Introduction
Breast cancer, which is considered to be the most
common type of cancer in women worldwide (1), is divided into four sub-types,
including luminal A-like, luminal B-like, human epidermal
receptor-2 (HER-2) positive and triple-negative breast cancer
(TNBC) (2). TNBC is characterized by
the loss of expression of progesterone receptor, estrogen receptor
and HER-2 gene expression (3). In
total, 15–20% of breast cancer cases are TNBC and the majority of
these are basal-like (4). TNBC,
particularly the basal-like type, has higher rates of metastasis
and poorer survival rates compared with other breast cancer
sub-types (4). Previously, great
improvements have been made in the early detection and treatment of
breast cancer. However, due to sub-optimal hormonal therapy and a
lack of more specific and effective therapeutic targets, TNBC
treatment remains a challenge (5,6). Thus,
identifying biological markers of TNBC progress is necessary and
could provide novel therapeutic strategies for TNBC treatment.
MicroRNAs (miRs) are a class of endogenous,
non-coding, single-stranded RNAs that are widely expressed in
eukaryotes and are 18–22 nucleotides in length. They serve critical
roles in the regulation of gene expression. Furthermore, they are
involved in a series of pathological and physiological processes,
including tumor proliferation, differentiation and apoptosis
(7–9). Furthermore, miRs can transcriptionally
inhibit the expression of target genes by binding to the
3′-untranslated region (3′-UTR), acting as oncogenes or tumor
inhibitors (10). A growing number
of studies have indicated that abnormal expression of miRs is
associated with human breast cancer (11–16).
Furthermore, mounting evidence indicates that miRs serve key
functions in the development of TNBC (17–19).
However, the exact molecular mechanism of miRs in TNBC is not yet
understood.
miR-146a-5p has been identified as a tumor
suppressor in various types of cancer. Zhang et al (20) reported that miR-146a-5p was
downregulated in hepatocellular carcinoma and acted as a tumor
suppressor. Wang et al (21)
revealed that miR-146a-5p promoted esophageal squamous cell
carcinoma progression by regulating epithelial-mesenchymal
transition (EMT) via targeting Notch2. Furthermore, miR-146a-5p has
been confirmed to inhibit non-small cell lung cancer (NSCLC) cell
proliferation and cell cycle progression (22). Another study indicated that
miR-146a-5p could promote prostate cancer cell apoptosis by
targeting ROCK1 (23). However, the
function of miR-146a-5p in the development of TNBC remains unclear.
Therefore, the present study aimed to investigate the function of
miR-146a-5p in TNBC and explore its underlying molecular
mechanism.
Materials and methods
Clinical specimens
A total of 20 paired TNBC and adjacent normal breast
tissues were identified and collected during biopsies from 20
female patients (age, 27–58 years) with TNBC who were diagnosed by
clinical symptoms and imaging examination at the Nanjing Medical
University Affiliated Jiangsu Cancer Hospital (Nanjing, China) from
January 2015 to December 2016. No patient received preoperative
radiotherapy or chemotherapy. All tissue samples were immediately
flash-frozen in liquid nitrogen and stored at −80°C. The present
study was approved by the Human Ethics Committee Review Board at
the Nanjing Medical University Affiliated Jiangsu Cancer Hospital
(Nanjing, China). Informed consent was provided by every
patient.
Cell culture
The non-malignant breast epithelial cell line,
MCF-10A, and TNBC cell lines, MDA-MB-231, MDA-MB-468, BT549 and
Hs578T, were obtained from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). All cell lines were
grown in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal
bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and 1% penicillin/streptomycin. All cell lines were incubated
in a 5% CO2 incubator at 37°C for ~48 h.
Cell transfection
For cell transfection, Hs578T cells were seeded into
a 6-well plate (5×104 cells/well) the day prior to
transfection. Next, cells were transiently transfected with 50 nM
miR-146a-5p mimics (sense, 5′-UGAGAACUGAAUUCCAUGGGUU-3′ and
antisense, 5′-CCCAUGGAAUUCAGUUCUCAUU-3′), 50 nM negative control
miR (sense, 5′-UUCUCCGAACGUGUCACGUdTdT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAAdTdT-3′; GenePharma Co., Ltd., Shanghai,
China) or miR-146a-5p mimics + SOX5-plamids (GenScript, Piscataway,
NJ, USA) using Lipofectamine 2000 transfection reagent (Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
At 24 h after cell transfection, the cells were collected and used
for the following analyses.
Dual-luciferase reporter assay
TargetScan bioinformatics software (www.targetscan.org/vert_71) was used to predict
the target genes of miR-146a-5p, and SOX5 was identified as a
potential target of miR-146a-5p. In order to explore whether
miR-146a-5p targets the 3′-UTRs of SOX5, vectors termed
SOX5-3′-UTR-WT and SOX5-3′-UTR-MUT with wild type and mutated
3′-UTR of SOX5 mRNA, respectively, were constructed. The sense and
anti-sense strands of the oligonucleotides of the SOX5-3′-UTR
containing the miR-146a-5p binding site were generated, annealed
and then sub-cloned into the pMIR-REPORT vector (GeneCopoeia, Inc.,
Rockville, MD, USA). The negative control was established by
sub-cloning scrambled sequences into the same vector. Hs578T cells
were seeded into a 24-well plate and then co-transfected with
SOX5-3′UTR-WT or SOX5-3′UTR-MUT and miR-146a-5p or its negative
control (hsamiR-NC) vector using Lipofectamine 2000 reagent,
following the manufacturer's protocol. Following transfection for
48 h the Dual-Luciferase® Reporter Assay system (Promega
Corporation, Madison, WI, USA) was used to determine the luciferase
activity according to the manufacturer's protocols. Renilla
luciferase activity was normalized to firefly luciferase activity.
Every experiment was repeated at least three times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from TNBC tissue samples and Hs578T cells
was extracted by TRIzol reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The RevertAid First
Strand cDNA synthesis kit (Fermentas; Thermo Fisher Scientific,
Inc.) was applied for cDNA generation. qPCR was performed using
SYBR-Green qPCR mix (Toyobo Co., Ltd., Osaka, Japan) in a Thermal
Cycler Dice Real-Time system III TP950 1 Set (Takara Bio, Inc.,
Otsu, Japan). GAPDH (for mRNA) or U6 (for miR) acted as the
internal controls. The primer sequences used for qPCR were obtained
from GenScript and listed in Table
I. The thermocycling conditions for qPCR were as follows: 95°C
for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec
and annealing/elongation at 60°C for 30 sec. The 2−ΔΔCq
method (24) was used to calculate
the relative quantities of each gene. All tests were repeated at
least three times.
 | Table I.Primer sequence for polymerase chain
reaction. |
Table I.
Primer sequence for polymerase chain
reaction.
| Gene name | Direction | Sequence (5′-3′) |
|---|
| miR-146a-5p | F |
GCGAGGTCAAGTCACTAGTGGT |
| miR-146a-5p | R |
CGAGAAGCTTGCATCACCAGAGAACG |
| SOX5 | F |
CAGCCAGAGTTAGCACAATAGG |
| SOX5 | R |
CTGTTGTTCCCGTCGGAGTT |
| E-cadherin | F |
CGAGAGCTACACGTTCACGG |
| E-cadherin | R |
GGGTGTCGAGGGAAAAATAGG |
| N-cadherin | F |
TTTGATGGAGGTCTCCTAACACC |
| N-cadherin | R |
ACGTTTAACACGTTGGAAATGTG |
| Vimentin | F |
GACGCCATCAACACCGAGTT |
| Vimentin | R |
CTTTGTCGTTGGTTAGCTGGT |
| Fibronectin | F |
GAACCACGCCGAACTACGAT |
| Fibronectin | R |
ATGCGATACATGACCCCTTCA |
| Twist1 | F |
GGACAAGCTGAGCAAGATTCA |
| Twist1 | R |
CGGAGAAGGCGTAGCTGAG |
| U6 | F |
CTCGCTTCGGCAGCACA |
| U6 | R |
AACGCTTCACGAATTTGCGT |
| GAPDH | F |
CTTTGGTATCGTGGAAGGACTC |
| GAPDH | R |
GTAGAGGCAGGGATGATGTTCT |
Western blotting
Total protein was collected from tissues and cells
using RIPA lysis buffer (Auragene, Changsha, China). A
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology, Haimen, China) was applied for protein
quantification. Equal amount of protein samples (25 µg) were
resolved by 10% SDS-PAGE and then transferred onto a nitrocellulose
membrane (EMD Millipore, Billerica, MA, USA). Following blocking
with 5% skimmed milk for 2 h at room temperature, the membranes
were incubated with primary antibodies against SOX5 (cat. no.
ab94396), fibronectin (cat. no. ab23750) (both Abcam, Cambridge,
UK) Twist1 (cat. no. 46702), N-cadherin (cat. no. 13116) vimentin
(cat. no. 5741), E-cadherin (cat. no. 3195) and β-actin (cat. no.
4970) (all Cell Signaling Technology, Inc., Danvers, MA, USA) at
4°C overnight. All primary antibodies were used at a dilution ratio
of 1:1,000. The membranes were subsequently incubated with
anti-rabbit immunoglobulin G horseradish peroxidase-linked
secondary antibodies (cat. no. 7074; dilution ratio, 1:5,000; Cell
Signaling Technology, Inc.) at room temperature for 2 h. For
protein band observation, an enhanced chemiluminescence kit
(Applygen Technologies, Inc., Beijing, China) and the ChemiDoc
XRS+system with Image Lab™ software (cat. no. 170-8265; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used according to the
manufacturer's protocol.
MTT assay
At 24 h after cell transfection, 2.0×103
Hs578T cells/well were plated into a 96-well plate (Corning, Inc.,
Corning, NY, USA) and incubated for ~24 h at 37°C before treatment.
Subsequently, 20 µl MTT (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) solution (5 mg/ml) was added to each well, and then
incubated for another 4 h at 37°C. Intracellular formazan crystals
were dissolved using dimethyl sulfoxide (Sigma-Aldrich; Merck
KGaA). At the end of the test, cell proliferation ability was
determined by detecting the absorbance at 490 nm using a microplate
reader.
Transwell assay
To determine cell invasion and migration ability, a
Transwell assay was performed 24 h after cell transfection using
Transwell inserts (Corning Incorporated, Corning, NY, USA). For the
invasion assay, a membrane coated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA) was used to construct a matrix barrier.
Next, 5×104 Hs578T cells were seeded into the upper
chamber containing 200 µl serum-free DMEM medium and 0.1% bovine
serum albumin (Thermo Fisher Scientific, Inc.). In total, 500 µl
medium supplemented with 15% FBS was added into the lower chamber.
For the migration assay, Hs578T cells were incubated for 24 h at
37°C, and for the invasion assay, the cells were incubated for 48 h
at 37°C. Cells on the upper membranes were wiped away, and the
migrated or invasive cells on the lower membranes were firstly
fixed with 95% ethyl alcohol for 15 min at room temperature and
then stained with 0.1% crystal violet for 15 min at 37°C. At the
end of the experiment, the cells were counted under an inverted
light microscope (Olympus Corporation, Tokyo, Japan) using ImageJ
software version 1.48u (National Institutes of Health, Bethesda,
MD, USA).
Statistical analysis
Statistical analysis was applied using SPSS version
21.0 (IBM Corp., Armonk, NY, USA). Each experiment was repeated in
triplicate. Data are presented as the mean ± standard deviation.
Comparisons between two groups were performed using Student's
t-test. Comparisons between multiple groups were performed using
one-way analysis of variance followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-146a-5p is downregulated in TNBC
specimens and cell lines
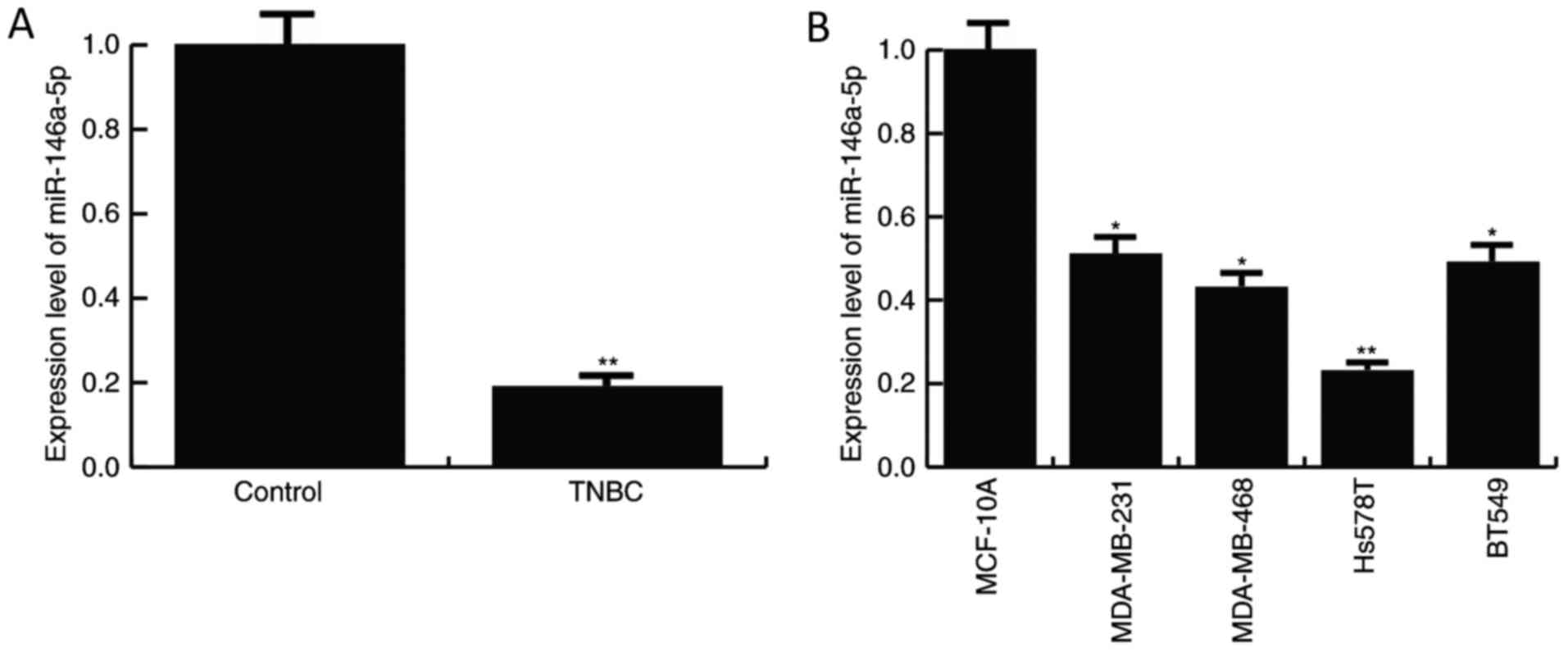
The expression level of miR-146a-5p was detected in
TNBC specimens and the TNBC cell lines, MDA-MB-231, MDA-MB-468,
BT549 and Hs578T, by RT-qPCR. As depicted in Fig. 1, compared with the normal control,
the level of miR-146a-5p was significantly decreased in TNBC
tissues and the TNBC cell lines, MDA-MB-231, MDA-MB-468, BT549 and
Hs578T. The data indicated that miR-146a-5p may be involved in TNBC
progression. Since the TNBC cell line Hs578T demonstrated a more
marked decrease in miR-146a-5p expression compared with the
control, Hs578T cells were selected for further experiments.
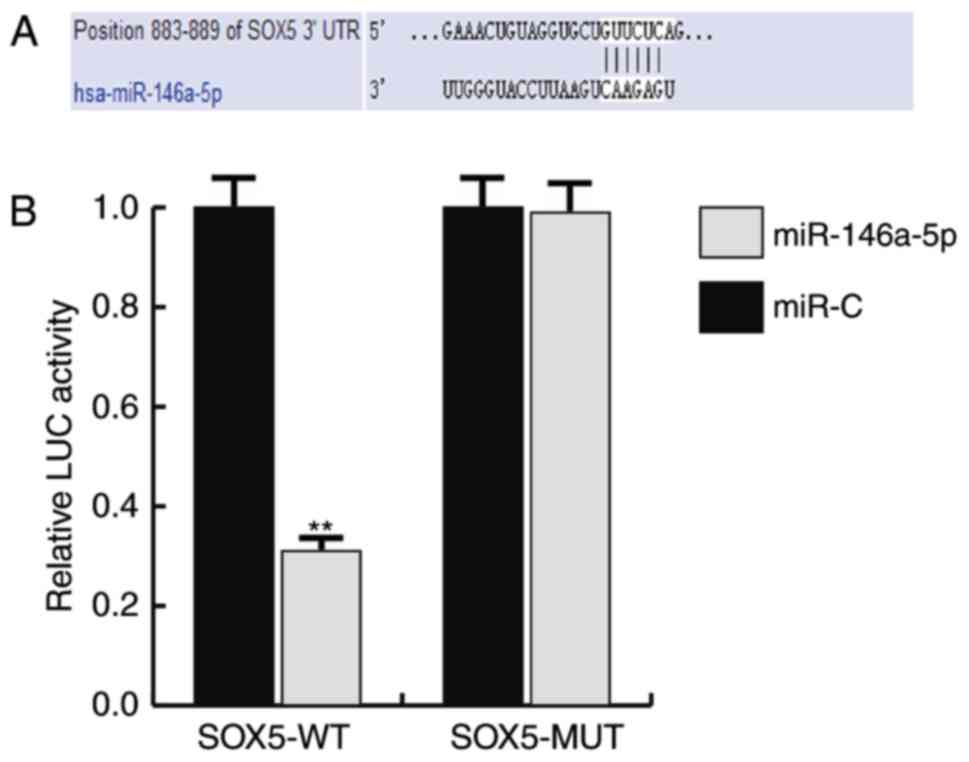
miR-146a-5p targets SOX5
To investigate the mechanisms of the function that
miR-146a-5p serves in TNBC, the target gene of miR-146a-5p was
predicted using TargetScan, and a dual luciferase assay was
performed to verify the prediction. As predicted, it was revealed
that miR-146a-5p directly targets SOX5 (Fig. 2).
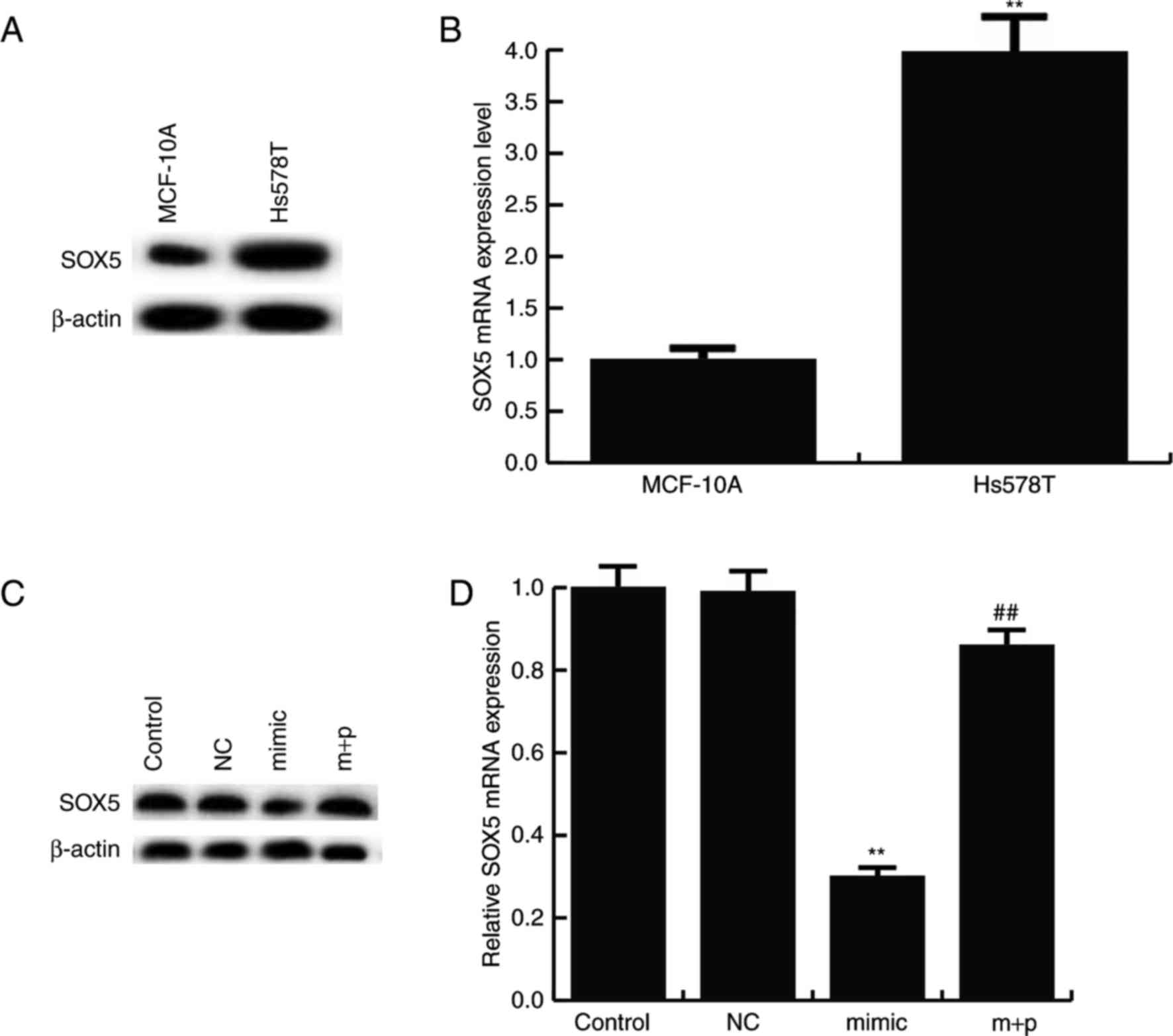
Furthermore, it was revealed that SOX5 protein was
expressed at notably higher levels in the TNBC cell line, Hs578T,
compared with MCF-10A cells (Fig.
3A). Compared with MCF-10A cells, the mRNA level of SOX5 was
significantly increased in Hs578T cells (Fig. 3B). Additionally, it was revealed that
miR-146a-5p negatively regulated the protein and mRNA expression of
SOX5 in Hs578T cells. miR-146a-5p mimics markedly decreased the
SOX5 protein expression, and this decrease was reversed by
SOX5-plasmids (Fig. 3C). It was also
observed that the mRNA level of SOX5 significantly decreased in
miR-146a-5p mimics treated Hs578T cells and this reduction was
significantly reversed by SOX5-plasmids (Fig. 3D).
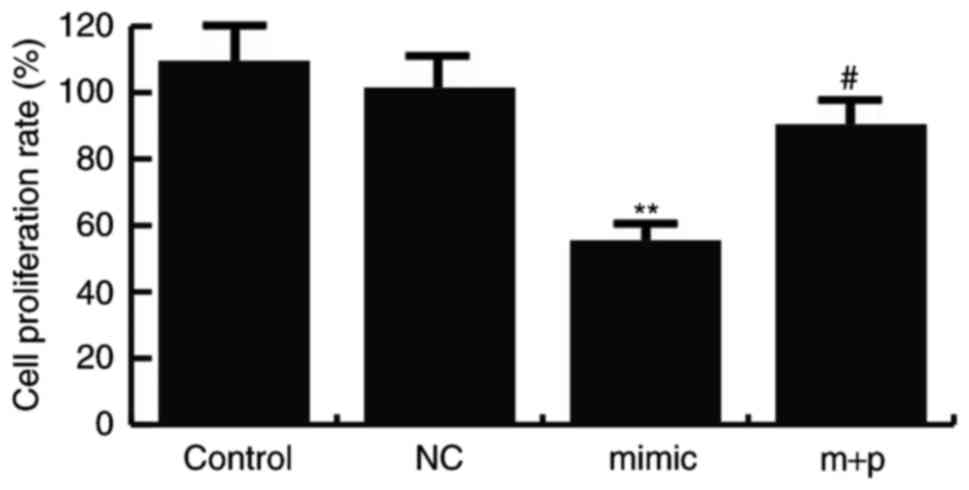
miR-146a-5p inhibits TNBC cell
proliferation
To investigate the effect of miR-146a-5p on TNBC
cell proliferation, an MTT assay was performed. Hs578T cells were
transfected with miR-146a-5p mimics, its negative control or
miR-146a-5p mimics + SOX5-plamids, and the MTT assay was performed
24 h after cell transfection. The results indicated that compared
with the controls, the Hs578T cell proliferation ability was
significantly decreased in the cells transfected with miR-146a-5p
mimics, and SOX5-plamids significantly reverse this effect
(Fig. 4).
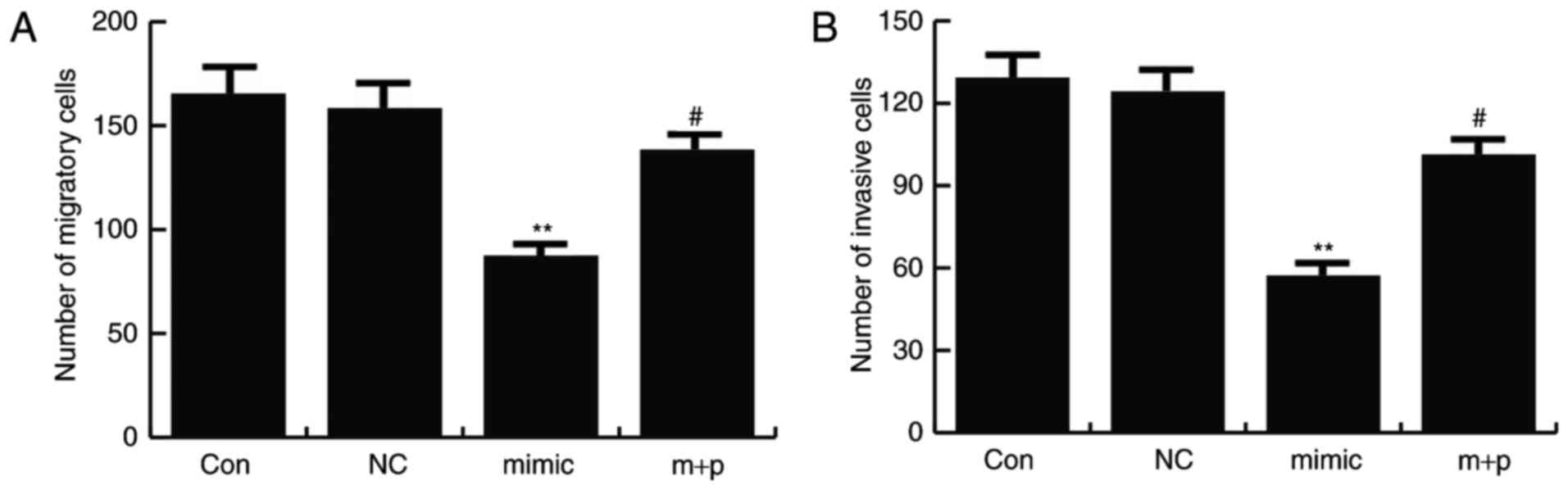
miR-146a-5p decreases migration and
invasion capacities of TNBC cells
After 24 h of cell transfection, Transwell assays
were performed to determine cell migration and invasion rates. It
was revealed that miR-146a-5p mimics significantly inhibited Hs578T
cell migration and invasion compared with the controls, and this
effect could be significantly reversed by SOX5 overexpression
(Fig. 5).
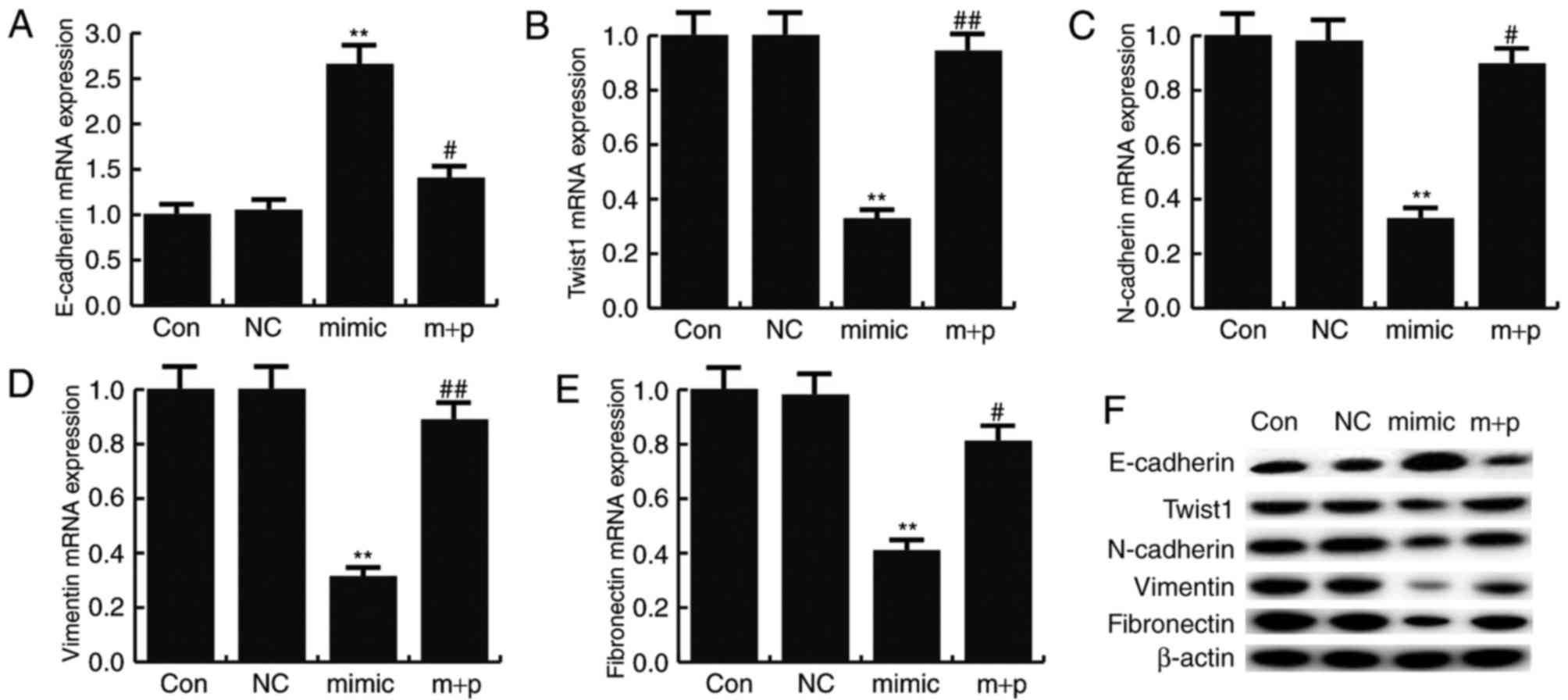
miR-146a-5p inhibits EMT
To determine the effect of miR-146a-5p on TNBC cell
EMT, the expression levels of mesenchymal markers (N-cadherin,
vimentin and fibronectin) and an epithelial marker (E-cadherin)
were detected by RT-qPCR and western blotting. The results revealed
that miR-146a-5p mimics significantly reduced the mRNA expression
of N-cadherin, vimentin and fibronectin compared with the control,
but significantly enhanced the mRNA level of E-cadherin (Fig. 6A-E). Similar results were obtained
from western blot analysis (Fig.
6F). These effects could be significantly reversed by SOX5
overexpression. A previous study revealed that SOX5 could induce
EMT by regulating Twist1 expression (25). In the present study, the data
indicated that miR-146a-5p mimics significantly decreased Twist1
expression compared with the controls, and this decrease could be
significantly reversed by SOX5 overexpression (Fig. 6).
Discussion
TNBC has a strong potential to metastasize, and the
majority of patients succumb to the disease due to distant
metastasis (26). Recently, EMT has
become the focal point of research into the metastatic process
(27,28). Due to a lack of reliable markers and
effective therapeutic targets, TNBC has increasingly attracted the
attention of researchers.
Abnormal expression of miRNAs is known to be
involved in the development of a variety of types of cancer, and
studies have revealed potential of miRNAs as biomarkers for
diagnosis and prognosis (29,30).
Recently, a number of studies have indicated that miRNAs serve
critical roles in the development of TNBC. Jia et al
(31) reported that miR-490-3p could
suppress the growth and invasion of TNBC by inhibiting tankyrase-2
expression. Choi et al (32)
indicated that miR-141/200c was involved in TNBC migration and
invasion via activating the focal adhesion kinase and
phosphoinositide 3-kinase/AKT signaling pathways. miRNA-454 was
revealed to be associated with poor prognosis in TNBC (17) and miRNA-200b could suppress TNBC
metastasis by regulating protein kinase C α (19). Furthermore, miR-211-5p served a tumor
suppressor role in TNBC progression by targeting SETBP1 (33) and miRNA-21 could promote TNBC cell
proliferation and invasion by regulating phosphatase and tensin
homolog (34).
miR-146a-5p functions as a tumor suppressor in
various types of cancer, including hepatocellular carcinoma,
esophageal squamous cell carcinoma, NSCLC and prostate cancer
(20–23). However, the role of miR-146a-5p in
the development of TNBC remains unclear. The present study
investigated the expression and role of miR-146a-5p in TNBC, and it
was revealed that miR-146a-5p was downregulated in TNBC tissues and
cell lines (MDA-MB-231 MDA-MB-468, BT549 and Hs578T). SOX5 was
identified as a target gene of miR-146a-5p and was overexpressed in
TNBC cells. As miR-146a-5p was more evidently decreased in the TNBC
cell line Hs578T, the Hs578T cell line was selected to perform
further investigations. Additionally, the present study
demonstrated that miR-146a-5p could prevent TNBC Hs578T cell
proliferation, migration, invasion and EMT.
To the best of our knowledge the present study was
the first to demonstrate that miR-146a-5p was downregulated in TNBC
tissues and cells, and miR-146a-5p could repress TNBC cell
proliferation, migration, invasion and EMT by targeting SOX5.
Furthermore, miR-146a-5p and SOX5 may potentially be used as novel
targets for TNBC treatment, and the current study provides basis
for exploring novel therapies for the clinical treatment of
TNBC.
Acknowledgements
The present study was supported by Jiangsu
Provincial Medical Youth Talent and the Project of Invigorating
Health Care through Science, Technology and Education (grant no.
QNRC2016661).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang H, Tan G, Dong L, Cheng L, Li K, Wang
Z and Luo H: Circulating MiR-125b as a marker predicting
chemoresistance in breast cancer. PLoS One. 7:e342102012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B and Senn HJ: Panel
Members: Tailoring therapies-improving the management of early
breast cancer: St Gallen international expert consensus on the
primary therapy of early breast cancer 2015. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Y, Zhu P, Wang Y, Wei Z, Tao L, Zhu Z,
Sheng X, Wang S, Ruan J, Liu Z, et al: Antimetastatic therapies of
the polysulfide diallyl trisulfide against triple-negative breast
cancer (TNBC) via suppressing MMP2/9 by blocking NF-kB and ERK/MAPK
signaling pathways. PLoS One. 10:e01237812015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gucalp A and Traina TA: Triple-negative
breast cancer: Role of the androgen receptor. Cancer J. 16:62–65.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sandhu S and Garzon R: Potential
applications of microRNAs in cancer diagnosis, prognosis, and
treatment. Semin Oncol. 38:781–787. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoo B, Kavishwar A, Ross A, Wang P,
Tabassum DP, Polyak K, Barteneva N, Petkova V, Pantazopoulos P,
Tena A, et al: Combining miR-10b-targeted nanotherapy with low-dose
doxorubicin elicits durable regressions of metastatic breast
cancer. Cancer Res. 75:4407–4415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Leeneer K and Claes K: Non coding RNA
molecules as potential biomarkers in breast cancer. Adv Exp Med
Biol. 867:263–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yuan Y, Anbalagan D, Lee LH, Samy RP,
Shanmugam MK, Kumar AP, Sethi G, Lobie PE and Lim LH: ANXA1
inhibits miRNA-196a in a negative feedback loop through NF-κB and
c-Myc to reduce breast cancer proliferation. Oncotarget.
7:27007–27020. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoo JO, Kwak SY, An HJ, Bae IH, Park MJ
and Han YH: miR-181b-3p promotes epithelial-mesenchymal transition
in breast cancer cells through Snail stabilization by directly
targeting YWHAG. Biochim Biophys Acta. 1863:1601–1611. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh
F, Asghari F and Yousefi M: The role of oncomirs in the
pathogenesis and treatment of breast cancer. Biomed Pharmacother.
78:129–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu
X, Song CG and Shao ZM: High expression of microRNA-454 is
associated with poor prognosis in triple-negative breast cancer.
Oncotarget. 7:64900–64909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang L, Wei D and Yan F: MicroRNA-145
functions as a tumor suppressor by targeting matrix
metalloproteinase 11 and Rab GTPase family 27a in triple-negative
breast cancer. Cancer Gene Ther. 23:258–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Humphries B, Wang Z, Oom AL, Fisher T, Tan
D, Cui Y, Jiang Y and Yang C: MicroRNA-200b targets protein kinase
Cα and suppresses triple-negative breast cancer metastasis.
Carcinogenesis. 35:2254–2263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Ye ZH, Liang HW, Ren FH, Li P,
Dang YW and Chen G: Down-regulation of miR-146a-5p and its
potential targets in hepatocellular carcinoma validated by a TCGA-
and GEO-based study. FEBS Open Bio. 7:504–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang C, Zhang W, Zhang L, Chen X, Liu F,
Zhang J, Guan S, Sun Y, Chen P, Wang D, et al: miR-146a-5p mediates
epithelial-mesenchymal transition of oesophageal squamous cell
carcinoma via targeting Notch2. Br J Cancer. 115:1548–1554. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li YL, Wang J, Zhang CY, Shen YQ, Wang HM,
Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL and Jin YX: MiR-146a-5p
inhibits cell proliferation and cell cycle progression in NSCLC
cell lines by targeting CCND1 and CCND2. Oncotarget. 7:59287–59298.
2016.PubMed/NCBI
|
|
23
|
Xu B, Huang Y, Niu X, Tao T, Jiang L, Tong
N, Chen S, Liu N, Zhu W and Chen M: Hsa-miR-146a-5p modulates
androgen-independent prostate cancer cells apoptosis by targeting
ROCK1. Prostate. 75:1896–1903. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pei XH, Lv XQ and Li HX: Sox5 induces
epithelial to mesenchymal transition by transactivation of Twist1.
Biochem Biophys Res Commun. 446:322–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tseng LM, Hsu NC, Chen SC, Lu YS, Lin CH,
Chang DY, Li H, Lin YC, Chang HK, Chao TC, et al: Distant
metastasis in triple-negative breast cancer. Neoplasma. 60:290–294.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han Y, Zhang L, Wang W, Li J and Song M:
Livin promotes the progression and metastasis of breast cancer
through the regulation of epithelial-mesenchymal transition via the
p38/GSK3β pathway. Oncol Rep. 38:3574–3582. 2017.PubMed/NCBI
|
|
28
|
Okita Y, Kimura M, Xie R, Chen C, Shen LT,
Kojima Y, Suzuki H, Muratani M, Saitoh M, Semba K, et al: The
transcription factor MAFK induces EMT and malignant progression of
triple-negative breast cancer cells through its target GPNMB. Sci
Signal 10: pii: eaak9397. 2017. View Article : Google Scholar
|
|
29
|
van Schooneveld E, Wildiers H, Vergote I,
Vermeulen PB, Dirix LY and Van Laere SJ: Dysregulation of microRNAs
in breast cancer and their potential role as prognostic and
predictive biomarkers in patient management. Breast Cancer Res.
17:212015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McGuire A, Brown JA and Kerin MJ:
Metastatic breast cancer: The potential of miRNA for diagnosis and
treatment monitoring. Cancer Metastasis Rev. 34:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jia Z, Liu Y, Gao Q, Han Y, Zhang G, Xu S,
Cheng K and Zou W: miR-490-3p inhibits the growth and invasiveness
in triple-negative breast cancer by repressing the expression of
TNKS2. Gene. 593:41–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi SK, Kim HS, Jin T, Hwang EH, Jung M
and Moon WK: Overexpression of the miR-141/200c cluster promotes
the migratory and invasive ability of triple-negative breast cancer
cells through the activation of the FAK and PI3K/AKT signaling
pathways by secreting VEGF-A. BMC Cancer. 16:5702016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen LL, Zhang ZJ, Yi ZB and Li JJ:
MicroRNA-211-5p suppresses tumour cell proliferation, invasion,
migration and metastasis in triple-negative breast cancer by
directly targeting SETBP1. Br J Cancer. 117:78–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|