Introduction
Cervical cancer, ovarian cancer and endometrial
cancer are the three most common malignant tumor types of the
female genital system (1,2). Ovarian cancer accounts for only 4% of
malignant tumors in women, but it has the highest mortality rate
among all gynecological malignant tumor types (3,4).
Recurrence and early metastasis of tumor cells are the primary
reasons for the poor prognosis of patients with ovarian cancer
(5). Ovarian tissue is located deep
within the female pelvic cavity and the symptoms of ovarian cancer
are usually hard to detect. Due to the lack of specific early
diagnosis, the majority of patients who are diagnosed with ovarian
cancer are in the middle or advanced stages of the disease, or have
distant metastasis, leading to poor prognosis (6,7). In
recent years, the combination of surgery and chemotherapy has
significantly improved the 5-year survival rate of patients with
ovarian cancer, but the overall prognosis is still poor (8,9). It is
reported that the occurrence and development of ovarian cancer is a
complex process that involves multiple genes, steps and stages, and
the molecular mechanism of ovarian cancer is still unclear
(10). Therefore, screening key
genes that are associated with the malignant biological behavior of
ovarian cancer will be valuable in improving the early diagnosis
and treatment of ovarian cancer.
MicroRNA (miRNA or miR) molecules are non-coding
small RNA molecules consisting of 18–22 nucleotides (11). miRNAs are able to regulate mRNA
translation by binding with the 3′-untranslated region (3′-UTR) of
target genes (12). miRNAs are
involved in nearly all pathophysiological processes and stably
exist in body fluids, including peripheral blood, urine and saliva,
which makes them key biomarkers and therapeutic targets (13). A previous study demonstrated that
expression of miRNAs is disordered in multiple tumor tissues, and
miRNAs serve oncogene and tumor-suppressor gene functions (14). For example, miR-590-5p activates the
Akt/extracellular signal-regulated kinase signaling pathway by
downregulating expression of reversion-inducing-cysteine-rich
protein with kazal motifs, resulting in the promotion of
proliferation and migration in gastric cancer cells (15). In addition, miR-138 acts as a
tumor-suppressor gene by regulating the expression of different
genes in multiple tumor types. For example, miR-138 inhibits the
invasion and metastasis of non-small cell lung cancer (NSCLC) cells
by decreasing the expression of LIMK1 gene (16). Furthermore, miR-138 suppresses the
proliferation, invasion and metastasis of hepatocellular carcinoma
by downregulating SOX9 gene expression (17). In colon cancer, miR-138 regulates
programmed cell death protein 1 expression and inhibits distant
metastasis of tumor cells (18). It
has also been reported that miR-138 inhibits the proliferation,
invasion and migration of ovarian cancer cells via targeted
inhibition of SOX4 and hypoxia-inducible factor-1α (19). Therefore, miR-138 has a variety of
downstream target genes that are involved in numerous signaling
pathways and biological functions. However, the expression of
miR-138 in ovarian cancer tissues and its clinical importance are
not yet clear. The present study aimed to investigate the
expression and biological functions of miR-138 in ovarian
cancer.
Materials and methods
Patients
A total of 47 female patients (age range, 32–63
years; mean age, 41.5 years) with ovarian cancer who received
surgical resection at Department of Obstetrics, The Affiliated
Hospital of Jining Medical University (Jining, China) between
February 2014 and October 2015 were included in the present study.
Patients with any other tumors, chronic basic diseases, autoimmune
diseases or a history of long-term medicine use were excluded.
Resected primary ovarian cancer tissues were diagnosed and
classified by two individual pathologists as epithelial ovarian
cancer. Contralateral normal ovarian tissues were also resected as
controls. According to the FIGO 2000 criteria (20), ovarian cancer tissues were subjected
to staging classification. Among all cases, 11 were at stage I, 13
were at stage II, 13 were at stage III and 10 were at stage IV. In
addition, 25 cases had lymphatic metastasis, while the other 22
cases had no lymphatic metastasis. All tissues were frozen in
liquid nitrogen and stored at −80°C. None of the patients had
history of other tumors or complications, or received
chemoradiotherapy or any other anti-tumor therapies. The clinical
information and pathological data of all subjects were collected.
All procedures were approved by the Ethics Committee of Jining
Medical University. Written informed consent was obtained from all
patients or their families.
Cells
Ovarian cancer A2780 cells (Type Culture Collection
of the Chinese Academy of Sciences, Shanghai, China) were cultured
in RPMI-1640 medium supplemented with 10% fetal bovine serum (both
Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C. On the
day before transfection, A2780 cells (2×105) with
log-phase growth were seeded onto 24-well plates containing
RPMI-1640 without fetal bovine serum or antibiotics. The cells were
divided into negative control (NC) and miR-138 mimic groups. When
the cells reached 70–80% confluence, 1.5 µl miR-138 mimic
(5′-AGCUGGUGUUGUGAAUCAGGCCG-3; 25 pmol/µl) or miR-NC (cat. no.
miR01201-1-5; both Guangzhou RiboBio Co., Ltd., Guangzhou, China)
and 1 µl liposome (Lipofectamine® 2000) were mixed with
50 µl OptiMEM medium (both Thermo Fisher Scientific, Inc.), in
individual Eppendorf tubes. Following standing for 5 min, the
contents of the two Eppendorf tubes were mixed and kept at room
temperature for 20 min, followed by addition into each culture well
of a 24-well plate (1×105 cells/well). After an
incubation at 37°C for 6 h, the medium was replaced with fresh
RPMI-1640 medium supplemented with 10% fetal bovine serum, and the
cells were cultured under 37°C and 5% CO2 for 48 h prior
to use.
For the target gene function analysis, the cells
were transfected with small interfering RNA (siR)-SOX12
(5′-CATGGCGGATTACCCGGACTA-3′) and siR-NC
(5′-UUTCCUCCGAACGUGUCACGUtt-3′; Hanbio Biotechnology Co., Ltd.,
Shanghai, China) instead of miR-138 mimic and miR-NC, using the
same transfection procedure as described above.
For the rescue study, the cells were infected with
lentiviral vector with overexpression of SOX12 (LV-GFP-Puro-sox12;
Hanbio Biotechnology Co., Ltd.) with multiplicity of infection of
20. Following incubation at 37°C for 72 h, the cells were harvested
for further use.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Ovarian cancer and control tissues (100 mg) were
ground into powder in liquid nitrogen and mixed with 1 ml TRIzol
(Thermo Fisher Scientific, Inc.) for lysis. Then, total RNA was
extracted using the phenol chloroform method (21). The purity of RNA was determined by
A260/A280 using ultraviolet spectrophotometry (Nanodrop 2000
Spectrophotometer; Thermo Fisher Scientific, Inc.). Then, cDNA was
obtained by RT at 37°C for 1 h using a Reverse Transcription system
(Takara Biotechnology Co., Ltd., Dalian, China) from 1 µg RNA and
stored at −20°C.
The expression of miR-138 was determined using an
SYBR PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd.), using
U6 as internal reference. The reaction system (20 µl) contained 10
µl qPCR mix, 0.5 µl upstream primer (miR-138,
5′-AGCTGGUGTTGTGAATCAGGCCG-3′; U6, 5′-CTCGCTTCGGCAGCACA-3′), 0.5 µl
downstream universal primer (miR-138, provided by the kit; U6,
5′-AACGCTTCACGAATTTGCGT-3′), 1 µl cDNA and 8 µl ddH2O.
The reaction protocol was: Initial denaturation at 95°C for 10 min,
followed by 40 cycles of 95°C for 1 min and 60°C for 30 sec (iQ5;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
2−ΔΔCt method (22) was
used to calculate the relative expression of miR-138 against the
internal reference. Each sample was tested in triplicate.
Cell Counting Kit 8 (CCK-8) assay
Cells were seeded at 2,000 cells/well in 96-well
plates for transfection. At 48 h after transfection, the cells were
subjected to CCK-8 assay in order to evaluate proliferation rates.
At 24, 48 and 72 h after the initial 48 h of transfection, the
RPMI-1640 medium was discarded, and the cells were washed with
phosphate-buffered saline twice, followed by the addition of 10%
CCK-8 reaction reagent (Beyotime Institute of Biotechnology,
Shanghai, China) diluted in RPMI-1640 medium at 37°C. After
incubation at 37°C for 1 h, the absorbance of each well was
measured at 490 nm for plotting cell viability curves. Each group
was tested in three replicate wells and the values were
averaged.
Flow cytometry
Cells (1×106) were washed with
phosphate-buffered saline three times before centrifugation at 500
× g for 5 min at room temperature. After discarding the
supernatant, the cells were subjected to cell cycle determination
using a BD Pharmingen Cell Cycle kit (BD Biosciences, Franklin
Lakes, NJ, USA) according to the manufacturer's protocol. Briefly,
the cells were mixed with 150 µl A solution before gentle mixing
and standing for 10 min at room temperature. Then, 150 µl B
solution was added before gentle mixing and standing for 10 min at
room temperature. After addition of 120 µl C solution, the cells
were incubated in the dark for 15 min before flow cytometry
(FACSVerse™; BD Biosciences). ModFit 3.1 software (BD
Biosciences) was used to analyze the data.
Western blotting
Ovarian cancer and control tissues were ground into
powder in liquid nitrogen and 100 mg of the powder was mixed with
100 µl precooled radio-immunoprecipitation assay lysis buffer
(Beyotime Institute of Biotechnology) containing 1%
phenylmethylsulfonyl fluoride for lysis overnight at 4°C. Then, the
mixture was centrifuged at 12,000 × g and 4°C for 15 min. The
supernatant was used to determine protein concentration with a
bicinchoninic acid protein concentration determination kit
(RTP7102; Real-Times Biotechnology Co., Ltd., Beijing, China).
Protein samples (50 µg) were then mixed with 5× sodium dodecyl
sulfate (SDS) loading buffer before denaturation in a boiling water
bath for 10 min. Afterwards, the samples (20 µg/lane) were
subjected to 10% SDS-PAGE at 100 V. The resolved proteins were
transferred to polyvinylidene difluoride membranes on ice (100 V, 1
h) and blocked with 50 g/L skimmed milk at room temperature for 1
h. Then, the membranes were incubated with mouse anti-human SOX12
(1:1,000; cat. no. SAB1409702; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and GAPDH (1:5,000; cat. no. ab8245; Abcam,
Cambridge, UK) polyclonal primary antibodies at 4°C overnight.
Following five washes with phosphate-buffered saline with Tween-20
(5 min/wash), the membranes were incubated with polyclonal goat
anti-mouse horseradish peroxidase-conjugated secondary antibody
(1:10,000; cat. no. ab6789; Abcam) for 1 h at room temperature. The
membranes were washed five times with phosphate-buffered saline
with Tween-20 (5 min/wash), then developed using an enhanced
chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA) for
imaging. Image Lab v3.0 software (Bio-Rad Laboratories, Inc.) was
used to acquire and analyze imaging signals. The relative
expression of SOX12 protein was calculated against GAPDH.
Transwell assay
For invasion assay, Matrigel was thawed at 4°C
overnight and diluted with serum-free RPMI-1640 medium (dilution
1:1). The mixture (20 µl) was evenly smeared into the upper chamber
of a Transwell insert (Merck KGaA) and incubated at 37°C for 1 h.
For migration assay, Matrigel was not added. After solidification,
1×105 cells from each group were seeded into the upper
chamber containing 200 µl serum-free RPMI-1640 medium. In addition,
500 µl RPMI-1640 medium supplemented with 10% fetal bovine serum
was added into the lower chamber. After 48 h, the chamber was
removed and the cells in the upper chamber were wiped off. After
being fixed with 4% formaldehyde for 10 min, the membrane was
stained at room temperature for 2 h using the Giemsa method for
light microscopic observation of 5 random fields (magnification,
×200). The number of cells was calculated for the evaluation of
cell invasion and migration ability. All procedures were performed
on ice with pipetting tips being cooled at 4°C.
Dual luciferase reporter assay
Bioinformatics prediction is a powerful tool for the
study of the functions of miRNAs. To understand the regulatory
mechanism of SOX12 in ovarian cancer, miRanda (www.microrna.org/microrna/home.do),
TargetScan (www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
and PICTA (http://pictar.mdc-berlin.de/) databases were used to
predict miRNA molecules that may regulate SOX12. It was identified
that miR-138 was potentially able to regulate SOX12. According to
bioinformatics results, wild-type (WT) and mutant seed regions of
miR-138 in the 3′-UTR of SOX12 gene were chemically synthesized
in vitro, with SpeI and HindIII restriction
sites added. These were then cloned into pMIR-REPORT luciferase
reporter plasmids (Thermo Fisher Scientific, Inc.). Using
Lipofectamine 3000 (Thermo Fisher Scientific, Inc.), plasmids (0.8
µg) with WT or mutant 3′-UTR DNA sequences were co-transfected with
agomiR-138 (100 nM; Sangon Biotech, Shanghai, China) into 293T
cells (American Type Culture Collection, Manassas, VA, USA).
Following cultivation for 24 h, the cells were lysed using
Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, WI, USA) according to the manufacturer's
protocol, and fluorescence intensity was measured using a GloMax
20/20 luminometer (Promega Corporation). Using Renilla
fluorescence activity as an internal reference, the fluorescence
values of each group of cells were measured.
Statistical analysis
Statistical analysis was performed using SPSS 11.0
(SPSS, Inc., Chicago, IL, USA). Measurement data were expressed as
the mean ± standard deviation. Data were tested for normality. Two
groups of data were compared using t-tests. Multiple groups of data
were analyzed using one-way analysis of variance. In cases of
homogeneity of variance, the Student-Newman-Keuls post hoc test was
used for multiple comparisons. P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of miR-138 is associated
with the early occurrence and metastasis of ovarian cancer
To measure the expression of miR-138 in ovarian
cancer tissues, RT-qPCR was performed. The data indicated that the
level of miR-138 in ovarian cancer tissues was significantly lower
compared with the control tissues (P<0.05; Fig. 1A). In addition, the level of miR-138
in ovarian cancer patients with lymphatic metastasis was
significantly lower compared with in patients without lymphatic
metastasis (P<0.05; Fig. 1B).
Compared with the control group, the level of miR-138 in ovarian
cancer tissues at stages I, II, III and IV was significantly
reduced (P<0.05), but no significant differences were observed
among the four stages (Fig. 1C).
These results suggest that the expression of miR-138 is associated
with the early occurrence and metastasis of ovarian cancer.
Upregulation of miR-138 inhibits the
proliferation of A2780 cells
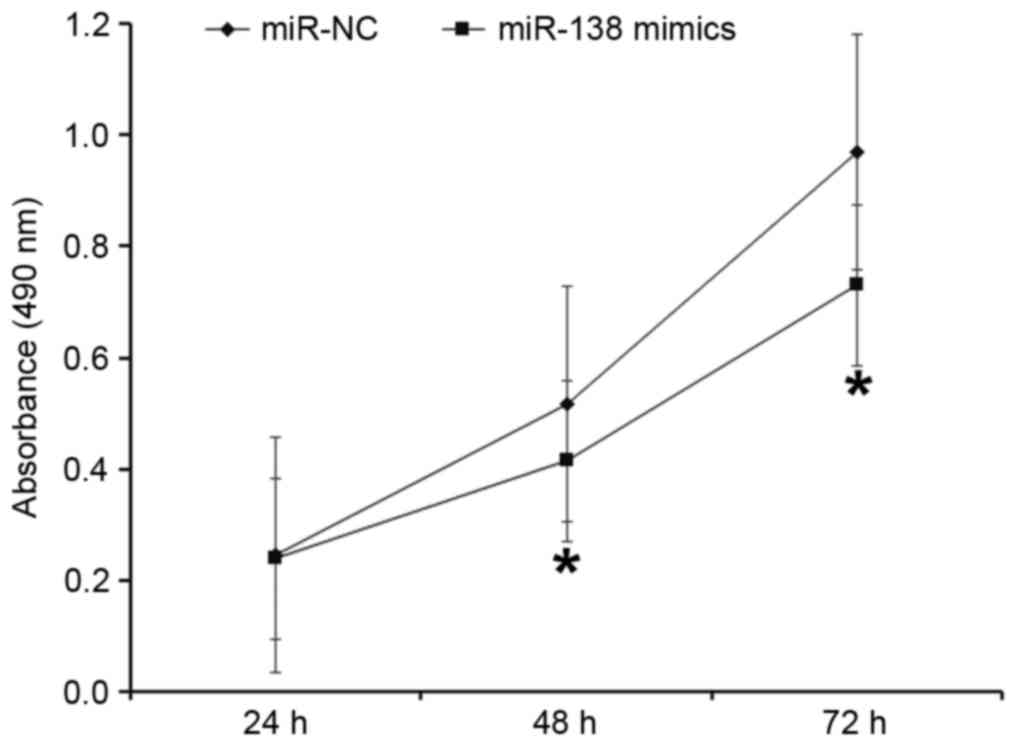
To evaluate the effect of miR-138 on the
proliferation of A2780 cells, a CCK-8 assay was performed. The data
indicated that the absorbance of cells transfected with miR-138
mimic was significantly lower compared with cells transfected with
miR-NC at 48 and 72 h (P<0.05; Fig.
2). These results indicate that upregulation of miR-138
inhibits the proliferation of A2780 cells.
Overexpression of miR-138 inhibits the
proliferation of A2780 cells by suppressing their G1/S phase
transition
To detect cell cycle distribution, flow cytometry
was performed. The data indicated that the percentage of
G1 phase cells in the miR-138 mimic group was
significantly higher compared with the miR-NC group (P<0.05),
while the percentage of S phase cells in the miR-138 mimic group
was significantly lower compared with the miR-NC group (P<0.05;
Fig. 3). These results suggest that
overexpression of miR-138 inhibits the proliferation of A2780 cells
by suppressing their G1/S phase transition.
miR-138 suppresses the invasion and
migration of A2780 cells
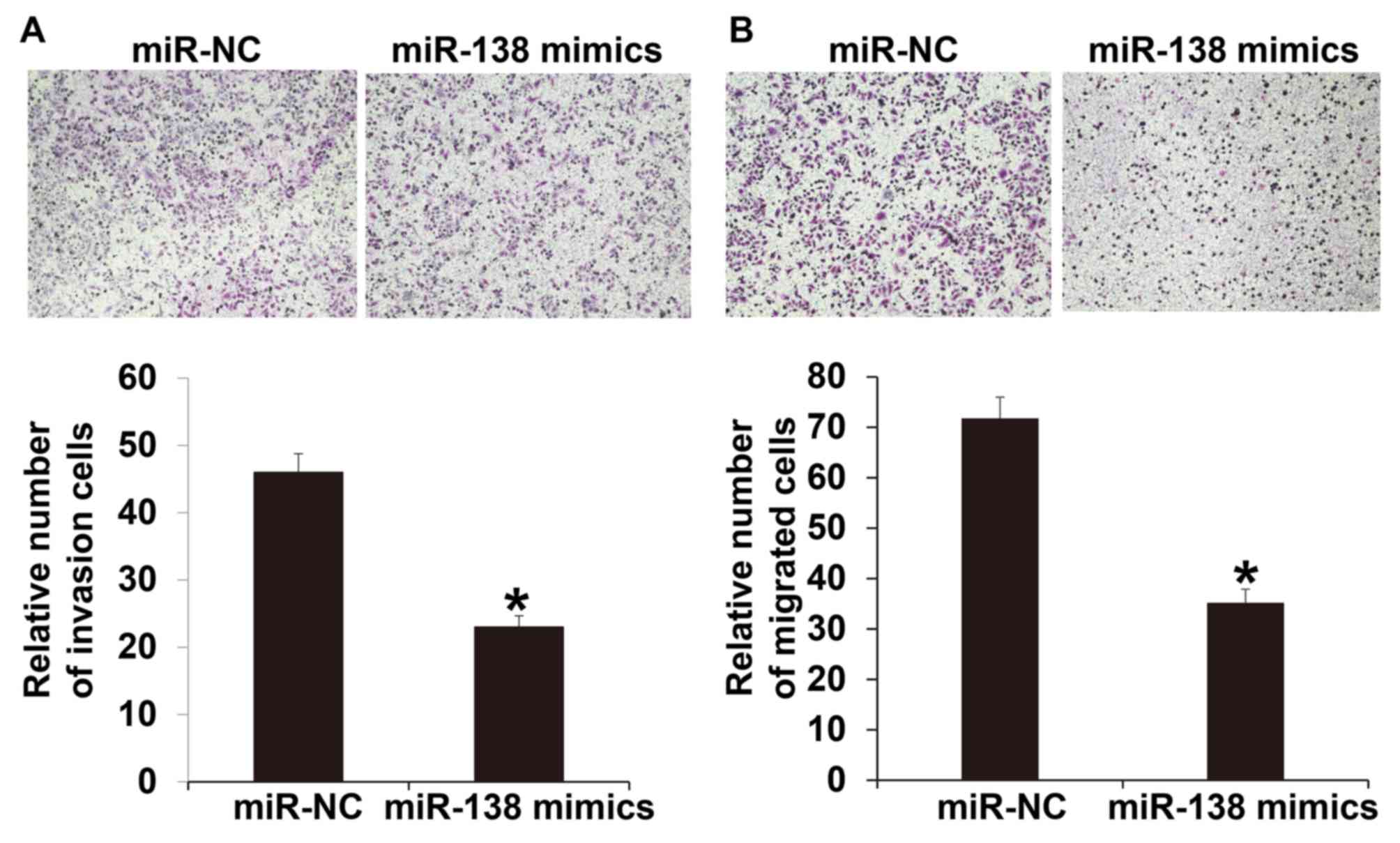
To investigate the invasion and migration abilities
of A2780 cells, Transwell assays were used. The invasion assay
indicated that the number of cells that crossed the Transwell
membrane in the miR-138 mimic group was significantly lower
compared with the miR-NC group (P<0.05; Fig. 4A). Similarly, the migration assay
indicated that the number of cells that crossed the Transwell
membrane in the miR-138 mimics group was significantly reduced
compared with the miR-NC group (P<0.05; Fig. 4B). These results suggest that miR-138
suppresses the invasion and migration of A2780 cells.
SOX12 promotes the proliferation,
invasion and migration of A2780 cells
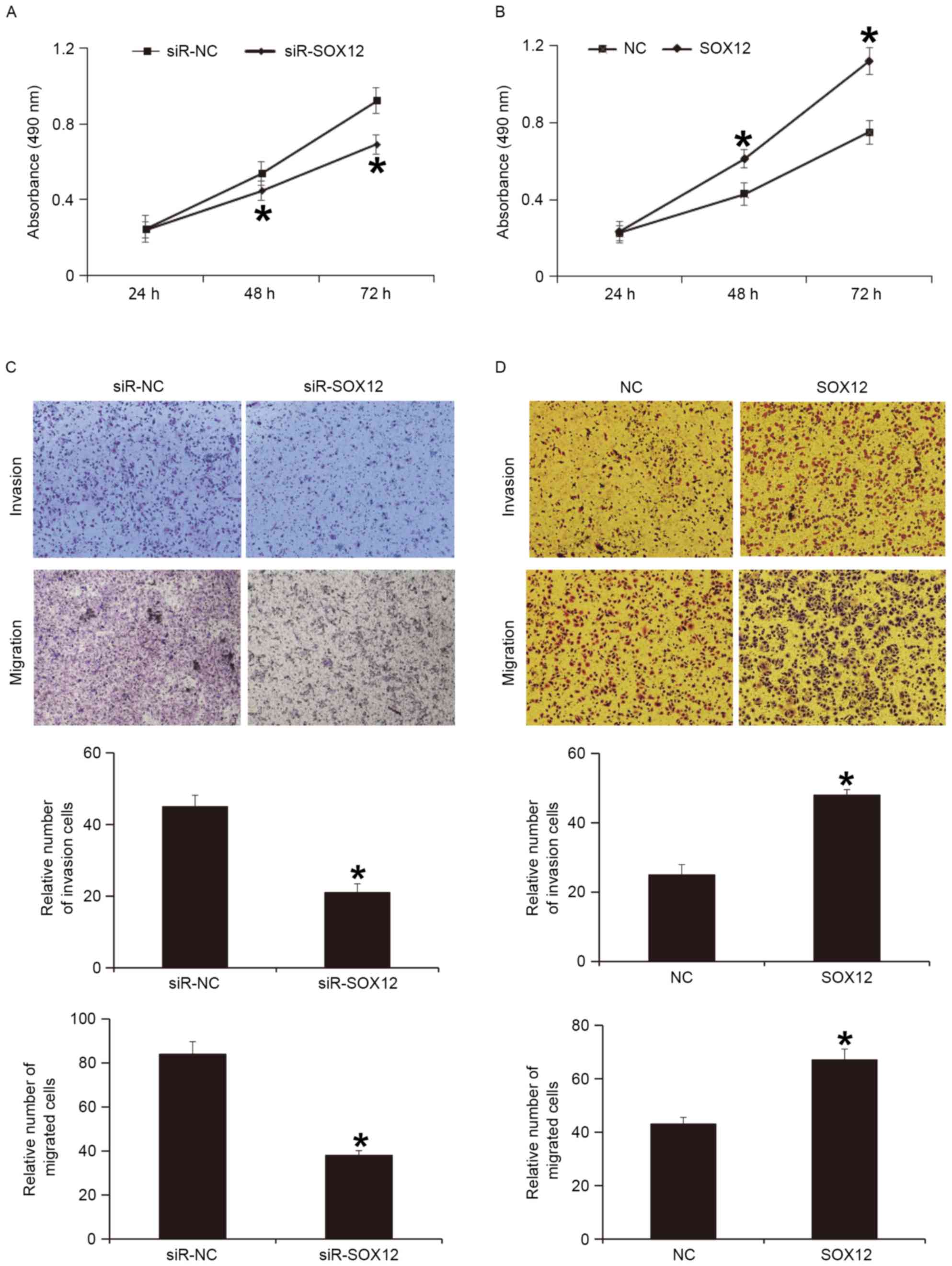
To examine whether the expression of SOX12 protein
regulates the biological functions of A2780 cells, SOX12 gene was
silenced or overexpressed in A2780 cells. A CCK-8 assay indicated
that silencing of SOX12 expression significantly reduced the
proliferation of A2780 cells at 47 and 72 h (P<0.05; Fig. 5A), while overexpression of SOX12
significantly enhanced the proliferation of A2780 cells at 48 and
72 h (P<0.05; Fig. 5B). A
Transwell assay indicated that silencing of SOX12 expression
significantly decreased the invasion and migration abilities of
A2780 cells (P<0.05; Fig. 5C),
while overexpression of SOX12 significantly increased the invasion
and migration abilities of A2780 cells (P<0.05; Fig. 5D). These results indicate that SOX12
promotes the proliferation, invasion and migration of A2780
cells.
miR-138 downregulates the protein
expression of SOX12
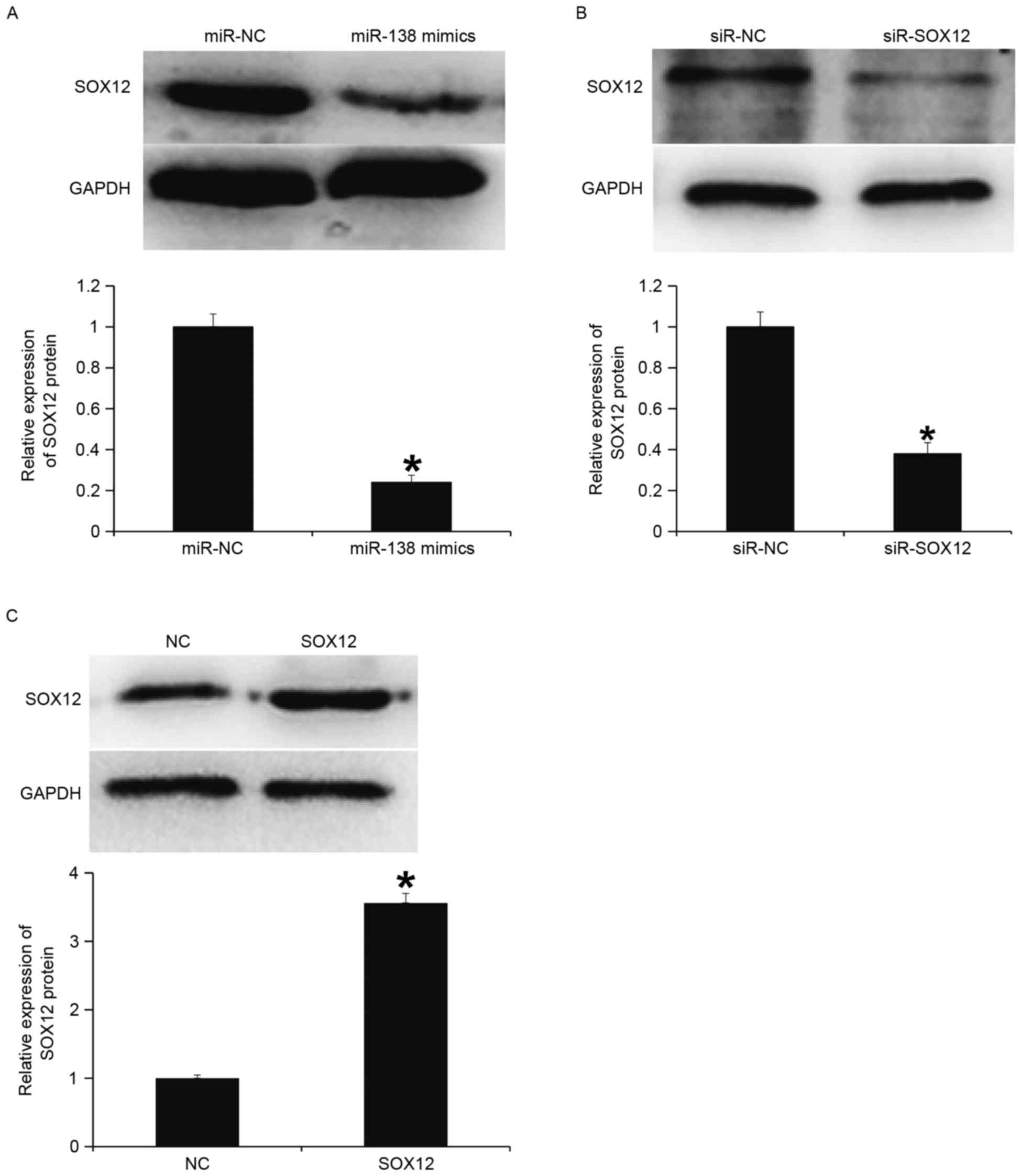
To determine protein expression, western blotting
was performed. The data indicated that the expression of SOX12
protein in the miR-138 mimics group was significantly lower
compared with the miR-NC group (P<0.05; Fig. 6A). Transfection with siR-SOX12
significantly reduced SOX12 protein expression compared with the
siR-NC group (P<0.05; Fig. 6B).
In addition, SOX12 protein expression in cells with overexpression
of SOX12 was significantly higher compared with the NC (P<0.05;
Fig. 6C). These results indicate
that miR-138 downregulates the protein expression of SOX12.
miR-138 downregulates the expression
of SOX12 by binding with the 3′-UTR of SOX12 gene
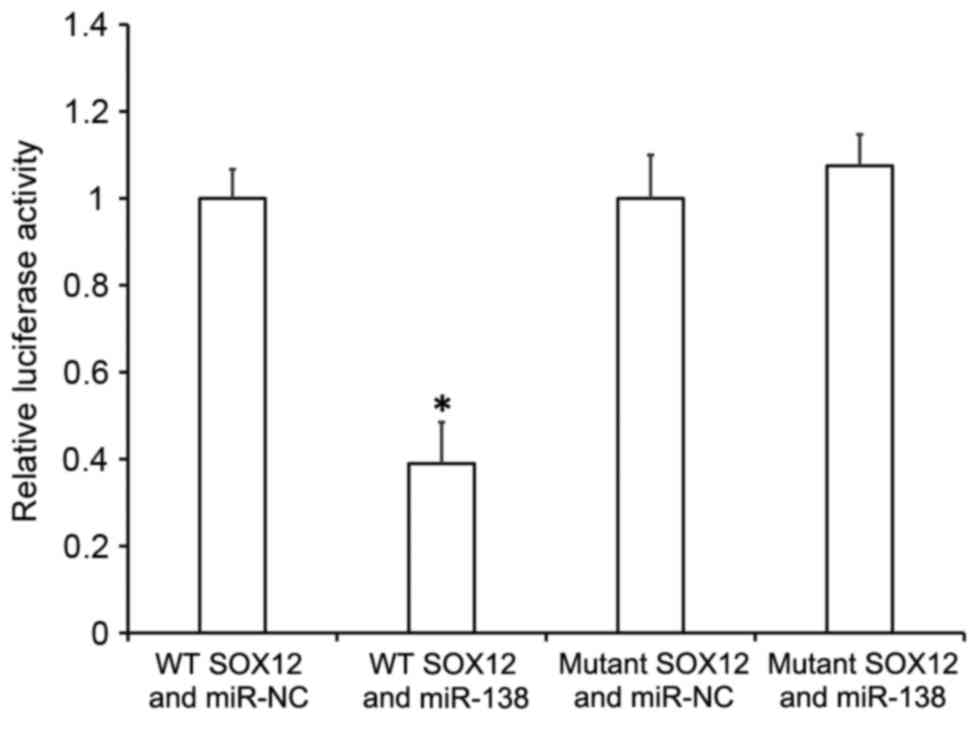
To understand whether miR-138 directly targets
SOX12, a dual luciferase reporter assay was performed. The data
indicated that transfection with miR-138 mimics and
pMIR-REPORT-wild type SOX12 led to significantly reduced
fluorescence intensity compared with the miR-NC group (P<0.05),
while transfection with miR-138 mimics and pMIR-REPORT-mutant SOX12
resulted in similar fluorescence intensity compared with the miR-NC
group (Fig. 7). These results
suggest that miR-138 downregulates the expression of SOX12 by
binding with the 3′-UTR of SOX12 gene.
Discussion
Previous studies have reported that miRNA molecules
have important functions in the occurrence and development of
tumors by widely participating in processes including
proliferation, differentiation, apoptosis and cell cycle (23,24). For
example, miR-590 inhibits the occurrence and metastasis of NSCLC
via targeted regulation of ADAM9 gene expression (25). In addition, miR-146a acts as a
tumor-suppressor gene by inhibiting the proliferation, invasion and
metastasis of cervical cancer and colon cancer cells (26). However, certain miRNA molecules may
promote the occurrence and development of tumors. For example,
miR-574-5 promotes the distant metastasis of NSCLC by targeting
PTPRU gene, while miR-10b facilitates the proliferation and
metastasis of hepatocellular carcinoma by downregulating the
expression of CUB and Sushi multiple domains 1 (27). In the present study, it was
identified that miR-138 expression is significantly reduced in
ovarian cancer cells, and the level of miR-138 in patients with
lymphatic metastasis is significantly lower compared with patients
without lymphatic metastasis, suggesting that miR-138 expression is
associated with the invasion and migration of ovarian cancer cells.
In addition, the levels of miR-138 in ovarian cancer patients at
stages I, II, III and IV are not significantly different from each
other, suggesting that miR-138 is associated with the early
occurrence of ovarian cancer. At the cellular level, it was
identified that overexpression of miR-138 inhibits the
proliferation, invasion and migration of A2780 cells. This suggests
that miR-138 acts as a tumor-suppressor gene in the occurrence and
development of ovarian cancer and the downregulation of miR-138
promotes the proliferation, invasion and migration of ovarian
cancer cells.
SOX genes, members of the high mobility group
superfamily, primarily encode transcription factors (28,29). SOX
genes have been demonstrated to be associated with the
differentiation and proliferation of cells (30), and have oncogene functions (31,32). It
is reported that SOX10 and SOX1 are tumor-associated antigens of
melanoma and small cell lung cancer cells, respectively (33,34). The
expression of SOX genes is not the same in different tumor types.
For example, SOX7 expression is upregulated in esophageal squamous
cell carcinoma and gastric carcinoma (35), but is downregulated in prostate
cancer, breast cancer and rectal cancer (36). SOX12 is a member of the SOX family,
but its function and mechanism of action in tumors is not yet
clear. A previous study demonstrated that SOX12 is upregulated in
breast cancer and promotes the invasion and migration of tumor
cells (37). Huang et al
(38) demonstrated that SOX12
directly regulates FoxQ1, upregulates the expression of Twist1 and
FGFBP1, and facilitates the invasion and metastasis of
hepatocellular carcinoma. In the present study, it was identified
that miR-138 regulates the SOX12 gene by directly binding with the
3′-UTR of SOX12 and inhibiting the expression of SOX12 protein. In
addition, overexpression of SOX12 gene in A2780 cells promoted the
proliferation, invasion and migration of A2780 cells, while
silencing of SOX12 gene reduced the proliferation, invasion and
migration of the cells, suggesting that SOX12 gene is an oncogene
in A2780 cells. Furthermore, miR-138 inhibits the occurrence and
development of ovarian cancer by downregulating the expression of
SOX12 gene.
In the present study, changes in miRNA molecules
were determined in tumor tissues. Due to the complexity of tumor
tissue components, the distribution of miRNA molecules in tissues
is not yet clearly known. One of the limitations of the present
study is the absence of in situ hybridization to determine
the expression of miRNA in specimens observed by microscopy.
In conclusion, miR-138 expression is downregulated
in ovarian cancer and thus, SOX12 gene expression is upregulated
and the occurrence and development of ovarian cancer is promoted.
Therefore, miR-138 is a potential therapeutic target and biomarker
for ovarian cancer.
Acknowledgements
The present study was supported by the Affiliated
Hospital of Jining Medical University. The authors would also like
to thank Dr Dongmei Man, Director of the Affiliated Hospital of
Jining Medical University.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The final version of the manuscript has been read
and approved by all authors, and each author believes that the
manuscript represents honest work. MQ and YZ collaborated to design
the study. MQ and MJ were responsible for experiments. MQ, MJ and
YZ analyzed the data. All authors collaborated to interpret results
and develop the manuscript.
Ethics approval and consent to
participate
All procedures performed in the current study were
approved by the Ethics Committee of Jining Medical University.
Written informed consent was obtained from all patients or their
families.
Patient consent for publication
Written informed consents for publication of any
associated data and accompanying images were obtained from all
patients or their parents, guardians or next of kin.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lam SS, Ip CK, Mak AS and Wong AS: A novel
p70 S6 kinase-microRNA biogenesis axis mediates multicellular
spheroid formation in ovarian cancer progression. Oncotarget.
7:38064–38077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Querleu D, Meurette J, Darai E, Morice P
and Planchamp F: Surgical management of ovarian cancer: Trends in
clinical practice. Bull Cancer. 103:935–940. 2016.(In French).
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen B, Campbell KR, Tilbury K, Nadiarnykh
O, Brewer MA, Patankar M, Singh V, Eliceiri KW and Campagnola PJ:
3D texture analysis for classification of second harmonic
generation images of human ovarian cancer. Sci Rep. 6:357342016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Z, Xu S, Jin P, Yang X, Li X, Wan D,
Zhang T, Long S, Wei X, Chen G, et al: MARCKS contributes to
stromal cancer-associated fibroblast activation and facilitates
ovarian cancer metastasis. Oncotarget. 7:37649–37663.
2016.PubMed/NCBI
|
|
5
|
Correa DD, Root JC, Kryza-Lacombe M, Mehta
M, Karimi S, Hensley ML and Relkin N: Brain structure and function
in patients with ovarian cancer treated with first-line
chemotherapy: Apilot study. Brain Imaging Behav. 11:1652–1663.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomar T, de Jong S, Alkema NG, Hoekman RL,
Meersma GJ, Klip HG, van der Zee AG and Wisman GB: Genome-wide
methylation profiling of ovarian cancer patient-derived xenografts
treated with the demethylating agent decitabine identifies novel
epigenetically regulated genes and pathways. Genome Med. 8:1072016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harada T, Nakamura Y, Sato K, Nagaya T,
Okuyama S, Ogata F, Choyke PL and Kobayashi H: Near-infrared
photoimmunotherapy with galactosyl serum albumin in a model of
diffuse peritoneal disseminated ovarian cancer. Oncotarget.
7:79408–79416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Halpern JA, Shoag JE, Mittal S, Oromendia
C, Ballman KV, Hershman DL, Wright JD, Shih YT, Nguyen PL and Hu
JC: Prognostic significance of digital rectal examination and
prostate specific antigen in the prostate, lung, colorectal, and
ovarian cancer screening arm. J Urol. 197:363–368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren F, Shen J, Shi H, Hornicek FJ, Kan Q
and Duan Z: Novel mechanisms and approaches to overcome multidrug
resistance in the treatment of ovarian cancer. Biochim Biophys
Acta. 1866:266–275. 2016.PubMed/NCBI
|
|
10
|
Salerno L, Marchetti C, Bevilacqua E,
Musella A, Riganelli L, Ruscito I, Perniola G, Muzii L and Panici
Benedetti P: Beyond the beyond: First case of 9 cytoreductive
surgeries in a long-surviving ovarian cancer patient: Case report.
Tumori. 102 Suppl 2:53012016.
|
|
11
|
Matikas A, Syrigos KN and Agelaki S:
Circulating biomarkers in non-small-cell lung cancer: Current
status and future challenges. Clin Lung Cancer. 17:507–516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Wainscott C and Xi Y: MicroRNA
provides insight into understanding esophageal cancer. Thorac
Cancer. 2:134–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giza DE, Fuentes-Mattei E, Bullock MD,
Tudor S, Goblirsch MJ, Fabbri M, Lupu F, Yeung SJ, Vasilescu C and
Calin GA: Cellular and viral microRNAs in sepsis: Mechanisms of
action and clinical applications. Cell Death Differ. 23:1906–1918.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer -
an emerging concept. EbioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen B, Yu S, Zhang Y, Yuan Y, Li X, Zhong
J and Feng J: miR-590-5p regulates gastric cancer cell growth and
chemosensitivity through RECK and the AKT/ERK pathway. Onco Targets
Ther. 9:6009–6019. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tan Y, Hu H, Tan W, Jin L, Liu J and Zhou
H: MicroRNA-138 inhibits migration and invasion of non-small cell
lung cancer cells by targeting LIMK1. Mol Med Rep. 14:4422–4428.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Zhang W, Liu K, Liu S, Ji B and
Wang Y: miR-138 suppresses cell proliferation and invasion by
inhibiting SOX9 in hepatocellular carcinoma. Am J Transl Res.
8:2159–2168. 2016.PubMed/NCBI
|
|
18
|
Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z,
Liu R, Tang A, Li X, Liu F and Shen S: The tumor suppressor
miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget.
7:45370–45384. 2016.PubMed/NCBI
|
|
19
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng Q, Duan P, Li L and Miao Y:
Expression of placenta growth factor is associated with unfavorable
prognosis of advanced-stage serous ovarian cancer. Tohoku J Exp
Med. 244:291–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brown RAM, Epis MR, Horsham JL, Kabir TD,
Richardson KL and Leedman PJ: Total RNA extraction from tissues for
microRNA and target gene expression analysis: Not all kits are
created equal. BMC Biotechnol. 18:162018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Paris PL, Chen J, Ngo V, Yao H,
Frazier ML, Killary AM, Liu CG, Liang H, Mathy C, et al: Next
generation sequencing of pancreatic cyst fluid microRNAs from low
grade-benign and high grade-invasive lesions. Cancer Lett.
356:404–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chao A, Lai CH, Chen HC, Lin CY, Tsai CL,
Tang YH, Huang HJ, Lin CT, Chen MY, Huang KG, et al: Serum
microRNAs in clear cell carcinoma of the ovary. Taiwan J Obstet
Gynecol. 53:536–541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang FF, Wang S, Xue WH and Cheng JL:
microRNA-590 suppresses the tumorigenesis and invasiveness of
non-small cell lung cancer cells by targeting ADAM9. Mol Cell
Biochem. 423:29–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sathyanarayanan A, Chandrasekaran KS and
Karunagaran D: microRNA-146a inhibits proliferation, migration and
invasion of human cervical and colorectal cancer cells. Biochem
Biophys Res Commun. 480:528–533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang
JR, Han XJ, Zhu SJ, Wang SM, Li YH and Zhang W: miR-10b exerts
oncogenic activity in human hepatocellular carcinoma cells by
targeting expression of CUB and sushi multiple domains 1 (CSMD1).
BMC Cancer. 16:8062016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagase S, Iyoda T, Kanno H, Akase T,
Arakawa I, Inoue T and Uetsuka Y: Comparison of the
cost-effectiveness of the sox and cox regimens in patients with
unresectable advanced and recurrent colorectal cancer using a
clinical decision analysis approach. Gan To Kagaku Ryoho.
43:1201–1205. 2016.(In Japanese). PubMed/NCBI
|
|
29
|
Righi S, Pileri S, Agostinelli C, Bacci F,
Spagnolo S and Sabattini E: Reproducibility of SOX-11 detection in
decalcified bone marrow tissue in mantle cell lymphoma patients.
Hum Pathol. 59:94–101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu L and Shi YB: The Sox transcriptional
factors: Functions during intestinal development in vertebrates.
Semin Cell Dev Biol. 63:58–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weina K, Wu H, Knappe N, Orouji E, Novak
D, Bernhardt M, Hüser L, Larribère L, Umansky V, Gebhardt C and
Utikal J: TGF-β induces SOX2 expression in a time-dependent manner
in human melanoma cells. Pigment Cell Melanoma Res. 29:453–458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song WS, Yang YP, Huang CS, Lu KH, Liu WH,
Wu WW, Lee YY, Lo WL, Lee SD, Chen YW, et al: Sox2, a stemness
gene, regulates tumor-initiating and drug-resistant properties in
CD133-positive glioblastoma stem cells. J Chin Med Assoc.
79:538–545. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li N and Li S: Epigenetic inactivation of
SOX1 promotes cell migration in lung cancer. Tumour Biol.
36:4603–4610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gambichler T, Petig AL, Stockfleth E and
Stucker M: Expression of SOX10, ABCB5 and CD271 in melanocytic
lesions and correlation with survival data of patients with
melanoma. Clin Exp Dermatol. 41:709–716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang M, Cui G, Ding M, Yang W, Liu Y, Dai
D and Chen L: miR-935 promotes gastric cancer cell proliferation by
targeting SOX7. Biomed Pharmacother. 79:153–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu H, Mastriani E, Yan ZQ, Yin SY, Zeng
Z, Wang H, Li QH, Liu HY, Wang X, Bao HX, et al: SOX7 co-regulates
Wnt/β-catenin signaling with Axin-2: Both expressed at low levels
in breast cancer. Sci Rep. 6:261362016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ding H, Quan H, Yan W and Han J: Silencing
of SOX12 by shRNA suppresses migration, invasion and proliferation
of breast cancer cells. Biosci Rep. 36:e003892016. View Article : Google Scholar
|
|
38
|
Huang W, Chen Z, Shang X, Tian D, Wang D,
Wu K, Fan D and Xia L: Sox12, a direct target of FoxQ1, promotes
hepatocellular carcinoma metastasis through up-regulating Twist1
and FGFBP1. Hepatology. 61:1920–1933. 2015. View Article : Google Scholar : PubMed/NCBI
|