Introduction
With the increase of competition pressure in modern
society, the number of people suffering from depression is growing
day by day (1). As a result,
research on depression has attracted widespread concerns (2). Pathophysiological studies show that the
volume and nerve density of hippocampus in patients with depression
are significantly decreased (3–5). Animal
model experiments also confirm that hippocampal neurons in
depressed rats are disordered and loose, the nuclei of hippocampal
neurons are shrunken, the nuclear membrane is irregular and vague,
and mitochondria are swelled and denatured (6,7). The
etiology of depression is dominated by monoamine hypothesis, which
suggests that the decrease of 5-hydroxy tryptamine, norepinephrine
and dopamine functions is its pathological mechanism (8). Some studies have focused on the role of
neuronal regeneration in the pathogenesis and treatment for
depression. According to the neurotrophic hypothesis of depression,
brain derived neurotrophic factor (BDNF), tumor necrosis factor-α
(TNF-α) (9), vascular endothelial
growth factor (VEGF) (10) or
fibroblast growth factor 2 (FGF-2) (11) may participate in the progression of
depression.
VEGFA is one of the most effective angiogenic growth
factors in human body, and its basic mechanism of action is to
promote angiogenesis and to increase blood supply (12). However, more and more studies show
that VEGFA can affect neurogenesis in a variety of ways, being a
multifunctional growth factor (13–15). It
is reported that after VEGFA signal transduction enters the cells,
it induces the dimerization of ligand and receptor, and then causes
phosphorylation of extracellular structure subunits with its
receptor to promote the survival, permeability, migration and
proliferation of hippocampal neurons (16).
MicroRNA (miRNA or miR) is single-stranded
small-molecule RNA (18–25 nucleotides) that exists in eukaryotic
cells and regulates gene expression at post-transcriptional levels
(17). miRNA plays important roles
in nerve cell development, differentiation, proliferation, and
apoptosis. It is reported that the expression of some miRNAs is
dysregulated in tissues from patients with depression, and
downstream targets of the miRNAs are associated with depression
symptoms (18). miRNA can modulate
central nervous activities, including reward feedback, circadian
rhythm and cognitive performance, and the abnormalities of these
activities are closely related with depression (19–21).
miR-27a is a miRNA with abundant functions. Studies show that
miR-27a can target epidermal growth factor receptor (EGFR), which
is activated in many types of tumors and promotes tumorigenesis
(22,23). However, it is never reported whether
miR-27a has regulatory effects on VEGFA.
In the present study, we investigate the behaviors
of depression rat model, measure the expression of relevant factors
in hippocampus and blood of the rats, and try to understand the
regulation between miR-27a and VEGFA.
Materials and methods
Animals
A total of 50 male rats (4–6 weeks old; 180–220 g)
were obtained from Chongqing Tengxin Biotech Company (Chongqing,
China; http://www.cqtx123.com/) with a
certificate no. SCXK(Yu)2016-0018. One week before experiments, the
rats had free access to food and water to adapt to the environment.
The Reduction, Replacement and Refinement Animal Welfare Principle
was followed during the experiments. The present study was approved
by the Ethics Committee of Zaozhuang Mental Health Center
(Zaozhuang, China).
The 50 rats were first raised in cages with
circadian rhythm (5 rats/cage) with free access to food and water
for three days. After that, body weight, daily intake of food,
tropism of syrup, and distance of spontaneous movement within 5 min
were recorded as baselines. Eight rats were excluded from the
experiments due to abnormal baseline activities. In the end, a
total of 40 rats with the closest scores were included in the
experiments, and randomly and evenly divided into control group and
depression model group (20 rats in each group). The rats in control
group were raised normally as described above at a density of 5
rats/cage. Rats in depression model group were raised at a density
of 1 rat/cage and used to construct depression model according to
methods reported by Willner (24).
The rats in model group were subjected to 14 types of stimulations,
including day/night reverse (24 and 48 h), repeated tilting of rat
cage (direction reverse every 4 for 12 and 24 h), water deprivation
(6 and 12 h), food deprivation (12 and 24 h), noise (metal clash
for 2 h and rat yawp for 2 h), damp padding (mild and sever for 12
and 24 h), foreign matter, no padding, suspension (6 h), tail
clamping (mild), cage sharing, horizontal oscillation, cage
exchange, and feeding environment change. Each rat was stimulated
with only one method on each day, and raised alone in a cage for 35
days. From the fourth week, the general indicators (weight, drink
amount and food amount), percentage of syrup consumption, and open
field behavior were observed. Body weight, tropism of syrup, and
open field behavior were tested for the two groups. After comparing
with baseline results, the rats with and without typical depressive
symptoms were noted.
To evaluate body weight, the food intake amount and
body weight changes of rats within 24 h (8:00 AM to 8:00 AM on the
next day) were recorded on the day before experiment, and days 28
and 35 of the experiment.
To carry out syrup consumption experiment (25), the syrup consumption percentage of
rats on the day before experiment, and day 28 and 35 of the
experiment were recorded. Total liquid consumption, sugar
consumption, pure water consumption, and syrup tropism percentage
(syrup consumption/total liquid consumption ×100%) were
calculated.
For open field test, changes in open field behavior
of the two groups of rats were recorded on the day before
experiment, and days 28 and 35 of the experiment. The dimension of
the open field behavior box was 100×100×40 cm (made by our research
group). There were 25 squares with equal sides in the field, and
the walls were black. The detailed method was as reported by Lin
et al (26). Horizontal score
was the number of squares being passed by the rats with four paws.
Vertical score was the times of vertical activities (front paws
leaving the field or climbing the walls). Total score in the open
field test was the sum of horizontal score and vertical score.
After fasting for 12 h following activity tests,
peripheral blood was collected from all rats and serum was
separated. Then, the rats were decapitated and hippocampal tissues
were collected. All animal experiments were conducted according to
the ethical guidelines of Zaozhuang Mental Health Center.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tissues (100 mg) were ground using liquid nitrogen
and mixed with 1 ml TRIzol (10606ES60; Yeasen, Shanghai, China) for
lysis. Serum samples (100 µl) were directly mixed with 1 ml TRIzol
(10606ES60; Yeasen, Shanghai, China) for lysis. Then, total RNA was
extracted using phenol chloroform method. The concentration and
quality of RNA was examined using ultraviolet spectrophotometry
(Nanodrop ND2000; Thermo Scientific, Waltham, MA, USA). Then, cDNA
was obtained by reverse transcription from 1 µg RNA and stored at
−20°C. Reverse transcription of mRNA was performed using TIANScript
II cDNA First Strand Synthesis kit (Tiangen, Beijing, China), and
reverse transcription of miRNA was carried out using miRcute miRNA
cDNA First Strand Synthesis kit (Tiangen).
SuperReal PreMix (SYBR-Green) RT-qPCR kit (Tiangen)
was used to detect mRNA expression of VEGFA. The sequences of VEGFA
were 5′-CCAGGAGTACCCCGATGAGATAG-3′ (upstream) and
5′-CTGGCTTTGGTGAGGTTTGATC-3′ (downstream); and those of β-actin
were 5′-ACCCCGTGCTGCTGACGGAG-3′ (upstream) and
5′-TCCCGGCCAGCCAGGTCCAT-3′ (downstream). PCR reaction system (20
µl) for VEGFA determination was composed of 10 µl RT-qPCR-Mix, 0.5
µl upstream primer, 0.5 µl downstream primer, 2 µl cDNA and 7 µl
ddH2O. PCR conditions for VEGFA determination were:
initial denaturation at 95°C for 2 min; 40 cycles of 95°C
denaturation for 30 sec, annealing at 58°C for 30 sec and
elongation at 72°C for 30 sec (iQ5; Bio-Rad, Hercules, CA, USA).
The 2−ΔΔCt method was used to calculate the relative
expression of VEGFA mRNA against β-actin. Each sample was tested in
triplicate.
Isolation of miR-27a was performed using miRcute
miRNA isolation kit (Tiangen). The expression of miR-27a was
determined by miRcute miRNA RT-PCR kit (Tiangen), using U6 as
internal reference. The sequences of miR-27a were
5′-GCGGCGGTTCACAGTGGCTAAG-3′ (upstream) and
5′-ATCCAGTGCAGGGTCCGAGG-3′ (downstream); and those of U6 were
5′-CTCGCTTCGGCAGCACATATACT-3′ (upstream) and
5′-ACGCTTCACGAATTTGCGTGTC-3′ (downstream). PCR reaction system (20
µl) for miR-27a determination was composed of 10 µl RT-qPCR-Mix,
0.5 µl upstream primer, 0.5 µl downstream primer, 2 µl cDNA and 7
µl ddH2O. PCR conditions for miR-27a determination were:
initial denaturation at 95°C for 30 sec; 45 cycles of 95°C
denaturation for 5 sec and annealing at 60°C for 30 sec (iQ5;
Bio-Rad, Hercules, CA, USA). The 2−ΔΔCq method was used
to calculate the relative expression of miR-27a against U6. Each
sample was tested in triplicate.
Western blot analysis
Precooled Radio-Immunoprecipitation Assay (RIPA)
lysis buffer (600 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1%
sodium dodecyl sulfate, 1% TritonX-100, 1% sodium deoxycholate;
Beyotime Institute of Biotechnology, Shanghai, China) was used to
lyse the samples. After lysis for 50 min on ice, the mixture was
centrifuged at 12,000 g/min and 4°C for 5 min. The supernatant was
used to determine protein concentration by bicinchoninic acid (BCA)
protein concentration determination kit (RTP7102; Real-Times
Biotechnology Co., Ltd., Beijing, China). Protein samples (20 µg)
were then mixed with sodium dodecyl sulfate loading buffer before
denaturation in boiling water bath for 5 min. Afterwards, the
samples were subjected to 10% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis. The resolved proteins were transferred to
polyvinylidene difluoride membranes on ice (100 V, 2 h) and blocked
with 5% skimmed milk at room temperature for 1 h. Then, the
membranes were incubated with rabbit anti-mouse VEGFA polyclonal
primary antibody (1:1,000; Abcam, Cambridge, UK) and rabbit
anti-mouse β-actin primary antibody (1:5,000; Abcam) at 4°C
overnight. After extensive washing with phosphate-buffered saline
with Tween-20 for 3 times of 15 min, the membranes were incubated
with goat anti-rabbit horseradish peroxidase-conjugated secondary
antibody (1:3,000; Abcam) for 1 h at room temperature before
washing with phosphate-buffered saline with Tween-20 for 3 times of
15 min. Then, the membrane was developed with enhanced
chemiluminescence detection kit (Abcam) for imaging. Image lab v3.0
software (Bio-Rad, Hercules, CA, USA) was used to acquire and
analyze imaging signals. The relative content of VEGFA protein was
expressed as VEGFA/β-actin ratio.
Enzyme-linked immunosorbent assay
(ELISA)
Serum was examined using VEGFA ELISA kit (Abcam). In
microplates, standards (50 µl), samples (10 µl sample liquid and 40
µl diluent) and blank were set into predefined wells. In the wells
for standards and samples, horseradish peroxidase-labelled
conjugates (100 µl) were added before sealing the plates for
incubation at 37°C for 1 h. After washing the plates 5 times,
substrates A (50 µl) and B (50 µl) were added into each well. After
incubation at 37°C for 15 min, stop solution (50 µl) was added into
each well, and absorbance of each well was measured at 450 nm
within 15 min.
Bioinformatics
Bioinformatics prediction is a powerful tool for the
study of the functions of miRNAs. To understand the regulatory
mechanism of VEGFA, we used miRanda (http://www.microrna.org/microrna/home.do), TargetScan
(http://www.targetscan.org), PiTa
(http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
and PICTA (http://pictar.mdc-berlin.de/) to predict miRNA
molecules that might regulate VEGFA, and found that miR-27a was
able to potentially regulate VEGFA (Fig.
1).
Dual luciferase reporter assay
According to bioinformatics results, wild-type (WT)
and mutant seed regions of miR-27a in the 3′-UTR of VEGFA gene were
chemically synthesized in vitro, added with Spe-1 and
HindIII restriction sites, and then cloned into pMIR-REPORT
luciferase reporter plasmids. Plasmids (0.8 µg) with WT or mutant
3′-UTR DNA sequences were co-transfected with agomiR-27a (100 nM;
Sangon Biotech, Shanghai, China) into 293T cells. After cultivation
for 24 h, the cells were lysed using dual luciferase reporter assay
kit (Promega, Fitchburg, WI, USA) according to the manufacturer's
manual, and fluorescence intensity was measured using GloMax 20/20
luminometer (Promega). Using Renilla fluorescence activity
as internal reference, the fluorescence values of each group of
cells were measured.
Statistical analysis
The results were analyzed using SPSS 18.0
statistical software (IBM, Armonk, NY, USA). The data were
expressed as means ± standard deviations. Data were tested for
normality. Multigroup measurement data were analyzed using one-way
ANOVA. In case of homogeneity of variance, Least Significant
Difference and Student-Newman-Keuls methods were used; in case of
heterogeneity of variance, Tamhane's T2 or Dunnett's T3 method was
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
Rats in depression model group
exhibited depression symptoms and the depression model was
successfully constructed
To confirm the successful construction of depression
rat model, the body weight, syrup consumption rates and open field
test scores of the two groups were compared. The data showed that
the body weight of the two groups was not significantly different
from each other before stimulations, but the body weight of rats in
depression model group was significantly lower than that in control
group on days 28 and 35 (P<0.01) (Table I). In addition, there was no
significant difference in syrup consumption rate between the two
groups before stimulations, but the syrup consumption rate in
depression model group was significantly reduced than that in
control group on days 28 and 35 (P<0.05) (Table II). Of note, open field test scores
of the two groups were not significantly different from each other
before stimulations. However, the total score of open field test in
depression model group was significantly decreased than that in
control group on days 28 and 35 (P<0.01) (Table III). These results suggest that the
rats in depression model group have shown manifest depression
symptoms and the depression model is successfully constructed.
 | Table I.Body weight. |
Table I.
Body weight.
| Groups | No. | Before (g) | Day 28 (g) | Day 35 (g) |
|---|
| Control | 20 |
205.21±23.02 |
372.58±35.23 |
419.59±36.95 |
| Depression
model | 20 |
199.69±21.63 |
315.47±29.16a |
349.78±31.03a |
 | Table II.Syrup consumption rate. |
Table II.
Syrup consumption rate.
| Groups | N | Before (%) | Day 28 (%) | Day 35 (%) |
|---|
| Control | 20 |
95.6±2.3 |
93.7±4.2 |
92.5±6.2 |
| Depression
model | 20 |
94.9±2.8 |
85.5±6.1a |
79.8±8.0a |
 | Table III.Open field test total score. |
Table III.
Open field test total score.
| Groups | N | Before | Day 28 | Day 35 |
|---|
| Control | 20 |
130.6±25.5 |
123.7±20.3 |
118.6±18.1 |
| Depression
model | 20 |
132.1±23.6 |
65.8±18.9a |
59.3±12.0a |
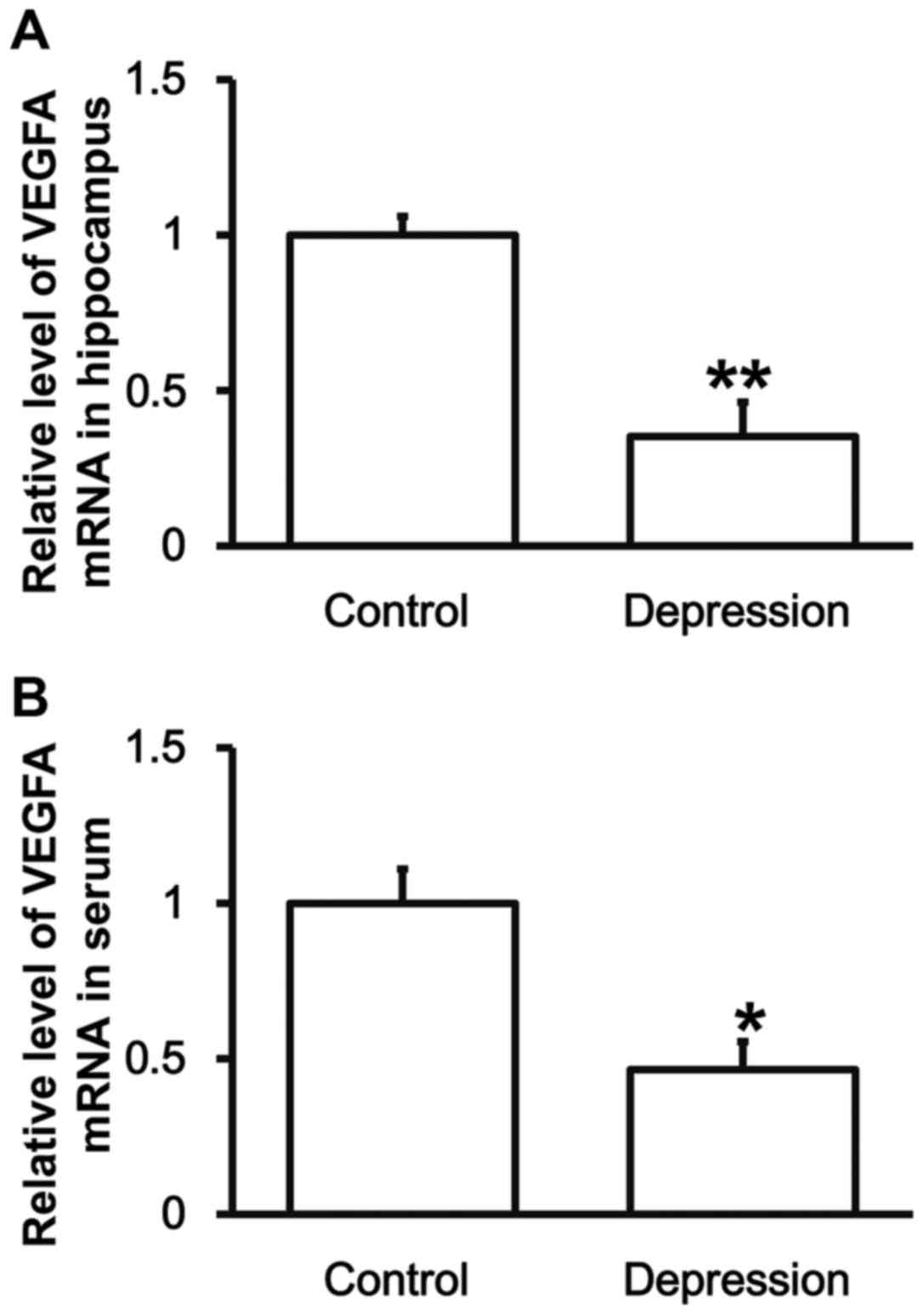
Rats with depression have lower VEGFA
mRNA expression in the hippocampus and peripheral blood than
control group
To determine the expression of VEGFA mRNA, RT-qPCR
was carried out. The data showed that the levels of VEGFA mRNA in
hippocampal tissues and serum from the rats in depression model
group were significantly lower than those in control group,
respectively (P<0.01) (Fig. 2).
The result indicates that rats with depression have lower VEGFA
mRNA expression in hippocampus and peripheral blood than control
group.
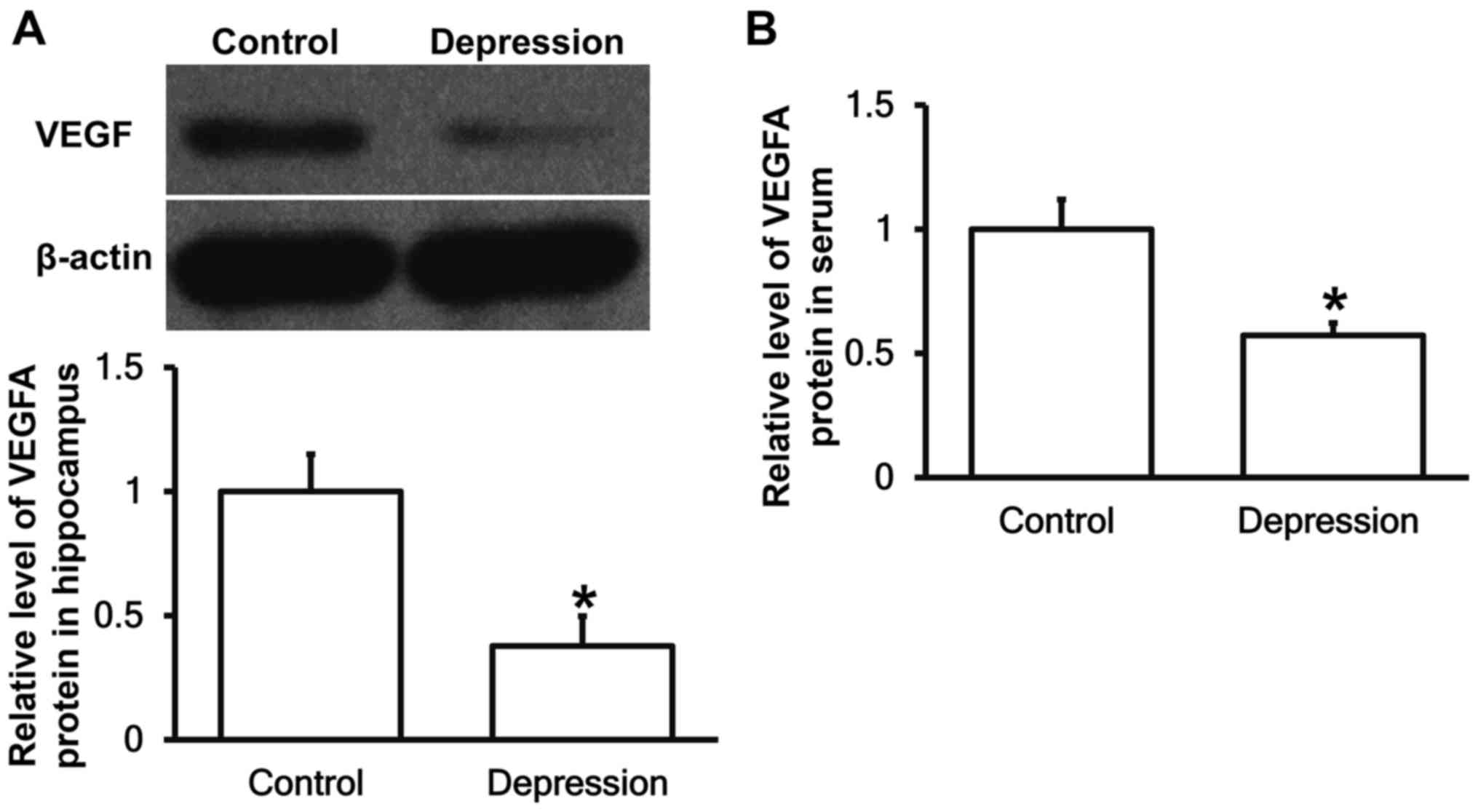
Depression model rats have decreased
VEGFA protein expression in the hippocampus and peripheral
blood
To measure the expression of VEGFA protein in
hippocampal tissues and serum, western blotting and ELISA were
performed, respectively. The data showed that VEGFA protein
expression in hippocampus and serum from depression model rats were
significantly reduced than that from control group (P<0.05)
(Fig. 3). The result suggests that
depression model rats have decreased VEGFA protein expression in
hippocampus and peripheral blood.
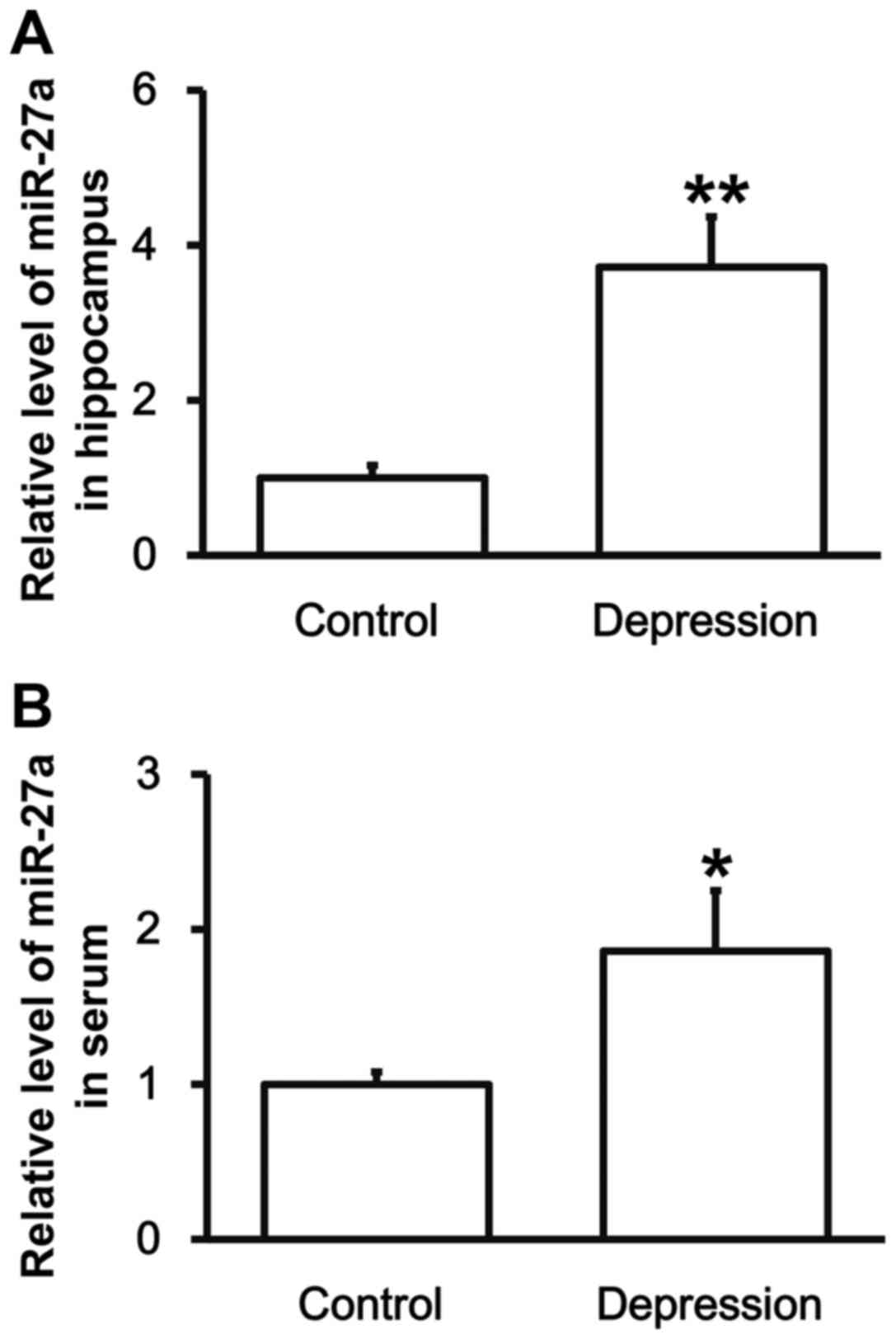
Rats with depression have higher
levels of miR-27a in the hippocampus and peripheral blood
To test the expression of miR-27a in hippocampal
tissues and serum, RT-qPCR was employed. The data showed that the
levels of miR-27a in hippocampal tissues and serum from depression
model rats were significantly upregulated compared with those from
control group (P<0.05) (Fig. 4).
The result indicates that rats with depression have higher levels
of miR-27a in hippocampus and peripheral blood, which may be
correlated with the levels of VEGFA.
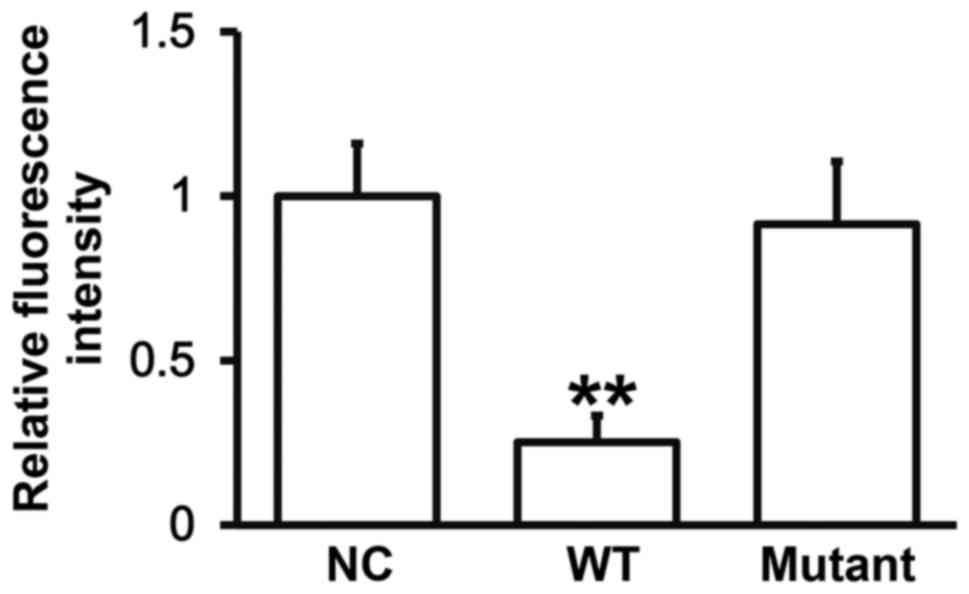
miR-27a can bind with the 3′-UTR
seeding region of VEGFA mRNA to regulate its expression
To identify the interaction between miR-27a and the
3′-UTR of VEGFA mRNA, dual luciferase reporter assay was performed.
The fluorescence value of cells co-transfected with agomiR-27a and
pMIR-REPORT-WT luciferase reporter plasmids was significantly lower
than that in negative control group (P<0.05). By contrast, the
fluorescence value of cells co-transfected with agomiR-27a and
pMIR-REPORT-mutant luciferase reporter plasmids was not
significantly different from that in negative control group
(P>0.05) (Fig. 5). The result
indicates that miR-27a can bind with the 3′-UTR seeding region of
VEGFA mRNA to regulate its expression.
Discussion
Emotional disorders are usually accompanied by
cognitive or behavioral changes or disorders. Cognitive impairment
associated with depression mainly includes declines in attention,
learning ability and memory. It is reported that learning ability
and memory of rats with depression are changed, and this may be
caused by damages of limbic system, in which hippocampus is an
important component (27).
Establishing animal models of central nervous diseases is one of
the most commonly used research methods (24). Chronic unpredictable mild stress
(CUMS) model proposed by Willner (24), and Willner and Mitchell (28) is one of the most widely used models
of depression, and we have used this method to construct depression
model in the present study. VEGFA receptor, fetal liver kinase-1
(Flk-1), is the major receptor that exerts the biological roles of
VEGFA. Flk-1 is mainly expressed in vascular endothelial cells and
neuron precursor cells of hippocampus. It is reported that lack of
Flk-1 receptors is important for the survival of neural stem cells
(29). VEGFA can affect the complex
processes of learning and memory (30), and play a role in regulating neurite
growth and maturation during brain development (31). The role of VEGFA in neurogenesis may
be mediated by its interaction with downstream effector genes
(32). In the present study, our
data show that VEGF mRNA and protein expression in hippocampal
tissues and serum are downregulated in depression model rats,
suggesting that downregulation of VEGFA plays a key role in the
depression of rats.
Regulation of mRNA transcription and expression is a
complex process of multiple factors. miRNAs cut mRNAs and inhibits
their translation to achieve negative feedback regulation (33,34).
miRNAs are important regulators in normal development and
physiology, and many miRNAs have become biomarkers of various
diseases (35,36). Using bioinformatics, we discover that
miR-27a is an upstream regulator of VEGFA. miR-27a has various
biological functions. It is discovered that miR-27a can regulate
the expression of tumor-suppressor gene FOX1 in breast cancer cells
(37). In addition, miR-27a inhibits
the expression of zinc finger protein ZBTB10, and promotes the
accumulation of SP protein in breast cancer cells (38,39),
leading to abnormal cell cycles. Single nucleotide polymorphism
occurring in miR-27a precursor prevents its transformation to
mature form, and families with this have reduced probability of
breast cancer (40). By contrast,
expression of miR-27 is decreased in several other types of tumors,
including acute promyelocytic leukemia (41), colorectal cancer (42,43),
malignant melanoma (44), oral
squamous cell carcinoma (45) and
prostatic carcinoma (46). These
reports suggest that miR-27a may also act as a tumor-suppressor in
these tumors. miR-27a promotes fibrosis in organs such as liver
(47) and lung (48), and is associated with myocardial
hypertrophy and heart failure (49,50). Our
results in the present study show that VEGFA mRNA and protein
expression in depression model rats are downregulated, while
miR-27a expression in depression rats is upregulated. Indeed, dual
luciferase reporter assay has shown the direct interaction between
miR-27a and the 3′-UTR of VEGFA mRNA. Considering the results in
open field tests, we discover that miR-27a, VEGFA, and changes in
learning ability and memory have regulatory connections with each
other.
In conclusion, the present study demonstrates that
the mechanism of depression in rats may be the upregulation of
miR-27a expression in hippocampal tissues and blood, which results
in the downregulation of VEGFA mRNA and protein expression. As a
regulator of VEGFA, miR-27a may become a target for the prevention
and amelioration of depression.
Acknowledgements
The present study was supported by Jining
Psychiatric Prevention (Jining, China) and Treatment Hospital and
Zaozhuang Mental Health Center.
References
|
1
|
Bortolato B, Carvalho AF, Soczynska JK,
Perini GI and McIntyre RS: The Involvement of TNF-α in cognitive
dysfunction associated with major depressive disorder: An
opportunity for domain specific treatments. Current Neuropharmacol.
13:558–576. 2015. View Article : Google Scholar
|
|
2
|
Kessler RC: The costs of depression.
Psychiatr Clin North Am. 35:1–14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie X, Shi Y and Zhang J: Structural
network connectivity impairment and depressive symptoms in cerebral
small vessel disease. J Affect Disord. 220:8–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suzuki H, Matsumoto Y, Ota H, Sugimura K,
Takahashi J, Ito K, Miyata S, Furukawa K, Arai H, Fukumoto Y, et
al: Hippocampal blood flow abnormality associated with depressive
symptoms and cognitive impairment in patients with chronic heart
failure. Circ J. 80:1773–1780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nabavi SM, Daglia M, Braidy N and Nabavi
SF: Natural products, micronutrients, and nutraceuticals for the
treatment of depression: A short review. Nutr Neurosci. 20:180–194.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Yan J, Zhu X, Zhu Y, Yao S, Xu Y and
Ju S: Dilated Virchow-Robin spaces in the hippocampus impact
behaviors and effects of anti-depressant treatment in model of
depressed rats. J Affect Disord. 219:17–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu X, Dong Y, Jin X, Zhang C, Zhang T,
Zhao J, Shi J and Li J: The novel and potent anti-depressive action
of triptolide and its influences on hippocampal neuroinflammation
in a rat model of depression comorbidity of chronic pain. Brain
Behav Immun. 64:180–194. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Liu Y, Li L, Qi Y, Zhang Y, Li L,
Teng L and Wang D: Dopamine and serotonin contribute to
Paecilomyces hepiali against chronic unpredictable mild stress
induced depressive behavior in Sprague Dawley rats. Mol Med Rep.
16:5675–5682. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta K, Gupta R, Bhatia MS, Tripathi AK
and Gupta LK: Effect of agomelatine and fluoxetine on HAM-D score,
serum brain-derived neurotrophic factor, and tumor necrosis
factor-α level in patients with major depressive disorder with
severe depression. J Clin Pharmacol. 57:1519–1526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi M, Lee SH, Chang HL and Son H:
Hippocampal VEGF is necessary for antidepressant-like behaviors but
not sufficient for antidepressant-like effects of ketamine in rats.
Biochim Biophys Acta. 1862:1247–1254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu CK, Tseng PT, Chen YW, Tu KY and Lin
PY: Significantly higher peripheral fibroblast growth factor-2
levels in patients with major depressive disorder: A preliminary
meta-analysis under MOOSE guidelines. Medicine (Baltimore).
95:e45632016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roberts E, Cossigny DA and Quan GM: The
role of vascular endothelial growth factor in metastatic prostate
cancer to the skeleton. Prostate Cancer. 2013:418–340. 2013.
View Article : Google Scholar
|
|
13
|
Lee BH and Kim YK: Increased plasma VEGFA
levels in major depressive or manic episodes in patients with mood
disorders. J Affect Disord. 136:181–184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nowacka MM and Obuchowicz E: Vascular
endothelial growth factor (VEGFA) and its role in the central
nervous system: A new element in the neurotrophic hypothesis of
antidepressant drug action. Neuropeptides. 46:1–10. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yasuhara T, Shingo T and Date I: The
potential role of vascular endothelial growth factor in the central
nervous system. Rev Neurosci. 15:293–307. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Encinas JM, Vaahtokari A and Enikolopov G:
Fluoxetine targets early progenitor cells in the adult brain. Proc
Natl Acad Sci USA. 103:8233–8238. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kato M and Slack FJ: microRNAs: Small
molecules with big roles-C. elegans to human cancer. Biol Cell.
100:71–81. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Z, Jiang Y, Huo X, Yang Y, Davies H,
Botchway BOA and Fang M3: Prospective role of MicroRNAs in
depression. Curr Med Chem. 24:3508–3521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansen KF, Karelina K, Sakamoto K, Wayman
GA, Impey S and Obrietan K: miRNA-132: A dynamic regulator of
cognitive capacity. Brain Struct Funct. 218:817–831. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hollander JA, Im HI, Amelio AL, et al:
Striatal microRNA controls cocaine intake through CREB signalling.
Nature. 466:197–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng HY, Papp JW, Varlamova O, Dziema H,
Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S
and Obrietan K: microRNA modulation of circadian-clock period and
entrainment. Neuron. 54:813–829. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Acunzo M, Romano G, Palmieri D, Laganá A,
Garofalo M, Balatti V, Drusco A, Chiariello M, Nana-Sinkam P and
Croce CM: Cross-talk between MET and EGFR in non-small cell lung
cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci USA.
110:8573–8578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang W, Cheng B, Miao L, Mei Y and Wu M:
Mutant p53-R273H gains new function in sustained activation of EGFR
signaling via suppressing miR-27a expression. Cell Death Dis.
4:e5742013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Willner P: Validity, reliability and
utility of the chronic mild stress model of depression: A 10-year
review and evaluation. Psychopharmacology (Berl). 134:319–329.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iñiguez SD, Warren BL, Parise EM,
Alcantara LF, Schuh B, Maffeo ML, Manojlovic Z and Bolaños-Guzmán
CA: Nicotine exposure during adolescence induces a depression-like
state in adulthood. Neuropsychopharmacology. 34:1609–1624. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin YH, Liu AH, Xu Y, Tie L, Yu HM and Li
XJ: Effect of chronic unpredictable mild stress on brain-pancreas
relative protein in rat brain and pancreas. Behav Brain Res.
165:63–71. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lambert KG: Rising rates of depression in
today's society: Consideration of the roles of effort-based rewards
and enhanced resilience in day-to-day functioning. Neurosci
Biobehav Rev. 30:497–510. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Willner P and Mitchell PJ: The validity of
animal models of predisposition to depression. Behav Pharmacol.
13:169–188. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wada T, Haigh JJ, Ema M, Hitoshi S,
Chaddah R, Rossant J, Nagy A and van der Kooy D: Vascular
endothelial growth factor directly inhibits primitive neural stem
cell survival but promotes definitive neural stem cell survival. J
Neurosci. 26:6803–6812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao L, Jiao X, Zuzga DS, Liu Y, Fong DM,
Young D and During MJ: VEGFA links hippocampal activity with
neurogenesis, learning and memory. Nat Genet. 36:827–835. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang WY, Dong JH, Liu X, Wang Y, Ying GX,
Ni ZM and Zhou CF: Vascular endothelial growth factor and its
receptor Flk-1 are expressed in the hippocampus following
entorhinal deafferentation. Neuroscience. 134:1167–1178. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harms KM, Li L and Cunningham LA: Murine
neural stem/progenitor cells protect neurons against ischemia by
HIF-1α-regulated VEGFA signaling. PLoS One. 5:e97672010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Inoue K: MicroRNA function in animal
development. Tanpakushitsu Kakusan Koso. 52:197–204. 2007.(In
Japanese). PubMed/NCBI
|
|
34
|
Williams AE, Moschos SA, Perry MM, Barnes
PJ and Lindsay MA: Maternally imprinted microRNAs are
differentially expressed during mouse and human lung development.
Dev Dyn. 236:572–580. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao
R and Cui L: The tumor suppressor miR-124 inhibits cell
proliferation by targeting STAT3 and functions as a prognostic
marker for postoperative NSCLC patients. Int J Oncol. 46:798–808.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lv ZC, Fan YS, Chen HB and Zhao DW:
Investigation of microRNA-155 as a serum diagnostic and prognostic
biomarker for colorectal cancer. Tumour Biol. 36:1619–1625. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Mertens-Talcott SU, Zhang S, Kim K,
Ball J and Safe S: MicroRNA-27a indirectly regulates estrogen
receptor {alpha} expression and hormone responsiveness in MCF-7
breast cancer cells. Endocrinology. 151:2462–2473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang R, Schlehe B, Hemminki K, Sutter C,
Bugert P, Wappenschmidt B, Volkmann J, Varon R, Weber BH,
Niederacher D, et al: A genetic variant in the pre-miR-27a oncogene
is associated with a reduced familial breast cancer risk. Breast
Cancer Res Treat. 121:693–702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saumet A, Vetter G, Bouttier M,
Portales-Casamar E, Wasserman WW, Maurin T, Mari B, Barbry P,
Vallar L, Friederich E, et al: Transcriptional repression of
microRNA genes by PML-RARA increases expression of key cancer
proteins in acute promyelocytic leukemia. Blood. 113:412–421. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xi Y, Shalgi R, Fodstad O, Pilpel Y and Ju
J: Differentially regulated micro-RNAs and actively translated
messenger RNA transcripts by tumor suppressor p53 in colon cancer.
Clin Cancer Res. 12:2014–2024. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dai Y, Sui W, Lan H, Yan Q, Huang H and
Huang Y: Comprehensive analysis of microRNA expression patterns in
renal biopsies of lupus nephritis patients. Rheumatol Int.
29:749–754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Prueitt RL, Yi M, Hudson RS, Wallace TA,
Howe TM, Yfantis HG, Lee DH, Stephens RM, Liu CG, Calin GA, et al:
Expression of microRNAs and protein-coding genes associated with
perineural invasion in prostate cancer. Prostate. 68:1152–1164.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ji J, Zhang J, Huang G, Qian J, Wang X and
Mei S: Over-expressed microRNA-27a and 27b influence fat
accumulation and cell proliferation during rat hepatic stellate
cell activation. FEBS Lett. 583:759–766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kang BY, Park KK, Green DE, Bijli KM,
Searles CD, Sutliff RL and Hart CM: Hypoxia mediates mutual
repression between microRNA-27a and PPARγ in the pulmonary
vasculature. PLoS One. 8:e795032013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hernandez-Torres F, Aranega AE and Franco
D: Identification of regulatory elements directing
miR-23a-miR-27a-miR-24-2 transcriptional regulation in response to
muscle hypertrophic stimuli. Biochim Biophys Acta. 1839:885–897.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Roncarati R, Viviani Anselmi C, Losi MA,
Papa L, Cavarretta E, Da Costa Martins P, Contaldi C, Saccani Jotti
G, Franzone A, Galastri L, et al: Circulating miR-29a, among other
up-regulated microRNAs, is the only biomarker for both hypertrophy
and fibrosis in patients with hypertrophic cardiomyopathy. J Am
Coll Cardiol. 63:920–927. 2014. View Article : Google Scholar : PubMed/NCBI
|