Introduction
Cancer represents the second greatest cause of
mortality in developing countries, for which the chemotherapy is
the most effective treatment. However, its efficiency is limited by
the ability of cancer cells to acquire resistance to the treatment
regiment, which accounts for ~90% of failures (1,2). Drug
resistance is currently considered the most notable challenge in
cancer therapy. This resistance may be determined by genetic and
epigenetic mechanisms as a response to different biochemical
processes, and also by changes in drug levels during the course of
treatment (3). One potential
approach to overcome this, while improving the prognostic and
outcome of patients, is the use of adjuvant therapy in addition to
the standard chemotherapy regimen.
Deuterium is a natural and stable isotope of
hydrogen, which is found in the Earth's atmosphere at a
concentration of 144-155 ppm (4). An
increase in deuterium concentration leads to the formation of heavy
water, which has different physical and chemical proprieties
compared with normal water (5). This
heavy water has detrimental effects on living organisms; ingestion
can cause sterility, while high concentrations of deuterium can
result in mortality via modification of the mitotic cycle and cell
membrane function (6). Therefore it
is presumable that deuterium-depleted water (DDW) may have
beneficial effects by interfering cell metabolism and the overall
function of living cells (7,8).
A number of previous studies have explored the
effects of deuterium-depletion on cell proliferation, on both in
vitro and in vivo cancer models. A study on the A549
lung carcinoma cell line demonstrated that deuterium-depleted water
leads to a reduction in cell proliferation between 10 and 72 h of
exposure, with a peak of cellular structural changes occurring at
72 h of exposure (9). This effect of
tumor inhibition has also been confirmed on orthotopic models of
BALB/c mice (9) and human patients
with lung cancer (10). This tumor
regression may be correlated with a reduction in the expression
levels of several oncogenes, such as KRAS, B cell lymphoma 2 (Bcl2)
or c-Myc, as previously reported (10), suggesting apoptosis as the potential
mechanism of tumor inhibition by DDW in cell lines that overexpress
Bcl2, a process also observed in pancreatic cell lines (11). Besides the pro-apoptotic effect, DDW
seems to have an inhibitory effect on migration and invasion by
downregulating proliferating cell nuclear antigen and matrix
metalloproteinase 9 previous in nasopharyngeal cell carcinoma
(12). Additionally, this study
indicated an induction of NAD(P)H quinone dehydrogenase 1
expression, which is a protein that regulates different cell cycle
factors, such as cyclin D1, p21 and c-Myc.
Free or exosome-released microRNA (miRNA or miR)
patterns may be used as valuable biomarkers for defining
physiological and pathological processes of cell subpopulation
(13–16). Previous studies have emphasized the
important role of the manipulation of miRNA profiles (17,18),
which is considered as a novel approach for colon cancer
prevention, chemotherapy and avoidance of drug resistance related
mechanisms (19,20). Therefore, in the present study the
impact of DDW on the capacity to alter released miRNA patterns was
evaluated in colorectal cancer cells, along with a set of
preliminary functionality tests in order to evaluate the utility of
DDW as an adjuvant in colorectal cancer therapy.
Materials and methods
Materials and cell lines
Powdered RPMI-1640 medium supplemented with
glutamine, liquid RPMI-1640, fetal bovine serum (FBS), PBS,
penicillin-streptomycin 100X and trypsin-EDTA solutions were
obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Dimethyl sulfoxide (DMSO) and MTT were obtained from Sigma-Aldrich;
Merck KGaA (Darmstadt, Germany). DLD-1 (colorectal carcinoma) cell
line was obtained from American Type Culture Collection (Manassas,
VA, USA). Oxaliplatin was obtained from Fresenius Kabi
Asia-Pacific, Ltd. (Wanchai, Hong Kong) and 5-fluorouracil (5-FU)
was obtained from Ebewe Pharma GmbH (Unterach, Austria). DDW was
provided by Qlarivia; Mecro System SRL (Bucharest, Romania) with
25±5 ppm D/(D+H), obtained by vacuum distillation.
MTT cell viability assay
For assessing the cytotoxicity of selected
chemotherapeutic agents, DLD-1 cell line was maintained for 64
passages in standard conditions (SC) and for 66 passages in medium
with low concentration of deuterium (DDW). A total of 10,000 DLD-1
colorectal cancer cells/well were plated in 96-well plates in
RPMI-1640 medium supplemented with 10% FBS, 2 mM glutamine and 1X
penicillin-streptomycin for SC cell culture, and 10,000 DLD-1
colorectal cancer cells/well were plated in 96-well plates in
filter-sterilized RMPI-1640 medium powder (supplemented with
glutamine) prepared with DDW supplemented with 10% FBS and 1X
penicillin-streptomycin for DDW cell culture. Cells were incubated
at 37°C in a 5% CO2 atmosphere for 24 h and the cytostatic agents
were subsequently applied separately in triplicate at
concentrations of 1, 2, 3, 4, 5, 8, 10 and 12 µM for 5-FU, and 2,
4, 6, 8, 10, 12, 14 and 20 µM for oxaliplatin. At 24, 48 and 72 h
following treatment, the medium was discarded, and the adherent
cells were treated with 150 µl MTT solution (1 mg/ml). Following 1
h incubation at 37°C, the formazan crystals were solubilized with
100 µl DMSO and the viability of the cells was assessed at 492 nm
wavelength using a microplate reader (BioTek Instruments, Inc.,
Winooski, VT, USA).
Computation of inhibition constant 50
(IC50)
The response curves to cytostatic drugs were
generated from the MTT data and computed using MasterPlex Readerfit
software (trial version 2.0.0.73; MiraiBio Group of Hitachi
Solutions America, Ltd., San Francisco, CA, USA), and IC50 values
were determined based on the response curves.
Total RNA extraction from
exosome-released medium
The exosomes released from cells cultured in SC and
DDW cell culture medium without 5-FU or oxaliplatin were
precipitated from 4 ml cell culture media using Total Exosome
Isolation Reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Subsequently the precipitated exosomal
fraction was used for extraction of total RNA using a Plasma/Serum
Circulating and Exosomal RNA Purification kit (Slurry Format;
Norgen Biotek Corp., Thorold, ON, Canada) for the cells maintained
in SC and those maintained in DDW-prepared medium.
Microarray evaluation, data analysis
and identification of differentially expressed miRNAs released in
cell culture medium
The microarray experiment was conducted using the
Human miRNA Microarray Kit (release 21.0; format, 8×60K slides;
Agilent Technologies, Inc., Santa Clara, CA, USA). The miRNA
Labeling and Hybridization kit was used according to the
manufacturer's protocol, together with the Complete Labeling and
Hybridization kit (both Agilent Technologies, Inc.) using 100 ng
total RNA from 2 samples of cells grown in SC and 2 samples
cultivated in DDW-prepared medium, labeled with pCp-Cy3 and
purified with MicroBioSpin 6 Columns (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Slides were hybridized for 20 h at 54°C, washed
with Agilent washing solution 1 and 2, and scanned using the
Agilent Microarray Scanner G2565BA (Agilent Technologies,
Inc.).
Samples were grouped according to duplicates, and
preliminary data analysis was performed using Feature Extraction
Software (version 10.7; Agilent Technologies, Inc.). Differential
analysis of miRNA expression was performed using GeneSpring GX
version 12.6.1 software (Agilent Technologies, Inc.), using P≤0.05
and −1.5≤ fold change (FC) ≥1.5. For the integration of the altered
miRNA patterns as an effect of DDW exposure in biological
mechanisms, the NCBI Database was used (www.ncbi.nlm.nih.gov).
Ingenuity Pathway Analysis (IPA®; Qiagen,
Inc., Valencia, CA, USA) is a valuable bioinformatics tool that was
used for the identification of the most relevant altered pathways
based on the identified altered transcriptomic profile in the cells
cultured in DDW conditions compared with those in SC conditions. It
contains a database with >302 metabolic networks and 360
signaling pathways used for the altered miRNA signature, the miRNAs
were scored on the significance of their overlap with these
networks and pathways.
The Cancer Genome Atlas (TCGA) data
analysis for colorectal cancer
The extent to which the expression of microRNAs is
prone to modification as a result of treatment was subsequently
evaluated. TCGA was used and differential analysis was performed
using the miRNA expression data for patients with colorectal
cancer, which was downloaded from the University of California
Santa Cruz genome browser (https://genome.ucsc.edu). The data for colon
adenocarcinoma, which consisted of 433 tumor samples and 8 normal
samples, was combined with expression data for rectum
adenocarcinoma, which consisted of 162 tumor tissues and 3 normal
tissues, to obtain the colorectal cancer cohort. Using GeneSpring
GX software (version 14.9; Agilent Technologies, Inc.),
differential expression analysis was performed, and by applying
filters of using P≤0.05 and −1.5≤FC≥1.5, a list containing the most
upregulated and downregulated microRNAs between tumors and normal
tissues was generated, as described previously (21–23).
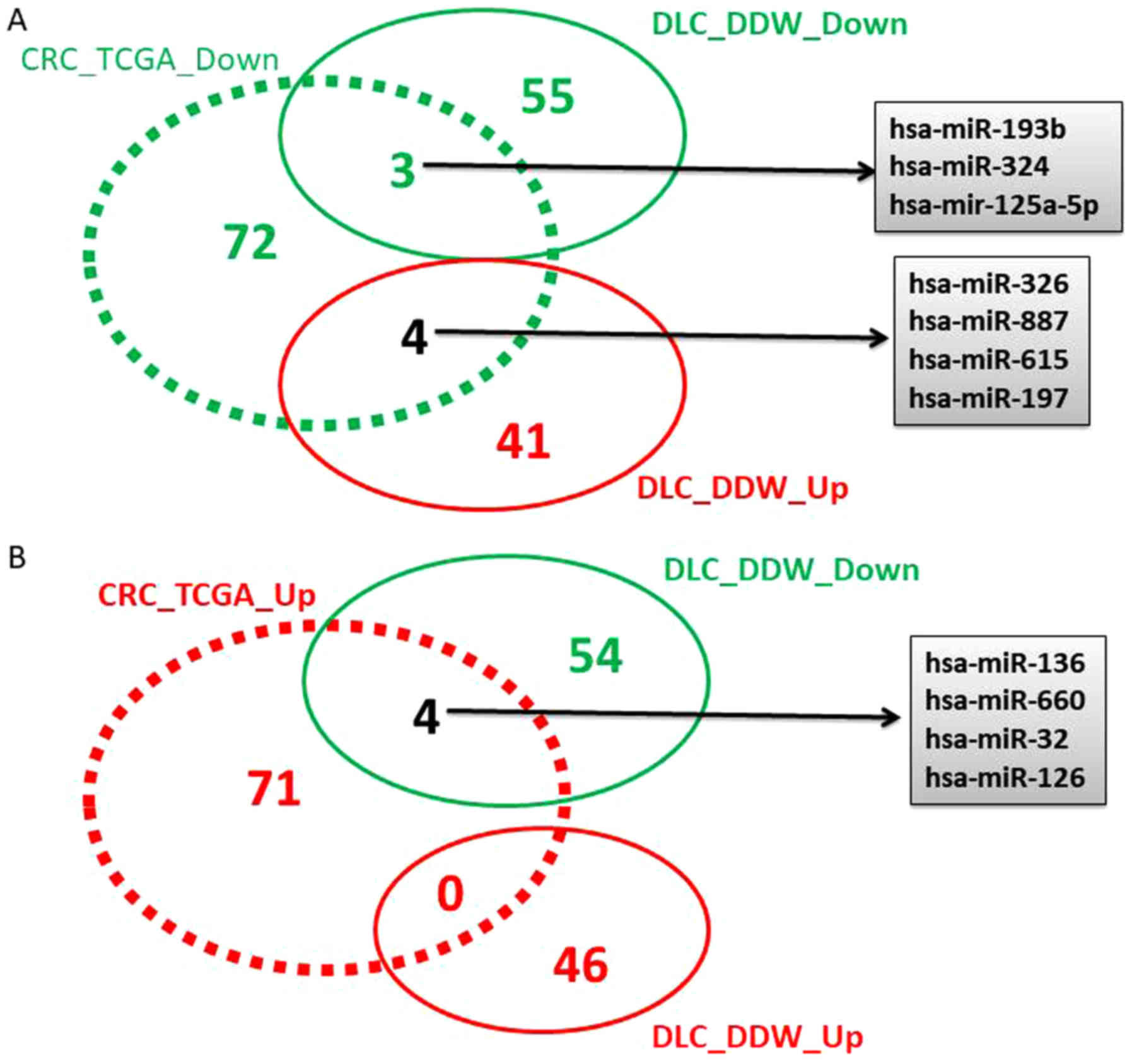
Generation of Venn diagram
Using the obtained TCGA miRNA data the Venn diagram
was generated using the online tool Venny version 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/) and
indicated the commonly or inversely regulated transcripts in
DDW-treated cells.
Assessing apoptosis via fluorescence microscopy
tetramethylrhodamine, ethyl ester (TMRE)/Hoechst double
staining. Evaluation of apoptosis was performed using a
Multi-Parameter Apoptosis kit (Cayman Chemical Company, Ann Arbor,
MI, USA), according to the manufacturer's protocol. Cells from the
SC and DDW groups were analyzed at UV wavelength (560/595 nm) for
Hoechst and TMRE staining using a Mitochondrial Membrane Potential
Assay kit (cat no. 701310; Cayman Chemical Company) according to
the manufacturer's protocol, and Olympus IX71 microscope (Olympus
Corporation, Tokyo, Japan) at a magnification of ×20. Hoechst is a
cell permeable dye for the nuclei, independent of the cell membrane
status, whereas TMRE dye allows the evaluation of mitochondrial
function by assessing the membrane potential, which is lost in
cells entering apoptosis (24–26).
Cell apoptosis was evaluated at 48 h of exposure to 5-FU and
oxaliplatin at the IC50 concentration.
Evaluation of senescence
For senescence evaluation of cells cultured in SC
and DDW conditions, the Senescence Detection kit (Abcam, Cambridge,
UK) was used according to the manufacturer's protocol. The kit
detects β-galactosidase, which is overexpressed in senescent cells
(27). Cells were visualized in both
cases using an Olympus IX71 microscope (magnification, ×20).
Results
Evaluation of the altered miRNA
expression profiles
Differential miRNA expression profiles in the case
of colorectal cells maintained in DDW vs. those maintained in SC
were evaluated using miRNA labeling and hybridization, and
microarray evaluation. During data analysis using the specialized
GeneSpring GX software, a cut-off value of −1.5≤FC≥1.5 and
P<0.05 were considered, and were able to identify several
transcripts with altered expression levels. In total, 105
transcripts with modified expression levels were identified, from
which 46 were upregulated and 59 downregulated (Table I).
 | Table I.The altered miRNA pattern for the
case of deuterium-depleted water-maintained cells vs. those
maintained in standard conditions. |
Table I.
The altered miRNA pattern for the
case of deuterium-depleted water-maintained cells vs. those
maintained in standard conditions.
| Systematic
name | Fold change | Corrected
P-value |
|---|
| hsa-miR-4261 | −22.7086 | 2.16E-04 |
|
hsa-miR-3912-5p | −22.0231 | 2.16E-04 |
|
hsa-miR-4711-3p | −18.467 | 2.16E-04 |
| hsa-miR-362-5p | −16.8774 | 0.001651 |
| hsa-miR-3923 | −16.6997 | 0.001231 |
|
hsa-miR-664a-3p | −16.3672 | 0.001651 |
| hsa-miR-34b-5p | −16.0323 | 0.008314 |
| hsa-miR-136-5p | −14.7967 | 0.002438 |
|
hsa-miR-5007-5p | −13.0422 | 2.16E-04 |
| hsa-miR-4317 | −13.0337 | 6.44E-04 |
|
hsa-miR-193b-5p | −12.6629 | 6.44E-04 |
|
hsa-miR-4700-5p | −12.6546 | 0.021906 |
|
hsa-miR-125a-5p | −12.1643 | 0.001253 |
| hsa-miR-936 | −11.01 | 0.014825 |
|
hsa-miR-6516-5p | −10.4295 | 0.012692 |
| hsa-miR-4488 | −10.3318 | 0.012692 |
| hsa-miR-545-3p | −9.28502 | 0.013747 |
|
hsa-miR-4715-5p | −9.23226 | 0.008749 |
| hsa-miR-4468 | −7.60702 | 3.74E-04 |
| hsa-miR-32-5p | −5.34052 | 0.011851 |
|
hsa-miR-6872-3p | −4.00105 | 0.017607 |
| hsa-miR-4291 | −2.81713 | 0.03264 |
| hsa-miR-1260b | −2.52161 | 0.006656 |
|
hsa-miR-1233-5p | −2.2066 | 0.022847 |
| hsa-miR-5684 | −2.15719 | 0.013747 |
| hsa-miR-3976 | −2.12003 | 0.042139 |
| hsa-miR-182-5p | −2.11261 | 0.010321 |
| hsa-miR-5100 | −2.11064 | 0.014468 |
| hsa-miR-1260a | −2.08488 | 0.00429 |
| hsa-miR-660-3p | −2.06228 | 0.03624 |
|
hsa-miR-3064-5p | −2.03183 | 0.021906 |
|
hsa-miR-6717-5p | −2.0278 | 0.017607 |
|
hsa-miR-6788-5p | −1.96362 | 0.009607 |
|
hsa-miR-7114-5p | −1.95488 | 0.014468 |
| hsa-miR-18b-5p | −1.9179 | 0.012687 |
| hsa-miR-324-5p | −1.8771 | 0.034038 |
|
hsa-miR-1273g-3p | −1.87346 | 0.033338 |
| hsa-miR-6131 | −1.8712 | 0.019085 |
| hsa-miR-921 | −1.87005 | 0.037554 |
|
hsa-miR-6826-5p | −1.85113 | 0.014468 |
|
hsa-miR-7159-5p | −1.83784 | 0.03967 |
|
hsa-miR-4753-5p | −1.80366 | 0.014468 |
| hsa-miR-320b | −1.74454 | 0.019579 |
| hsa-miR-126-3p | −1.73136 | 0.045633 |
|
hsa-miR-5196-5p | −1.72965 | 0.033735 |
|
hsa-miR-6872-5p | −1.65159 | 0.024325 |
| hsa-miR-211-3p | −1.61236 | 0.021906 |
|
hsa-miR-6875-5p | −1.60393 | 0.014468 |
| hsa-miR-320e | −1.57832 | 0.012692 |
|
hsa-miR-146a-5p | −1.57515 | 0.033735 |
| hsa-miR-23a-3p | −1.57461 | 0.034732 |
| hsa-let-7d-5p | −1.57108 | 0.037564 |
| hsa-miR-6085 | −1.57099 | 0.012896 |
| hsa-miR-23b-3p | −1.55252 | 0.034038 |
| hsa-miR-3907 | −1.53592 | 0.012692 |
| hsa-miR-3650 | −1.52958 | 0.013747 |
|
hsa-miR-3689b-3p | −1.52255 | 0.033715 |
| hsa-miR-4286 | −1.52084 | 0.034127 |
| hsa-miR-539-5p | −1.51706 | 0.026802 |
|
hsa-miR-6777-3p | 12.35759 | 0.001651 |
|
hsa-miR-1247-5p | 10.66176 | 0.012687 |
|
hsa-miR-5008-3p | 10.36788 | 0.017607 |
|
hsa-miR-6870-3p | 10.21028 | 0.010321 |
| hsa-miR-4508 | 10.0584 | 0.014468 |
| hsa-miR-373-5p | 8.170245 | 0.04109 |
|
hsa-miR-6812-3p | 7.939796 | 0.042139 |
| hsa-let-7b-3p | 7.565567 | 0.034732 |
| hsa-miR-326 | 7.435405 | 0.027191 |
| hsa-miR-375 | 3.686158 | 0.012687 |
|
hsa-miR-6726-5p | 3.444442 | 0.001231 |
|
hsa-miR-3934-5p | 3.160403 | 0.009522 |
|
hsa-miR-1236-5p | 2.984447 | 0.001231 |
| hsa-miR-887-3p | 2.868279 | 0.033735 |
|
hsa-miR-135a-3p | 2.717766 | 0.03624 |
| hsa-miR-4634 | 2.264031 | 0.039541 |
| hsa-miR-718 | 2.204079 | 0.011618 |
| hsa-miR-422a | 2.180337 | 0.033338 |
|
hsa-miR-5585-3p | 2.140601 | 0.024133 |
| hsa-miR-8089 | 2.08933 | 0.008314 |
|
hsa-miR-6760-3p | 2.02642 | 0.014468 |
| hsa-miR-615-3p | 2.022371 | 0.033735 |
| hsa-miR-1181 | 1.971624 | 0.012687 |
| hsa-miR-4476 | 1.904539 | 0.026744 |
| hsa-miR-197-5p | 1.886881 | 0.033715 |
| hsa-miR-6133 | 1.845085 | 0.012687 |
|
hsa-miR-7846-3p | 1.83585 | 0.017607 |
| hsa-miR-5787 | 1.829065 | 0.017607 |
|
hsa-miR-6858-3p | 1.826295 | 0.028224 |
|
hsa-miR-4646-5p | 1.779853 | 0.012687 |
| hsa-miR-630 | 1.766601 | 0.012687 |
| hsa-miR-3646 | 1.748758 | 0.011618 |
| hsa-miR-583 | 1.748449 | 0.017607 |
|
hsa-miR-3679-5p | 1.736775 | 0.033735 |
|
hsa-miR-3622b-5p | 1.725858 | 0.014468 |
|
hsa-miR-1185-2-3p | 1.654606 | 0.013747 |
|
hsa-miR-4769-5p | 1.653051 | 0.027191 |
| hsa-miR-6125 | 1.63833 | 0.013747 |
|
hsa-miR-125a-3p | 1.638016 | 0.033735 |
| hsa-miR-6068 | 1.636307 | 0.012687 |
| hsa-miR-4530 | 1.627722 | 0.029275 |
|
hsa-miR-7107-5p | 1.625659 | 0.033735 |
| hsa-miR-601 | 1.610057 | 0.012687 |
| hsa-miR-5703 | 1.598128 | 0.012692 |
| hsa-miR-3960 | 1.593793 | 0.021971 |
| hsa-miR-2861 | 1.537284 | 0.026802 |
IPA® Network analysis
One of the aims of the present study was to evaluate
the potential significance of altered miRNAs at the cellular and
molecular level, mediated by DDW. IPA® analysis was
performed for all miRNAs with an altered expression level for the
assessment of the potential consequences by evaluating the
networks; the principle of the gene network interactions were
discussed in a previous study (28).
Fig. 1 presents the integration of
the altered miRNAs transcripts in with the highest significance
score, including organismal injury and abnormalities, reproductive
system disease, cancer (Fig. 1A),
inflammatory diseases, the inflammatory response, organismal injury
and abnormalities (Fig. 1B), and
hematological diseases and hereditary disorders (Fig. 1C). The IPA® analysis also
classified the altered miRNAs transcripts in terms of the most
relevant diseases and biological function (Table II) or molecular functions (Table III) associated with DDW
exposure.
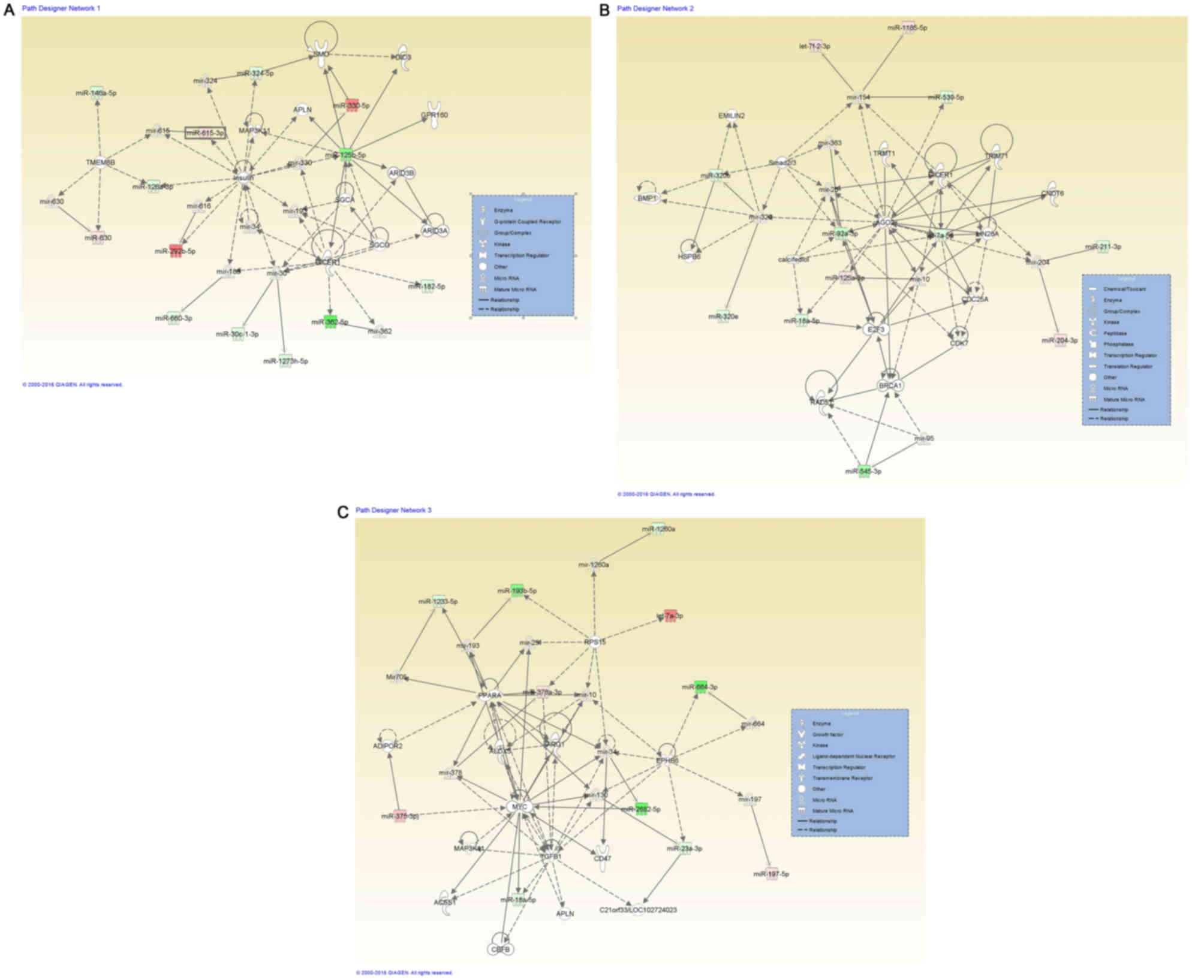 | Figure 1.miRNA-miRNA gene network interactions
generated using Ingenuity Pathway Analysis (Qiagen, Inc., Valencia,
CA, USA), for miRNAs altered in deuterium-depleted water-maintained
DLD-1 cells vs. those maintained in standard conditions. (A) A
representative network for organismal injury and abnormalities,
reproductive system disease and cancer, containing 13 miRNAs with
Dicer1 as a central molecule. (B) Network representative for
inflammatory diseases, inflammatory response, and organismal injury
and abnormalities, containing 12 miRNAs with Dicer1, Ago2 and E2F3
as key molecules. (C) Network representative for cancer containing
12 miRNAs, hematological diseases and hereditary disorders, with
Myc and TGFβ1 as central molecules. miRNAs expression changes are
depicted in red (upregulation) and green (downregulation), and the
direct target genes that interact with altered miRNAs are grey.
miRNA/miR, microRNA; Ago2, Argonaute 2; E2F3, transcription factor
E2F3; TGF, transforming growth factor. |
 | Table II.Most significant diseases and
functions altered in cells cultured in deuterium-depleted water vs.
those maintained in standard conditions. |
Table II.
Most significant diseases and
functions altered in cells cultured in deuterium-depleted water vs.
those maintained in standard conditions.
|
Disease/function | P-value | Molecules (n) |
|---|
| Inflammatory
disease |
1.90E-02-3.95E-12 | 13 |
| Inflammatory
response |
3.22E-02-3.95E-12 | 13 |
| Organismal injury
and abnormalities |
4.99E-02-3.95E-12 | 29 |
| Cancer |
4.86E-02-3.59E-11 | 18 |
 | Table III.Most significant molecular functions
from the miRNA pattern of cell cultured in deuterium-depleted water
vs. those maintained in standard conditions. |
Table III.
Most significant molecular functions
from the miRNA pattern of cell cultured in deuterium-depleted water
vs. those maintained in standard conditions.
| Name | P-value | Molecules (n) |
|---|
| Cell-to-cell
signaling and interaction |
4.26E-03-8.72E-05 | 3 |
| Cellular function
and maintenance |
3.77E-02-8.72E-05 | 3 |
| Cell cycle |
3.91E-02-1.00E-03 | 3 |
| Cellular
movement |
3.98E-02-1.62E-03 | 5 |
| Cell
morphology |
2.53E-02-2.84E-03 | 1 |
Overlapping the altered miRNA pattern
in DDW cells with TGCA data
Following TCGA data analysis, 163 miRNAs with
altered expression levels were identified, 82 downregulated and 81
overexpressed (data not shown), considering a cut-off value of
−1.5≤FC≥1.5 and P<0.05. A Venn diagram was constructed to
identify the overlap of differentially regulated miRNAs obtained
from the TCGA analysis for colorectal cancer, and the altered miRNA
pattern obtained in the case of DLD-1 cells maintained in DDW vs.
SC (Fig. 2).
DDW augmentation exhibits a weak
pro-apoptotic effect in 5-FU and oxaliplatin treated DLD-1
cells
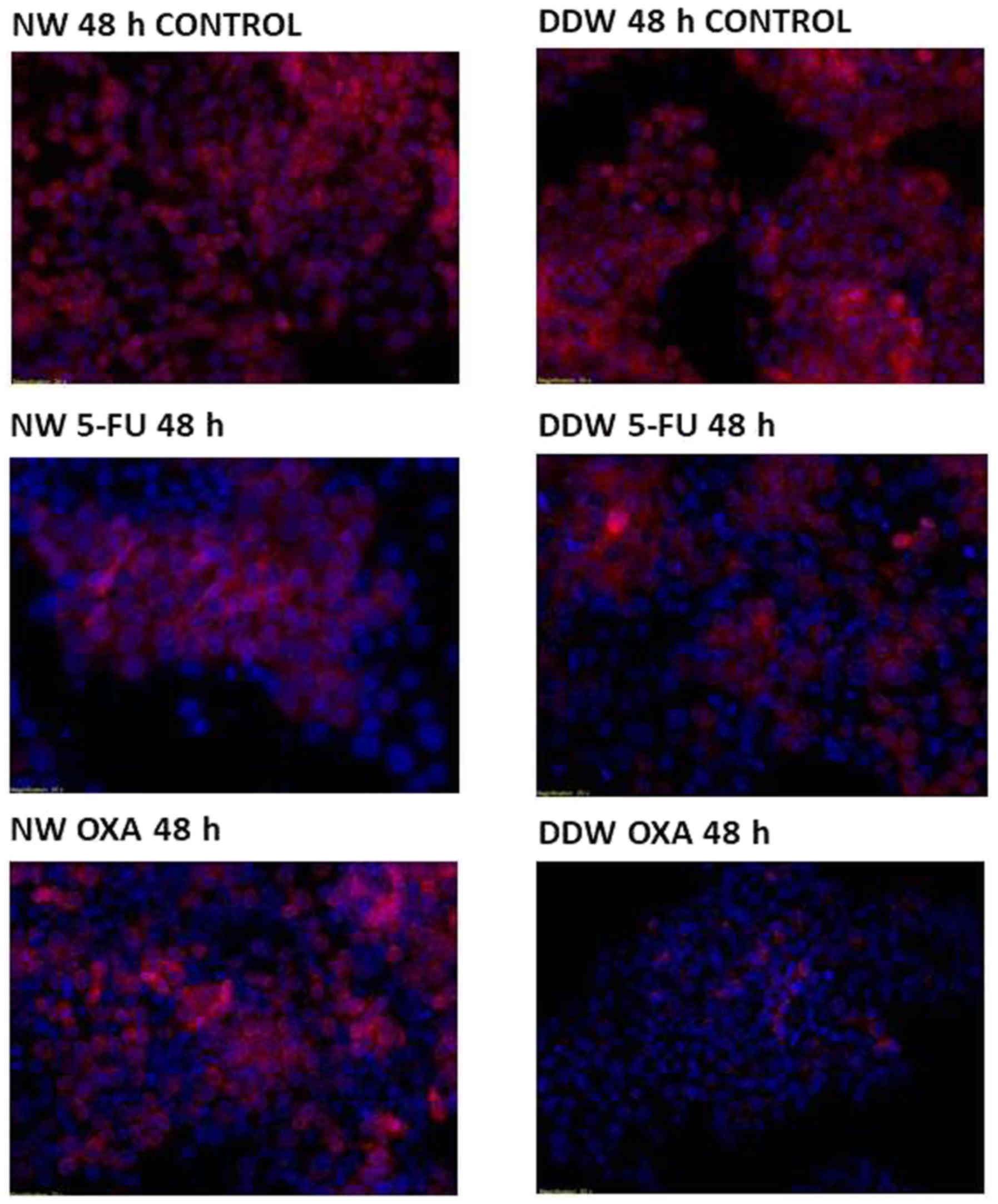
In order to determine whether DDW exhibits an
adjuvant effect with 5-FU and oxaliplatin treatment on DLD-1 cells,
cell viability was assessed via fluorescent microscopy using
TMRE/Hoechst double staining. A common hallmark of apoptosis is the
fragmentation and condensation of the nucleus.
In SC, no modifications of the nuclei were observed,
however 5-FU treatment appears to be complemented by a decreased
concentration of deuterium, with nuclei displaying slight
structural modifications at 48 h following 5-FU exposure in the
presence of DDW. The mitochondrial membrane potential remains
unaffected (Fig. 3), with cells
retaining their viability independent of deuterium concentrations
(Table IV). By constrast,
oxaliplatin treatment dissipates the mitochondrial membrane
potential, with cells preserving the nucleic integrity (Fig. 3).
 | Table IV.IC50 concentrations for 5-FU and
oxaliplatin obtained in DLD-1 cells. |
Table IV.
IC50 concentrations for 5-FU and
oxaliplatin obtained in DLD-1 cells.
| RPMI medium | Cytostatic
agent | IC50 24 h (µM) | IC50 48 h (µM) | IC50 72 h (µM) |
|---|
| SC | 5-FU |
7.64 | 4.64 | – |
|
| Oxaliplatin |
4.57 | 4.60 | 4.63 |
| DDW | 5-FU |
4.42 | 3.74 | 7.30 |
|
| Oxaliplatin | 21.04 | 4.92 | 6.09 |
Senescence in the cancer cell line
DLD-1 is activated by DDW
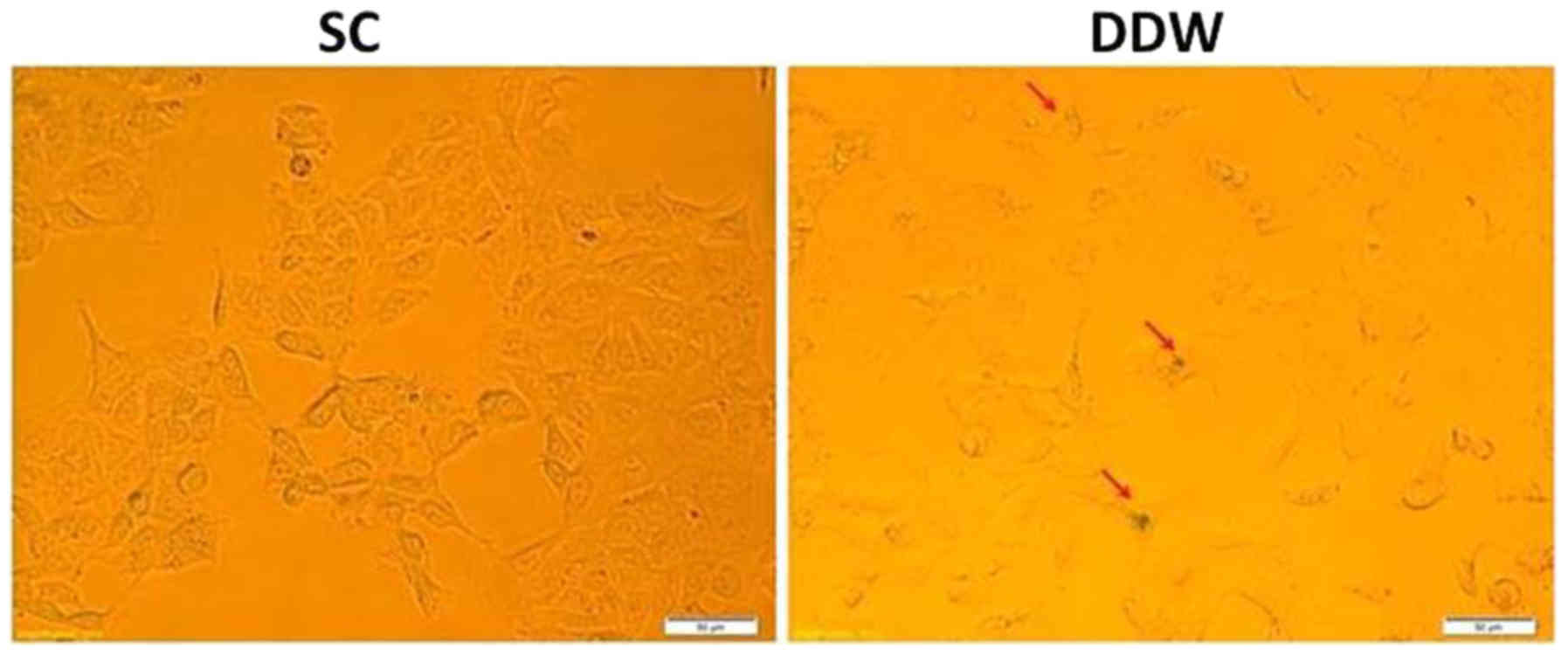
It was investigated whether maintaining the cells in
DDW was able to induce senescence in the DLD-1 cell line, and
therefore offer a novel platform for cancer therapeutics. DLD-1
cells were cultivated for 13 passages in SC or medium prepared with
DDW, and were assessed for senescence via detection of
β-galactosidase expression. The results indicated a low induction
of senescence in cells cultivated in DDW medium compared with those
maintained in SC (Fig. 4).
Discussion
Supplementation of cancer therapy with different
adjuvant systems have been reported to induce notable benefits in a
number cancer types, including colorectal cancer (29). The most well-known combination is
that of oxaliplatin and 5-FU (30,31).
These systems were developed to counteract the activated mechanisms
that sustain tumor development, and particularly to avoid
chemoresistance (32). Therefore,
there is an urgent demand for evaluating novel adjuvant systems
that may be implemented in clinical practice (29,32).
Chemoresistance has been associated with altered
enzyme functions that are associated with microRNA maturation and
primarily Dicer protein activity (19). Taking into account that
chemoresistance is the main obstacle to the success of cancer
treatment (1,2), the modulation of miRNA patterns may be
used to avoid activation of drug resistance mechanism and to
increase the therapeutic efficacy of classical chemotherapy
(17,33).
In previous studies, deuterium depletion has been
suggested to exhibit antiproliferative effects in cells exposed to
low concentrations of this natural isotope of hydrogen (9,10) and to
alter gene expression patterns in cancer cells (10). However, to the best of our knowledge,
whether deuterium depletion influences the response of cancer cells
to standard chemotherapy has not yet been investigated. Therefore,
the aim of the present study was to investigate if deuterium
concentration in the extracellular environment was able to modulate
the response of DLD-1 colorectal cancer cells to the standard
chemotherapeutic regimen. Oxaliplatin and 5-FU were selected as
chemotherapeutic agents following National Comprehensive Cancer
Network guidelines for colorectal cancer disease (34). In the present study, the effects of
deuterium depletion in DLD-1 cancer cells subjected to 5-FU and
Oxaliplatin treatment were assessed via microarray analysis to
observe the differences in the molecular profile of miRNAs and,
implicitly, of senescence and apoptosis genes modulated by these
miRNAs.
Senescence represents a growth-arrest aging process
that has been associated with degenerative pathologies in cells
subjected to stress stimuli, leading to loss of function (35,36).
Senescence has also been recognized as a potent antiproliferative
mechanism for suppressing tumor growth and progression, and has
been exploited as a potential anti-cancer target (37,38).
The capacity to regulate the miRNA pattern in
normal, physiological conditions may have implications on cell
fate. The modulation of released miRNA patterns may be considered
as a promising strategy for increased therapeutic efficacy and
particularly for avoiding the activation of drug resistance
mechanisms. The modulation of miRNA-processing enzymes, Dicer1 and
miRNAs may be important for senescence-related processes (39,40). The
network presented in Fig. 1A,
presents the key transcript Dicer1, which promotes senescence and
cell differentiation (39,40). IPA® networks revealed that
Dicer1 is targeted by several miRNAs, including miR-362-5p,
miR-182-5p and miR-125-5p. The second network (Fig. 1B) is centered on Dicer1, Argonaute 2
and transcription factor E2F3, which are associated with selective
autophagy (41), and are able to
target cell cycle regulators, cell proliferation and apoptosis
(42–44). Defective autophagy has been
demonstrated to accelerate senescence (45), whereas the stimulation of autophagy
may be an effective, novel therapeutic strategy, which may be used
to increase the response to classical chemotherapy. The miRNA
profiling data revealed two downregulated miRNAs (miR-23b and
miR-193b) and one overexpressed transcript (let-7b) that are known
to regulate autophagy, which was confirmed only partially by
apoptosis and senescence evaluation with a microscope.
A previous study has demonstrated that transforming
growth factor (TGF)β- and mitogen-activated protein
kinase-signaling are associated with oxidative stress and DNA
damage, and share a phenotype of drug-induced paracrine ‘bystander
senescence’ (46). Therefore, miRNAs
targeting TGFβ pathways may collaborate to induce and/or maintain
this bystander senescence (46). The
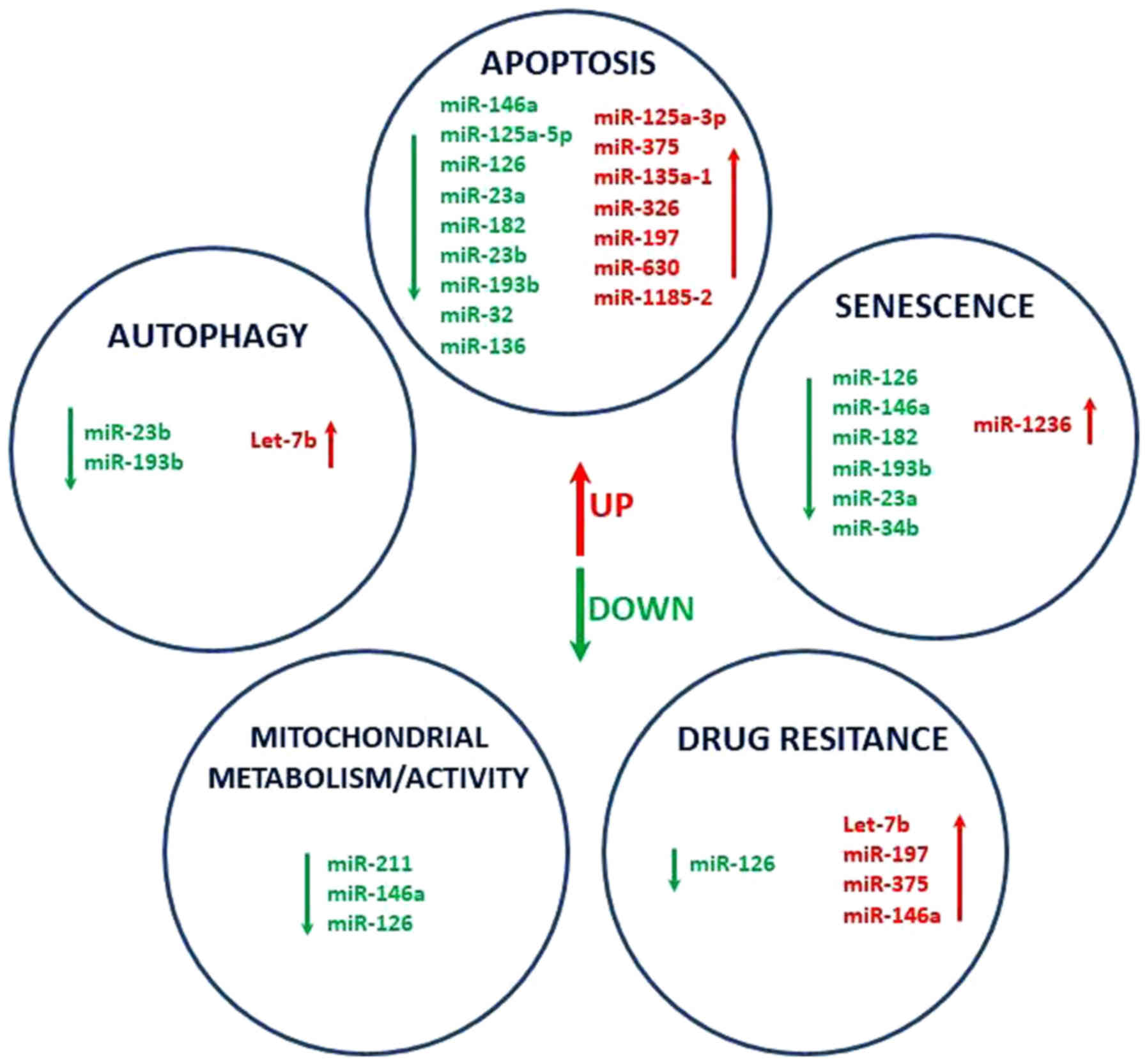
present miRNA profiling data reveals that DDW may contribute to
phenotypic alterations of senescent cells, autophagy, redox
homeostasis and mitochondrial metabolism by modulating the
expression of key regulatory transcripts (Fig. 5). Further investigation on the effect
of miRNA release patterns may elucidate the complex roles of miRNAs
in the response to therapy.
In conclusion, the establishment of adjuvant therapy
to allow premature senescence of tumor cells may be expressed by
the modulation of apoptosis, autophagy, senescent drug resistance
or mitochondrial activity miRNA transcripts, thus leading to the
activation of specific target genes with important roles in cell
fate. Exosomal-released miRNAs represent an effective method to
monitor the response to DDW as a possible adjuvant therapy, and to
identify novel activated mechanisms associated with drug
resistance. Furthermore, DDW may be an adjuvant treatment in
colorectal cancer, but a better comprehension of the associated
molecular mechanisms will be necessary for developing novel
efficient therapeutic strategies, where the transcriptomic pattern
serves an important role.
Acknowledgements
The authors would like to thank Mecro System SRL
(Bucharest, Romania) for providing Qlarivia deuterium-depleted
water as part of the GenCanD project.
Funding
The present study is part of the research grant ‘New
strategies for improving life quality and survival in cancer
patients: Molecular and clinical studies of the tumor genome in
deuterium-depleted water treatment augmentation-GenCanD’. The
research grant was supplied by Iuliu-Hatieganu University of
Medicine and Pharmacy (Cluj-Napoca, Romania; grant no. 128/2014;
PN-II-PT-PCCA-2013-4-2166).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SC, VP and AJ performed the cell culture
experiments. LR and LP performed the evaluations using the
microscope and were responsible for the RNA extraction and quality
control. CB performed the microarray study. RCP performed the
bioinformatics analysis. CI and IBN were responsible for the study
design, and supervising and revising of the manuscript. All the
authors contributed to the writing of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chabner BA and Roberts TG Jr: Timeline:
Chemotherapy and the war on cancer. Nat Rev Cancer. 5:65–72. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gibalová L, Sereš M, Rusnák A, Ditte P,
Labudová M, Uhrík B, Pastorek J, Sedlák J, Breier A and Sulová Z:
P-glycoprotein depresses cisplatin sensitivity in L1210 cells by
inhibiting cisplatin-induced caspase-3 activation. Toxicol In
Vitro. 26:435–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mahon KL, Henshall SM, Sutherland RL and
Horvath LG: Pathways of chemotherapy resistance in
castration-resistant prostate cancer. Endocr Relat Cancer.
18:R103–R123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qu B, Aho KS, Li C, Kang S, Sillanpää M,
Yan F and Raymond PA: Greenhouse gases emissions in rivers of the
Tibetan Plateau. Sci Rep. 7:165732017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goncharuk VV, Kavitskaya AA, Romanyukina
IY and Loboda OA: Revealing water's secrets: Deuterium depleted
water. Chem Cent J. 7:1032013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kushner DJ, Baker A and Dunstall TG:
Pharmacological uses and perspectives of heavy water and deuterated
compounds. Can J Physiol Pharmacol. 77:79–88. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Basov AA, Bykov IM, Baryshev MG, Dzhimak
SS and Bykov MI: Determination of deuterium concentration in foods
and influence of water with modified isotopic composition on
oxidation parameters and heavy hydrogen isotopes content in
experimental animals. Vopr Pitan. 83:43–50. 2014.(In Russian).
PubMed/NCBI
|
|
8
|
Rehakova R, Klimentova J, Cebova M, Barta
A, Matuskova Z, Labas P and Pechanova O: Effect of
deuterium-depleted water on selected cardiometabolic parameters in
fructose-treated rats. Physiol Res. 65 Supplementum 3:S401–S407.
2016.PubMed/NCBI
|
|
9
|
Cong FS, Zhang YR, Sheng HC, Ao ZH, Zhang
SY and Wang JY: Deuterium-depleted water inhibits human lung
carcinoma cell growth by apoptosis. Exp Ther Med. 1:277–283. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gyöngyi Z, Budán F, Szabó I, Ember I, Kiss
I, Krempels K, Somlyai I and Somlyai G: Deuterium depleted water
effects on survival of lung cancer patients and expression of Kras,
Bcl2, and Myc genes in mouse lung. Nutr Cancer. 65:240–246. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hartmann J, Bader Y, Horvath Z, Saiko P,
Grusch M, Illmer C, Madlener S, Fritzer-Szekeres M, Heller N, Alken
RG and Szekeres T: Effects of heavy water (D2O) on human pancreatic
tumor cells. Anticancer Res. 25:3407–3411. 2005.PubMed/NCBI
|
|
12
|
Wang H, Zhu B, He Z, Fu H, Dai Z, Huang G,
Li B, Qin D, Zhang X, Tian L, et al: Deuterium-depleted water (DDW)
inhibits the proliferation and migration of nasopharyngeal
carcinoma cells in vitro. Biomed Pharmacother. 67:489–496. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bu H, Wedel S, Cavinato M and Jansen-Dürr
P: MicroRNA regulation of oxidative stress-induced cellular
senescence. Oxid Med Cell Longev. 2017:23986962017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Braicu C, Cojocneanu-Petric R, Chira S,
Truta A, Floares A, Petrut B, Achimas-Cadariu P and Berindan-Neagoe
I: Clinical and pathological implications of miRNA in bladder
cancer. Int J Nanomedicine. 10:791–800. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Irimie AI, Sonea L, Jurj A, Mehterov N,
Zimta AA, Budisan L, Braicu C and Berindan-Neagoe I: Future trends
and emerging issues for nanodelivery systems in oral and
oropharyngeal cancer. Int J Nanomedicine. 12:4593–4606. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Braicu C, Tomuleasa C, Monroig P, Cucuianu
A, Berindan-Neagoe I and Calin GA: Exosomes as divine messengers:
Are they the Hermes of modern molecular oncology? Cell Death
Differ. 22:34–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ling H, Pickard K, Ivan C, Isella C, Ikuo
M, Mitter R, Spizzo R, Bullock M, Braicu C, Pileczki V, et al: The
clinical and biological significance of MIR-224 expression in
colorectal cancer metastasis. Gut. 65:977–989. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Braicu C, Calin GA and Berindan-Neagoe I:
MicroRNAs and cancer therapy-from bystanders to major players. Curr
Med Chem. 20:3561–3573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geretto M, Pulliero A, Rosano C, Zhabayeva
D, Bersimbaev R and Izzotti A: Resistance to cancer
chemotherapeutic drugs is determined by pivotal microRNA
regulators. Am J Cancer Res. 7:1350–1371. 2017.PubMed/NCBI
|
|
20
|
Ramalingam S, Subramaniam D and Anant S:
Manipulating miRNA expression: A novel approach for colon cancer
prevention and chemotherapy. Curr Pharmacol Rep. 1:141–153. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taranu I, Braicu C, Marin DE, Pistol GC,
Motiu M, Balacescu L, Beridan Neagoe I and Burlacu R: Exposure to
zearalenone mycotoxin alters in vitro porcine intestinal epithelial
cells by differential gene expression. Toxicol Lett. 232:310–325.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Braicu C, Cojocneanu-Petric R, Jurj A,
Gulei D, Taranu I, Gras AM, Marin DE and Berindan-Neagoe I:
Microarray based gene expression analysis of Sus Scrofa duodenum
exposed to zearalenone: Significance to human health. BMC Genomics.
17:6462016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pistol GC, Braicu C, Motiu M, Gras MA,
Marin DE, Stancu M, Calin L, Israel-Roming F, Berindan-Neagoe I and
Taranu I: Zearalenone mycotoxin affects immune mediators, MAPK
signalling molecules, nuclear receptors and genome-wide gene
expression in pig spleen. PLoS One. 10:e01275032015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Irimie AI, Braicu C, Pileczki V, Petrushev
B, Soritau O, Campian RS and Berindan-Neagoe I: Knocking down of
p53 triggers apoptosis and autophagy, concomitantly with inhibition
of migration on SSC-4 oral squamous carcinoma cells. Mol Cell
Biochem. 419:75–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Braicu C, Pileczki V, Irimie A and
Berindan-Neagoe I: p53siRNA therapy reduces cell proliferation,
migration and induces apoptosis in triple negative breast cancer
cells. Mol Cell Biochem. 381:61–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pileczki V, Braicu C, Gherman CD and
Berindan-Neagoe I: TNF-α gene knockout in triple negative breast
cancer cell line induces apoptosis. Int J Mol Sci. 14:411–420.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Piechota M, Sunderland P, Wysocka A,
Nalberczak M, Sliwinska MA, Radwanska K and Sikora E: Is
senescence-associated β-galactosidase a marker of neuronal
senescence? Oncotarget. 7:81099–81109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krämer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in ingenuity pathway
analysis. Bioinformatics. 30:523–530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carrato A: Adjuvant treatment of
colorectal cancer. Gastrointest Cancer Res. 2 4 Suppl:S42–S46.
2008.PubMed/NCBI
|
|
30
|
Berindan-Neagoe I, Braicu C, Pileczki V,
Cojocneanu Petric R, Miron N, Balacescu O, Iancu D and Ciuleanu T:
5-Fluorouracil potentiates the anti-cancer effect of oxaliplatin on
Colo320 colorectal adenocarcinoma cells. J Gastrointestin Liver
Dis. 22:37–43. 2013.PubMed/NCBI
|
|
31
|
Yoshikawa R, Kusunoki M, Yanagi H, Noda M,
Furuyama JI, Yamamura T and Hashimoto-Tamaoki T: Dual antitumor
effects of 5-fluorouracil on the cell cycle in colorectal carcinoma
cells: A novel target mechanism concept for pharmacokinetic
modulating chemotherapy. Cancer Res. 61:1029–1037. 2001.PubMed/NCBI
|
|
32
|
Braicu C, Tudoran O, Balacescu L, Catana
C, Neagoe E, Berindan-Neagoe I and Ionescu C: The significance of
PDGF expression in serum of colorectal carcinoma
patients-correlation with Duke's classification. Can PDGF become a
potential biomarker? Chirurgia (Bucur). 108:849–854. 2013.
|
|
33
|
Braicu C, Catana C, Calin GA and
Berindan-Neagoe I: NCRNA combined therapy as future treatment
option for cancer. Curr Pharm Des. 20:6565–6574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Benson AB III, Venook AP, Bekaii-Saab T,
Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ,
Fuchs CS, et al: Colon cancer, version 3.2014. J Natl Compr Canc
Netw. 12:1028–1059. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Campisi J: Aging, cellular senescence, and
cancer. Annu Rev Physiol. 75:685–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cerella C, Grandjenette C, Dicato M and
Diederich M: Roles of apoptosis and cellular senescence in cancer
and aging. Curr Drug Targets. 17:405–415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roninson IB: Tumor cell senescence in
cancer treatment. Cancer Res. 63:2705–2715. 2003.PubMed/NCBI
|
|
38
|
Ewald JA, Desotelle JA, Wilding G and
Jarrard DF: Therapy-induced senescence in cancer. J Natl Cancer
Inst. 102:1536–1546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao Y, Wu D, Fei C, Guo J, Gu S, Zhu Y,
Xu F, Zhang Z, Wu L, Li X and Chang C: Down-regulation of Dicer1
promotes cellular senescence and decreases the differentiation and
stem cell-supporting capacities of mesenchymal stromal cells in
patients with myelodysplastic syndrome. Haematologica. 100:194–204.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gorospe M and Abdelmohsen K:
MicroRegulators come of age in senescence. Trends Genet.
27:233–241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gibbings D, Mostowy S, Jay F, Schwab Y,
Cossart P and Voinnet O: Selective autophagy degrades DICER and
AGO2 and regulates miRNA activity. Nat Cell Biol. 14:1314–1321.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bian XJ, Zhang GM, Gu CY, Cai Y, Wang CF,
Shen YJ, Zhu Y, Zhang HL, Dai B and Ye DW: Down-regulation of Dicer
and Ago2 is associated with cell proliferation and apoptosis in
prostate cancer. Tumour Biol. 35:11571–11578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Prodromaki E, Korpetinou A, Giannopoulou
E, Vlotinou E, Chatziathanasiadou Μ, Papachristou NI, Scopa CD,
Papadaki H, Kalofonos HP and Papachristou DJ: Expression of the
microRNA regulators Drosha, Dicer and Ago2 in non-small cell lung
carcinomas. Cell Oncol (Dordr). 38:307–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gibbings D, Mostowy S and Voinnet O:
Autophagy selectively regulates miRNA homeostasis. Autophagy.
9:781–783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Grootaert MO, da Costa Martins PA, Bitsch
N, Pintelon I, De Meyer GR, Martinet W and Schrijvers DM: Defective
autophagy in vascular smooth muscle cells accelerates senescence
and promotes neointima formation and atherogenesis. Autophagy.
11:2014–2032. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hubackova S, Krejcikova K, Bartek J and
Hodny Z: IL1- and TGFβ-Nox4 signaling, oxidative stress and DNA
damage response are shared features of replicative,
oncogene-induced, and drug-induced paracrine ‘bystander
senescence’. Aging (Albany NY). 4:932–951. 2012. View Article : Google Scholar : PubMed/NCBI
|