Cell Int.
17:312017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Morey C and Avner P: Employment
opportunities for non-coding RNAs. FEBS Lett. 567:27–34. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dehury B, Panda D, Sahu J, Sahu M, Sarma
K, Barooah M, Sen P and Modi M: In silico identification and
characterization of conserved miRNAs and their target genes in
sweet potato (Ipomoea batatas L.) expressed sequence tags (ESTs).
Plant Signal Behav. 8:e265432013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cong D, He M, Chen S, Liu X, Liu X and Sun
H: Expression profiles of pivotal microRNAs and targets in thyroid
papillary carcinoma: An analysis of The Cancer Genome Atlas. Onco
Targets Ther. 8:2271–2277. 2015.PubMed/NCBI
|
|
13
|
Polioudakis D, Abell NS and Iyer VR:
MiR-191 regulates primary human fibroblast proliferation and
directly targets multiple oncogenes. PLoS One. 10:e01265352015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuan S, Tang C, Zhang Y, Wu J, Bao J,
Zheng H, Xu C and Yan W: mir-34b/c and mir-449a/b/c are required
for spermatogenesis, but not for the first cleavage division in
mice. Biol Open. 4:212–223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar P, Sharad S, Petrovics G, Mohamed A,
Dobi A, Sreenath TL, Srivastava S and Biswas R: Loss of miR-449a in
ERG-associated prostate cancer promotes the invasive phenotype by
inducing SIRT1. Oncotarget. 7:22791–22806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi J, Liu Y, Liu J and Zhou J:
Hsa-miR-449a genetic variant is associated with risk of gastric
cancer in a Chinese population. Int J Clin Exp Pathol.
8:13387–13392. 2015.PubMed/NCBI
|
|
17
|
You J, Zhang Y, Li Y, Fang N, Liu B, Zu L
and Zhou Q: MiR-449a suppresses cell invasion by inhibiting MAP2K1
in non-small cell lung cancer. Am J Cancer Res. 5:2730–2744.
2015.PubMed/NCBI
|
|
18
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo P, Huang ZL, Yu P and Li K: Trends in
cancer mortality in China: An update. Ann Oncol. 23:2755–2762.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Center MM and Jemal A: International
trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers
Prev. 20:2362–2368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CF, Hsu EC, Lin KT, Tu PH, Chang HW,
Lin CH, Chen YJ, Gu DL, Lin CH, Wu JY, et al: Overlapping
high-resolution copy number alterations in cancer genomes
identified putative cancer genes in hepatocellular carcinoma.
Hepatology. 52:1690–1701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takeshima N, Sozu T, Tajika A, Ogawa Y,
Hayasaka Y and Furukawa TA: Which is more generalizable, powerful
and interpretable in meta-analyses, mean difference or standardized
mean difference? BMC Med Res Methodol. 14:302014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Valera EM, Faraone SV, Murray KE and
Seidman LJ: Meta-analysis of structural imaging findings in
attention-deficit/hyperactivity disorder. Biol Psychiatry.
61:1361–1369. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sandbothe M, Buurman R, Reich N, Greiwe L,
Vajen B, Gürlevik E, Schäffer V, Eilers M, Kühnel F, Vaquero A, et
al: The microRNA-449 family inhibits TGF-β-mediated liver cancer
cell migration by targeting SOX4. J Hepatol. 66:1012–1021. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Tang W, Chen G, Ren F, Liang H,
Dang Y and Rong M: An encapsulation of gene signatures for
hepatocellular carcinoma, MicroRNA-132 predicted target genes and
the corresponding overlaps. PLoS One. 11:e01594982016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Huang CS, Yu W, et al:
MicroRNA-449a suppresses liver cancer migration and invasion
through targeting CXC chemokine ligand 5. Chin J Exp Surg.
34:228–230. 2017.
|
|
30
|
Liu S, Liu K, Zhang W, Wang Y, Jin Z, Jia
B and Liu Y: miR-449a inhibits proliferation and invasion by
regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res.
8:2609–2619. 2016.PubMed/NCBI
|
|
31
|
Chen SP, Liu BX, Xu J, Pei XF, Liao YJ,
Yuan F and Zheng F: MiR-449a suppresses the epithelial-mesenchymal
transition and metastasis of hepatocellular carcinoma by multiple
targets. BMC Cancer. 15:7062015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z
and Zha X: miR-449a promotes liver cancer cell apoptosis by
downregulation of Calpain 6 and POU2F1. Oncotarget. 7:13491–13501.
2016.PubMed/NCBI
|
|
33
|
Zhang X, Liu H, Xie Z, Deng W, Wu C, Qin
B, Hou J and Lu M: Epigenetically regulated miR-449a enhances
hepatitis B virus replication by targeting cAMP-responsive element
binding protein 5 and modulating hepatocytes phenotype. Sci Rep.
6:253892016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sarma NJ, Tiriveedhi V, Subramanian V,
Shenoy S, Crippin JS, Chapman WC and Mohanakumar T: Hepatitis C
virus mediated changes in miRNA-449a modulates inflammatory
biomarker YKL40 through components of the NOTCH signaling pathway.
PLoS One. 7:e508262012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
El-Serag HB, Kramer J, Duan Z and Kanwal
F: Racial differences in the progression to cirrhosis and
hepatocellular carcinoma in HCV-infected veterans. Am J
Gastroenterol. 109:1427–1435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Buurman R, Gürlevik E, Schäffer V, Eilers
M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F and
Skawran B: Histone deacetylases activate hepatocyte growth factor
signaling by repressing microRNA-449 in hepatocellular carcinoma
cells. Gastroenterology. 143:811–820.e15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Morita K, Shirabe K, Taketomi A, Soejima
Y, Yoshizumi T, Uchiyama H, Ikegami T, Yamashita Y, Sugimachi K,
Harimoto N, et al: Relevance of microRNA-18a and microRNA-199a-5p
to hepatocellular carcinoma recurrence after living donor liver
transplantation. Liver Transpl. 22:665–676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Diaz G, Melis M, Tice A, Kleiner DE,
Mishra L, Zamboni F and Farci P: Identification of microRNAs
specifically expressed in hepatitis C virus-associated
hepatocellular carcinoma. Int J Cancer. 133:816–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sato F, Hatano E, Kitamura K, Myomoto A,
Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S and
Shimizu K: MicroRNA profile predicts recurrence after resection in
patients with hepatocellular carcinoma within the Milan Criteria.
PLoS One. 6:e164352011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu AM, Yao TJ, Wang W, Wong KF, Lee NP,
Fan ST, Poon RT, Gao C and Luk JM: Circulating miR-15b and miR-130b
in serum as potential markers for detecting hepatocellular
carcinoma: A retrospective cohort study. BMJ Open. 2:e0008252012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Martinez-Quetglas I, Pinyol R, Dauch D,
Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A,
Rodriguez-Carunchio L, Solé M, et al: IGF2 Is Up-regulated by
epigenetic mechanisms in hepatocellular carcinomas and is an
actionable oncogene product in experimental models.
Gastroenterology. 151:1192–1205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Petrovic N, Ergün S and Isenovic ER:
Levels of MicroRNA heterogeneity in cancer biology. Mol Diagn Ther.
21:511–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo LJ, Zhang LP, Duan CY, Wang B, He NN,
Abulimiti P and Lin Y: The inhibition role of miR-22 in
hepatocellular carcinoma cell migration and invasion via targeting
CD147. Cancer Cell Int. 17:172017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xia W, Zhou J, Luo H, Liu Y, Peng C, Zheng
W and Ma W: MicroRNA-32 promotes cell proliferation, migration and
suppresses apoptosis in breast cancer cells by targeting FBXW7.
Cancer Cell Int. 17:142017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu C, Li G, Yang N, Su Z, Zhang S, Deng
T, Ren S, Lu S, Tian Y, Liu Y and Qiu Y: miR-324-3p suppresses
migration and invasion by targeting WNT2B in nasopharyngeal
carcinoma. Cancer Cell International. 17:22017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Towler BP, Jones CI and Newbury SF:
Mechanisms of regulation of mature miRNAs. Biochem Soc Trans.
43:1208–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tacke F, Zimmermann HW, Trautwein C and
Schnabl B: CXCL5 plasma levels decrease in patients with chronic
liver disease. J Gastroenterol Hepatol. 26:523–529. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang MH, Chen CL, Chau GY, Chiou SH, Su
CW, Chou TY, Peng WL and Wu JC: Comprehensive analysis of the
independent effect of twist and snail in promoting metastasis of
hepatocellular carcinoma. Hepatology. 50:1464–1474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Curtil C, Enache LS, Radreau P, Dron AG,
Scholtès C, Deloire A, Roche D, Lotteau V, André P and Ramière C:
The metabolic sensors FXRα, PGC-1α, and SIRT1 cooperatively
regulate hepatitis B virus transcription. FASEB J. 28:1454–1463.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schulze K, Imbeaud S, Letouze E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
ClinicalTrials.gov: A multicenter phase I
study of MRX34, microRNA miR-RX34 liposomal injection, NCT01829971.
2013.
|
|
54
|
Veals SA, Schindler C, Leonard D, Fu XY,
Aebersold R, Darnell JE Jr and Levy DE: Subunit of an
alpha-interferon-responsive transcription factor is related to
interferon regulatory factor and Myb families of DNA-binding
proteins. Mol Cell Biol. 12:3315–3324. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang H, Huang ZZ, Wang J and Lu SC: The
role of c-Myb and Sp1 in the up-regulation of methionine
adenosyltransferase 2A gene expression in human hepatocellular
carcinoma. FASEB J. 15:1507–1516. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D,
Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S and Young NS:
Human parvovirus B19 causes cell cycle arrest of human erythroid
progenitors via deregulation of the E2F family of transcription
factors. J Clin Invest. 120:3530–3544. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang Y, Yim SH, Xu HD, Jung SH, Yang SY,
Hu HJ, Jung CK and Chung YJ: A potential oncogenic role of the
commonly observed E2F5 overexpression in hepatocellular carcinoma.
World J Gastroenterol. 17:470–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Suryadinata R, Sadowski M and Sarcevic B:
Control of cell cycle progression by phosphorylation of
cyclin-dependent kinase (CDK) substrates. Biosci Rep. 30:243–255.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Iyirhiaro GO, Im DS, Boonying W, Callaghan
SM, During MJ, Slack RS and Park DS: Cdc25A Is a critical mediator
of ischemic neuronal death in vitro and in vivo. J Neurosci.
37:6729–6740. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shi Y, Qian ZR, Zhang S, Li W, Masugi Y,
Li T, Chan JA, Yang J, Da Silva A, Gu M, et al: Cell cycle protein
expression in neuroendocrine tumors: Association of CDK4/CDK6,
CCND1, and phosphorylated retinoblastoma protein with proliferative
index. Pancreas. 46:1347–1353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shang A, Lu WY, Yang M, Zhou C, Zhang H,
Cai ZX, Wang WW, Wang WX and Wu GQ: miR-9 induces cell arrest and
apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway.
Artif Cells Nanomed Biotechnol. 1–9. 2017. View Article : Google Scholar
|
|
63
|
Lulla AR, Slifker MJ, Zhou Y, Lev A,
Einarson MB, Dicker DT and El-Deiry WS: miR-6883 family miRNAs
target CDK4/6 to induce G1 phase cell cycle arrest in colon cancer
cells. Cancer Res. 77:6902–6913. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dall'Acqua A, Sonego M, Pellizzari I,
Pellarin I, Canzonieri V, D'Andrea S, Benevol S, Sorio R, Giorda G,
Califano D, et al: CDK6 protects epithelial ovarian cancer from
platinum-induced death via FOXO3 regulation. EMBO Mol Med.
9:1415–1433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhu H, Wang G, Zhou X, Song X, Gao H, Ma
C, Chang H, Li H, Liu FF, Lu J and Ma J: miR-1299 suppresses cell
proliferation of hepatocellular carcinoma (HCC) by targeting CDK6.
Biomed Pharmacother. 83:792–797. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wu H, Tao J, Li X, Zhang T, Zhao L, Wang
Y, Zhang L, Xiong J, Zeng Z, Zhan N, et al: MicroRNA-206 prevents
the pathogenesis of hepatocellular carcinoma by modulating
expression of met proto-oncogene and cyclin-dependent kinase 6 in
mice. Hepatology. 66:1952–1967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zou X, Tsutsui T, Ray D, Blomquist JF,
Ichijo H, Ucker DS and Kiyokawa H: The cell cycle-regulatory CDC25A
phosphatase inhibits apoptosis signal-regulating kinase 1. Mol Cell
Biol. 21:4818–4828. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sur S and Agrawal DK: Phosphatases and
kinases regulating CDC25 activity in the cell cycle: Clinical
implications of CDC25 overexpression and potential treatment
strategies. Mol Cell Biochem. 416:33–46. 2016. View Article : Google Scholar : PubMed/NCBI
|