Introduction
The canonical Wnt signalling pathway, also known as
the Wnt/β-catenin or the β-catenin/T-cell factor (TCF) pathway
(1), modulates diverse biological
processes via signal transduction (2–4). The key
step in Wnt/β-catenin signalling pathway is the stabilization and
accumulation of cytosolic β-catenin. Under normal conditions
without stimulation, β-catenin is constantly phosphorylated by the
glycogen synthase kinase-3β (GSK3β) complex (5). Phosphorylated β-catenin is subsequently
ubiquitinated and targeted for degradation by the 26s proteasome
(5,6). Wnt signalling activation leads to the
inhibition of GSK3β activity and β-catenin accumulation in the
cytosol and nucleus. In the nucleus, β-catenin forms a complex with
T-cell factor/lymphoid enhancer binding factor (TCF/LEF) and
induces the expression of downstream genes involved in Wnt
signalling (5). A genome-wide scan
of TCF/LEF binding sites revealed that TCF/LEF binds to the
putative cis-elements
(T/AC/GAAAG) in the
target gene promoters (7).
Hedgehog signalling plays important roles in both
vertebrate and invertebrate development (8). Hedgehog was first identified as a
secreted signalling protein whose expression is induced in the
Drosophila melanogaster embryonic segment (9). The three mammalian Hedgehog
(hh) genes, namely, Sonic hedgehog, Indian hedgehog, and
Desert hedgehog, are important in the patterning of many tissues
and biological structures (9). In
addition, abnormal activation of Hedgehog signalling is required
for nearly all basal cell carcinomas, medulloblastomas, and
rhabdomyosarcomas; however, overactivation of Hedgehog signalling
has also been observed in some tumours (8,10–12). In
the absence of Hedgehog stimulation, the transmembrane protein
Patched1 (Patch1) interacts with Smoothened (Smo), another
transmembrane protein that maintains inactivity of the Hedgehog
pathway. Sonic hedgehog binds to and inactivates Patch1 after
secretion, leading to Smo activation (11,13) and
subsequent transcription of downstream genes via the GLI-Kruppel
family transcription factors (13).
Among the three GLI-Kruppel family members (Glis), Gli1 and Gli2
are positive regulators, while Gli3 is a negative regulator
(14). Gli1 overexpression was
associated with promoting gastric initiation and progression in
patients (14). A previous study
identified the novel function of Gli1 in modulating
E-cadherin/β-catenin-regulated cancer cell properties. Gli1 was
demonstrated to interfere with membrane localization of E-cadherin
by upregulating MUC5AC, a gel-forming mucin, which in turn weakens
E-cadherin-dependent cell-cell adhesion and promotes cell migration
and invasiveness in pancreatic ductal adenocarcinoma (15). In addition, Gli3 deficient
showed prevention of premature fusion of calvarial suture in mice
via eliminate one allele of a key transcription factor Runx2
(16).
The extracellular matrix (ECM) is the largest
component of the dermal skin layer (17). Fibroblasts are known to participate
in ECM assembly and remodelling (18,19),
demonstrating their important roles in wound repair. More recently,
Wnt signalling was found to be involved in human fibroblast repair
(20). In our transcriptome study
using Wnt3a-stimulated cells, Wnt signalling activation was
observed to alter the expression patterns of a large number of
genes, including Hedgehog signalling genes (21). In addition, Hedgehog signalling genes
have been demonstrated to regulate fibroblast repair and are
controlled by β-catenin, a Wnt signalling regulator (22). However, the association between Wnt
and Hedgehog signalling pathways remain unclear.
In the present study, we further examined regulation
of Hedgehog signalling genes by Wnt signalling by examining the
gene expression profiles of Hedgehog signalling genes under Wnt3a
stimulation. In addition, promoter sequence analysis combined with
ChIP and yeast-one hybrid assays were performed to explore the
potential mechanisms that mediate the activation of Smo and
Gli1 transcription by β-catenin/TCF4 complex. Next,
TCF4 and β-catenin were transiently overexpressed in
fibroblasts, after which Smo and Gli1 expression
levels were determined. In addition, β-catenin expression
was suppressed via siRNA transfection, and Smo and
Gli1 levels were monitored. In conclusion, our analyses
indicated that the β-catenin/TCF4 complex directly regulates
Smo and Gli1 transcription, which provided direct
evidence for the link between the Wnt and Hedgehog signalling
pathways.
Materials and methods
Human foreskin fibroblast cell
culture
Human foreskin samples were isolated from 4 patients
in the Department of Dermatology, the First Affiliated Hospital,
Wenzhou Medical University (Wenzhou, China). The present study was
approved by the Ethics Committee of Wenzhou Medical University
(Wenzhou, Zhejiang, China) and written informed consent was
obtained from all of the patients involved (21). All the procedures followed for
purification and culture of human fibroblasts were described by
Xuan et al (23).
Cell culture
Human foreskin fibroblast cells were cultured for 12
h at 37°C in an incubator with 5% CO2 in Petri dishes
and subsequently cultured into a monolayer until reaching
confluence (23). Afterwards, cells
were cultured in DMEM containing 0.5% FBS and treated with Wnt3a
(100 ng/ml). Cells were harvested after 0, 1, and 3 h of Wnt3a
treatment.
Yeast-one hybrid assay
For the yeast one-hybrid assay, the 1.5-kb promoters
of Smo and Gli1 were synthesized by Sangon Biotech
(www.sangon.com/) and subsequently cloned into the
pHISi vector. The TCF4 ORF sequences were cloned into
the pGAD424 yeast expression vector. The pGAD424-TCF4
and pSmo-His, mpSmo-His, pGli1-His, or mpGli1-His
were transformed into the yeast one-hybrid bait strain (YM4271).
The growth of yeast cells was monitored on synthetic dropout Leu or
His.
ChIP assay
Chromatin immunoprecipitation (ChIP) assay was
performed using a chromatin immunoprecipitation assay kit (cat no.
17-295, Millipore, Billerica, MA) according to the manufacturer's
instructions. Immunoglobulin (1 µg; IgG; Abcam, ab172730),
anti-TCF4 antibody (Cell Signaling Technology, 2566), and
anti-β-catenin antibody (Abcam, ab32572) were used for
immunoprecipitation (24). The
immunoprecipitated DNA fragments were analysed via quantitative
PCR. Primer sequences used for ChIP-PCR are listed in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequences
(5′-3′) |
|---|
| Smo F |
ACCTATGCCTGGCACACTTC |
| Smo R |
AGGAAGTAGCCTCCCACGAT |
| PTCH F |
CAAACTCCTGGTGCAAACCG |
| PTCH R |
CCGGGATTCTCAGCCTTGTT |
| Gli1 F |
CCAGAGTTCAAGAGCCTGG |
| Gli1 R |
CCTCGCTCCATAAGGCTCAG |
| Gli2 F |
GTTCCAAGGCCTACTCTCGCCTG |
| Gli2 R |
CTTGAGCAGTGGAGCACGGACAT |
| Gli3 F |
GGGTGAACAGCATCAAAATGGAG |
| Gli3 R |
CCGATAGCCATGTTGGTGG |
| TCF4 F |
AGAGCGACAAGCCCCAGAC |
| TCF4 R |
ATTCGCTGCGTCTCCCATC |
| β-catenin F |
TCGCCAGGATGATCCCAGC |
| β-catenin R |
GCCCATCCATGAGGTCCTG |
| GAPDH F |
GACCTGCCGTCTAGAAAAAC |
| GAPDH R |
CTGTAGCCAAATTCGTTGTC |
| Smo F1 F |
CGTTGAGGGAGACTTGCTTA |
| Smo F1 R |
CTTGGATGAATACCTGTGGC3 |
| Smo F2 F |
CTCTGAGTGACTCCGAGGTTAT |
| Smo F2 R |
TAGTTGGTCCTAAGGTTGTTTG |
| Gli1 F3 F |
TGAAGTCTTATTCCCTCCCAC |
| Gli1 F3 R |
TCCCTCTACCATTTCTTGTTCT |
Western blot analysis
Cells were lysed in an ice-cold lysis solution [7 M
urea, 2 M thiourea, 2% CHAPS, 40 mM Trizma base, 40 mM
dithiothreitol (DTT), and 1% protease inhibitor]. After complete
lysis of the cells and centrifugation at 15,000 × g for 15 min, the
total protein concentration in the supernatant was measured using a
Bradford protein assay kit (Bio-Rad, Richmond, CA, USA). Proteins
were separated via SDS-PAGE and electrotransferred onto Immobilon-P
Transfer Membranes (Millipore, Tokyo, Japan). Membranes were
incubated in TBS containing 5% skim milk and 0.05% Tween-20 for 1 h
and subsequently blotted with primary antibodies at 4°C overnight.
Anti-Smo antibody (1:1,000, Abcam, ab72130), anti-Gli1 antibody
(1:2,000, Abcam, ab49314), and anti-glyceraldehyde 3-phosphate
dehydrogenase (GAPDH; Abcam, ab8245) were used as the primary
antibodies. Membranes were incubated for 1 h with an anti-mouse or
anti-rabbit HRP-linked secondary antibody (1:2,000; Cell Signaling
Technology).
Total RNA extraction, cDNA synthesis,
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA (1 µg) was extracted from human foreskin
fibroblast cells and reverse-transcribed using a GoScript Reverse
Transcription Kit (Reverse Transcription System, Promega) according
to the manufacturer's instructions. Gene expression was quantified
via RT-qPCR as previously described (25). A SYBR Green Master Mix kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used for performing qPCR
on an Illumina Eco 3.0 (Illumina, Inc., San Diego, CA, USA). A
typical reaction consisted of an initial denaturation at 95°C for 3
min, followed by 40 cycles of denaturation for 30 sec at 95°C,
annealing for 30 sec at 55°C and extension at 72°C for 30 sec,
followed by a final extension at 72°C for 5 min. The transcription
levels were normalized against those of GAPDH using the
2−ΔΔCq method (26).
Primer sequences used for RT-qPCR are listed in Table I.
Overexpression (OX) and RNA
interference
Open reading frame (ORF) regions of TCF4
(NM_013685.2, NCBI) and β-catenin (NM_001165902.1, NCBI)
were synthesized and cloned into the pcDNA3.1 (+) vector to
generate the TCF4 OX and β-catenin OX constructs.
siRNA targeting β-catenin (ON-TARGET plus SMART pool, L-004018),
siRNA targeting Gli1 (ON-TARGETplus SMART pool,
J-041026-05), and negative control siRNA (ON-TARGETplus
Non-targeting Control Pool, D-001810) were purchased from Dharmacon
RNA Technologies (Chicago, IL, USA). Fibroblast cells were seeded
for 12 h before transfection and allowed to reach 30 to 50%
confluence at the time of transfection. Afterwards, 30 nM siRNA
duplex and 2 µg of TCF4 OX or β-catenin OX plasmids
were transfected on day 0 using Lipofectamine 2000 (Invitrogen) and
Opti-MEM®I Reduced Serum Medium (Gibco) according to the
manufacturer's instructions. Cells reached confluence at 24 h after
transfection, and the OX or siRNA solutions were replaced with full
growth medium. The transfected cells were used for two experiments,
namely, cell migration assay (60–80% confluence at the time of cell
migration assay on day 2) and RT-qPCR (up to a density of 80–90%
confluence at the time of harvest for RNA preparation on day
3).
Cell proliferation assay
Cell proliferation was examined after siRNA
treatment. Proliferation ability was measured using a CCK-8 Kit
(Dojindo Bio., Japan) according to the manufacturer's instructions.
The cell densities of the β-catenin and Gli1
siRNA-transfected cells were analysed relative to those of the
control group as previously described (23).
Statistical analysis
Statistical calculations were performed using
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). Data are expressed as the mean ± standard deviation.
Significant differences between groups were analysed by one-way
analysis of variance, followed by Bonferroni's multiple comparison
post hoc tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Wnt3a induced the expression of
Hedgehog signalling genes
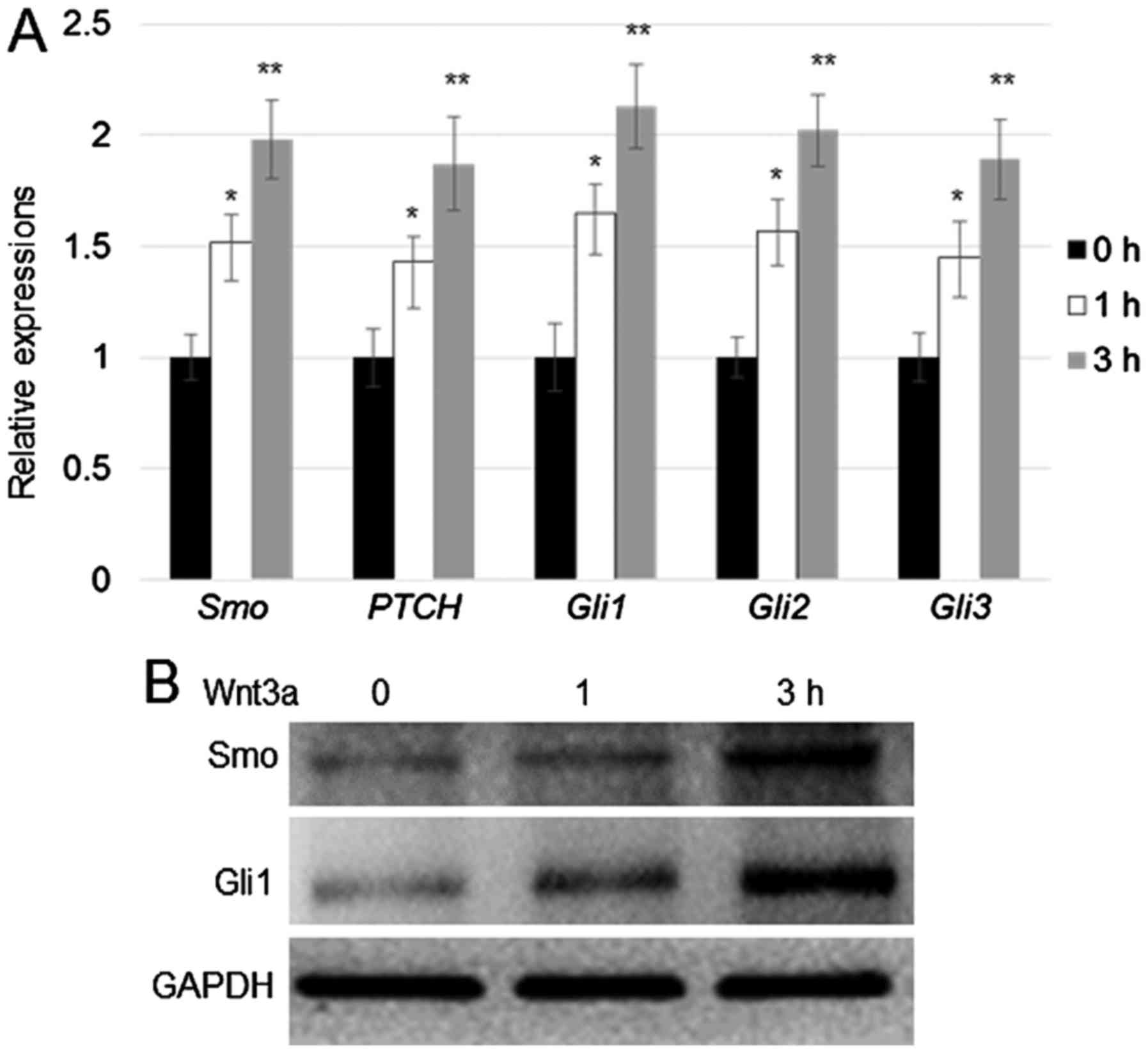
To verify the effect of Wnt3a on the expression of
hedgehog signalling genes, human fibroblast cells were treated with
Wnt3a (100 ng/ml) for 1 and 3 h. RT-qPCR assay was performed to
monitor the expression of Smo, PTCH, Gli1, Gli2, and
Gli3. The results indicated that expression of all the above
mentioned genes was induced by Wnt3a treatment, and peak expression
levels for all genes tested were detected after 3 h of Wnt3a
treatment (Fig. 1A). Western blot
analysis was performed to verify the results of RT-qPCR analysis.
Consistent with the results of RT-qPCR, western blotting results
revealed that Smo and Gli1 expression was induced after 3 h of
Wnt3a treatment (Fig. 1B).
The β-catenin/TCF4 complex directly
activates Smo and Gli1 transcription
Smo and Gli1 are induced by Wnt3a
treatment. Therefore, we next determined whether the key Wnt
signalling transcription factor complex β-catenin/TCF4 can directly
induce the transcription of Smo and Gli1. Promoter
sequence analysis revealed that two and one putative TCF/LEF
binding motifs (AGAAAG) (7) was
located within 1.5 kb of the Smo and Gli1 promoters,
respectively (Fig. 2A). To
investigate whether β-catenin/TCF4 directly binds to the putative
motifs located in the Smo and Gli1 promoters, ChIP
assays were performed using antibodies targeting TCF4 or β-catenin,
with IgG as the control. Immunoprecipitated DNA fragments were
amplified using the primer pairs targeting the F1, F2, and F3
regions of the Smo and Gli1 promoters (Fig. 2B). Data were normalized using the
input DNA as template. ChIP-PCR results showed that β-catenin and
TCF4 can bind to the F1, F2, and F3 regions (Fig. 2B). To verify the ChIP findings, yeast
one-hybrid assay was performed by co-expressing AD (activation
domain)-TCF4, and pSmo-His and mpSmo-His or
pGli1-His and mpGli1-His (Fig. 2C). The results indicated that cells
transfected with AD-TCF4 co-expressing pSmo-His or
pGli1-His were able to grow in the SD medium without
histidine, whereas cells transfected with AD-TCF4 co-expressing
mpSmo-His or mpGli1-His did not grow. All
transformants were normally grown in SD medium without leucine
(Fig. 2D). The above findings
indicated that TCF4 activates the transcription of Smo and
Gli1 by binding to their promoters in yeast cells.
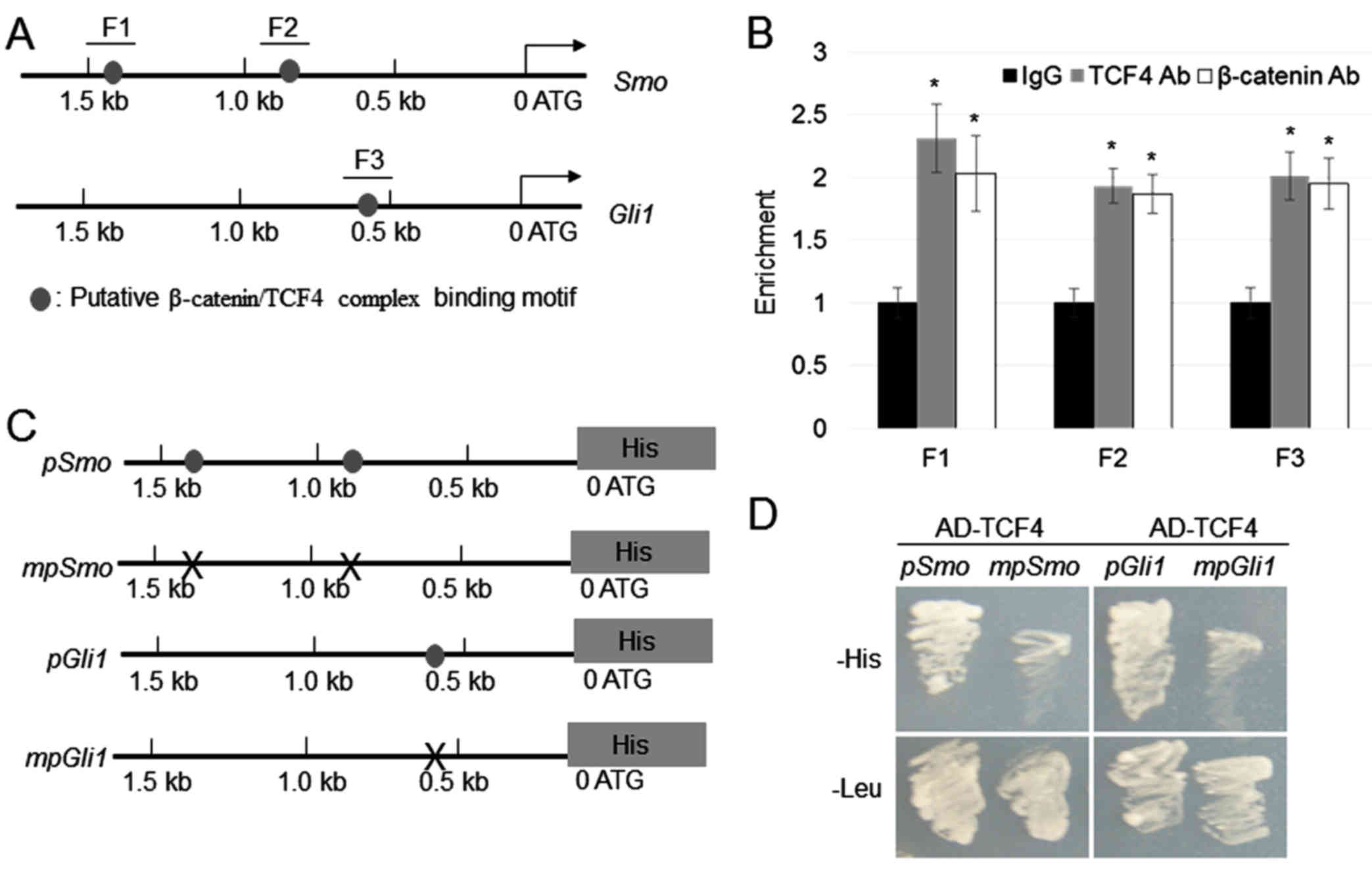 | Figure 2.β-catenin/TCF4 complex directly binds
to the Smo and Gli1 promoters. (A) Schematic diagram
indicating the locations of the putative TCF binding motifs (grey
circles) within 1.5 kb of the Smo and Gli1 promoters.
(B) ChIP assay was performed by amplifying immunoprecipitated DNA
from the F1, F2 and F3 regions; the relative ratios of
immunoprecipitated DNA and input DNA were determined via
ChIP-polymerase chain reaction. Data are expressed as means ±
standard error (n=3). *P<0.05 vs. IgG. (C) The 1.5-kb regions of
the Smo and Gli1 promoters with or without mutations
at the TCF4 binding motif were cloned into the pHISi vector,
which utilizes His as a reporter gene. Grey circles indicate TCF4
binding motifs, while ‘X’ marks indicate mutations in the TCF4
binding sequences. (D) Yeast one-hybrid assay was performed to
analyse the binding of tTCF4 with the Smo and Gli1
promoters. Yeast cells harbouring AD-TCF4 and pSmo-His and
mpSmo-His or pGli1-His and mpGli1-His were
grown on synthetic defined medium lacking Leu or His. TCF4,
antibody; Ab, antibody; IgG, immunoglobulin G; TCF4, T-cell factor
4; Smo, smoothened, frizzled class receptor; Gli, GLI family zinc
finger; ChIP, chromatin immunoprecipitation. |
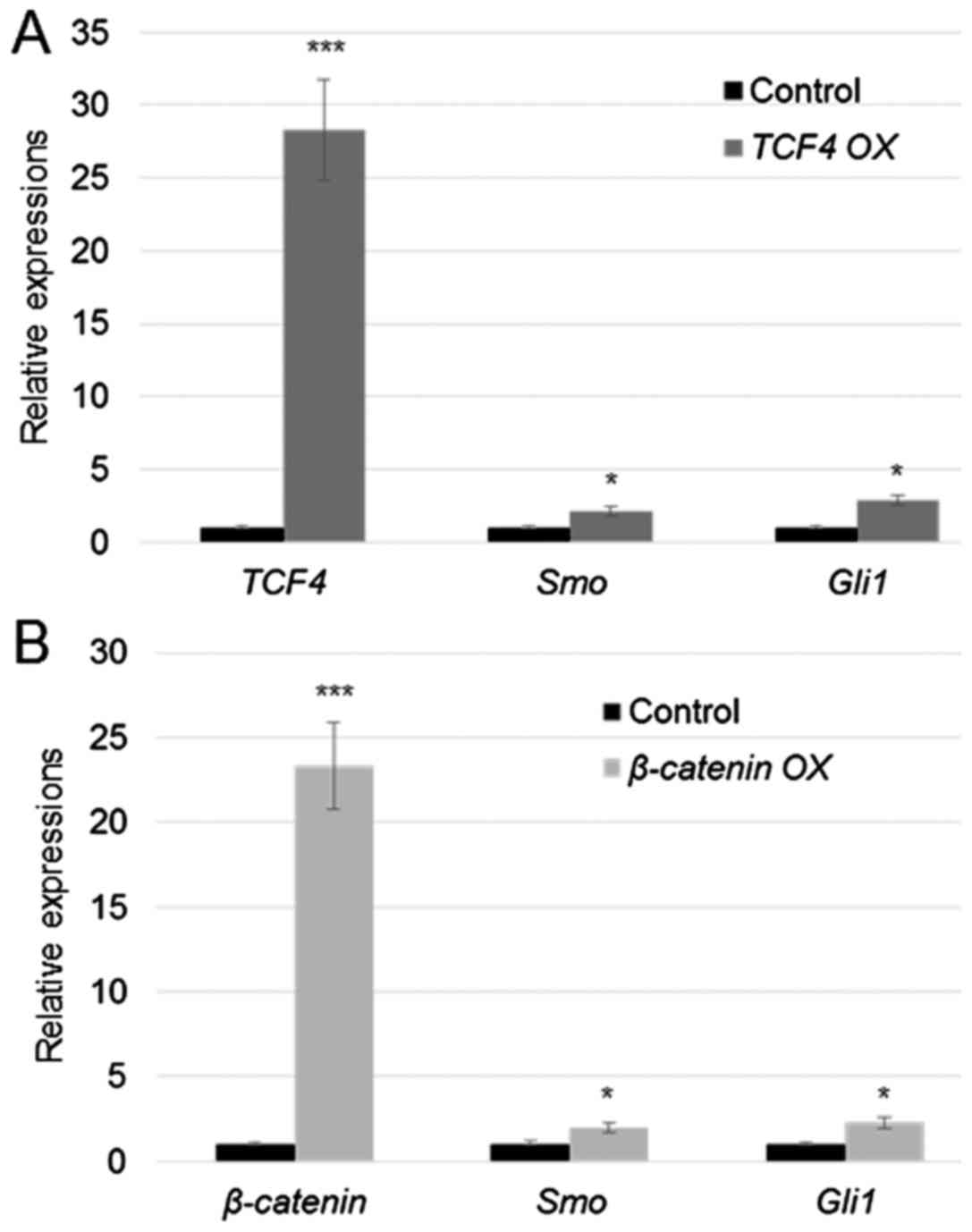
Overexpression of TCF4 and β-catenin
activated Smo and Gli1
Given that TCF4 and β-catenin bind to the Smo
and Gli1 promoters, their expression levels were further
analysed in the TCF4- and β-catenin-overexpressing
cells. TCF4 and β-catenin were transiently
overexpressed (TCF4 OX and β-catenin OX) in human
fibroblasts by transformation of the plasmids pcDNA3.1-TCF4
or pcDNA3.1-β-catenin using Lipofectamine 2000 reagent. Gene
expression analysis via RT-qPCR revealed that TCF4 and
β-catenin were significantly upregulated after 24 h of
transfection (Fig. 3). In addition,
Smo and Gli1 expression levels showed 2.2- and
2.8-fold upregulation in TCF4 OX cells relative to control
cells, respectively (Fig. 3A). In
addition, the results showed that Smo and Gli1
expression levels were upregulated by 2.2- and 2.6-fold in
β-catenin OX cells relative to control cells, respectively
(Fig. 3B).
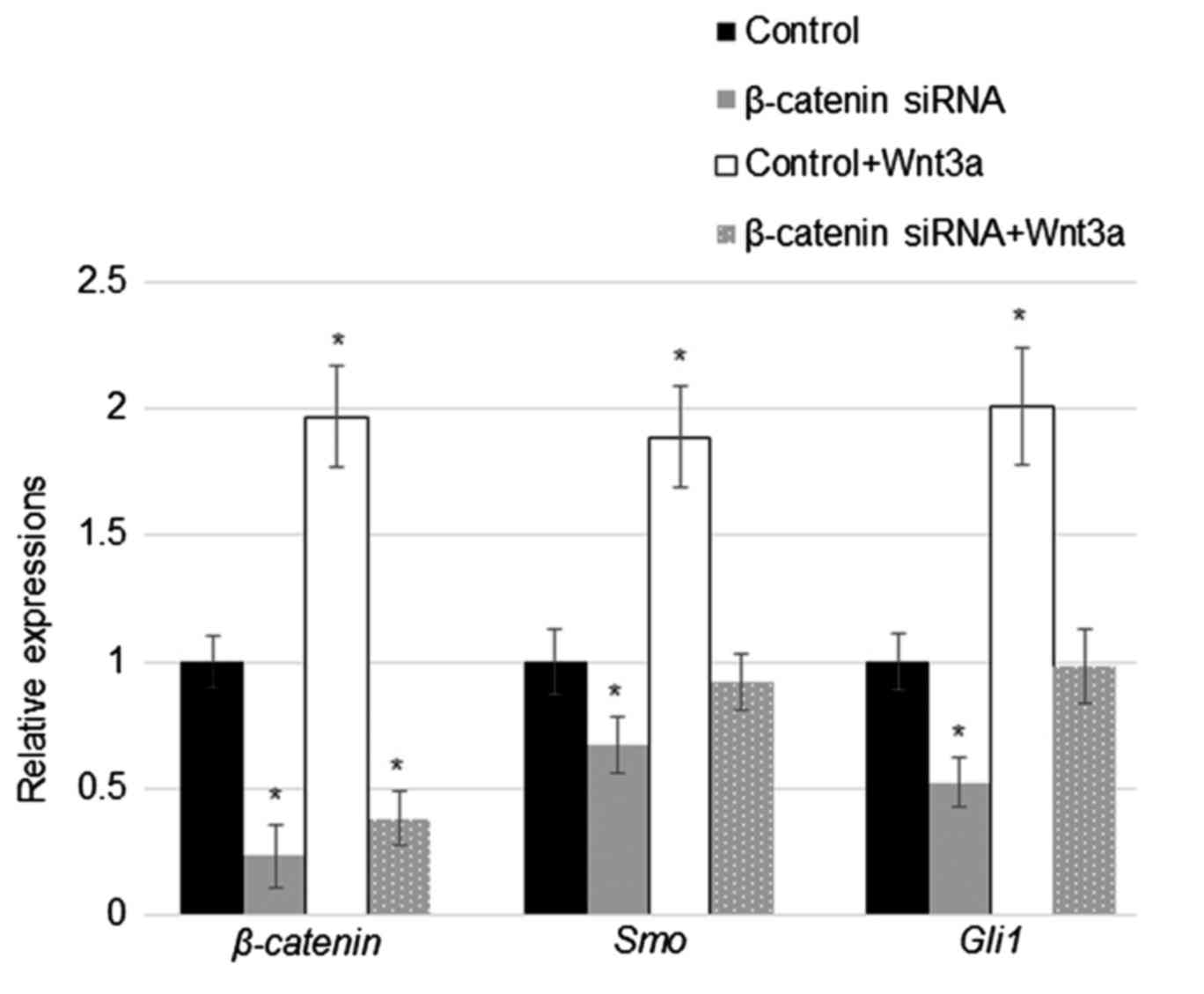
Suppression of β-catenin reduced Smo
and Gli1 levels with or without Wnt3a stimulation
Considering that overexpression of TCF4 and
β-catenin induced Smo and Gli1 expression,
TCF4 and β-catenin levels were further analysed in
the β-catenin siRNA-transfected human fibroblast cells with
or without Wnt3a treatment. RT-qPCR results indicated that
transfection with β-catenin-specific siRNA significantly
downregulated the expression of β-catenin independent of
Wnt3a treatment (about 60–70%), and Wnt3a treatment induced
β-catenin levels (Fig. 4).
Wnt3a treatment also upregulated Smo and Gli1
expression in cells with or without Wnt3a stimuli; however, the
relative increases in expression were lower in β-catenin
siRNA-transfected cells compared to those of control cells.
Furthermore, siRNA suppression of β-catenin expression
significantly reduced Smo and Gli1 levels in human
fibroblasts (Fig. 4).
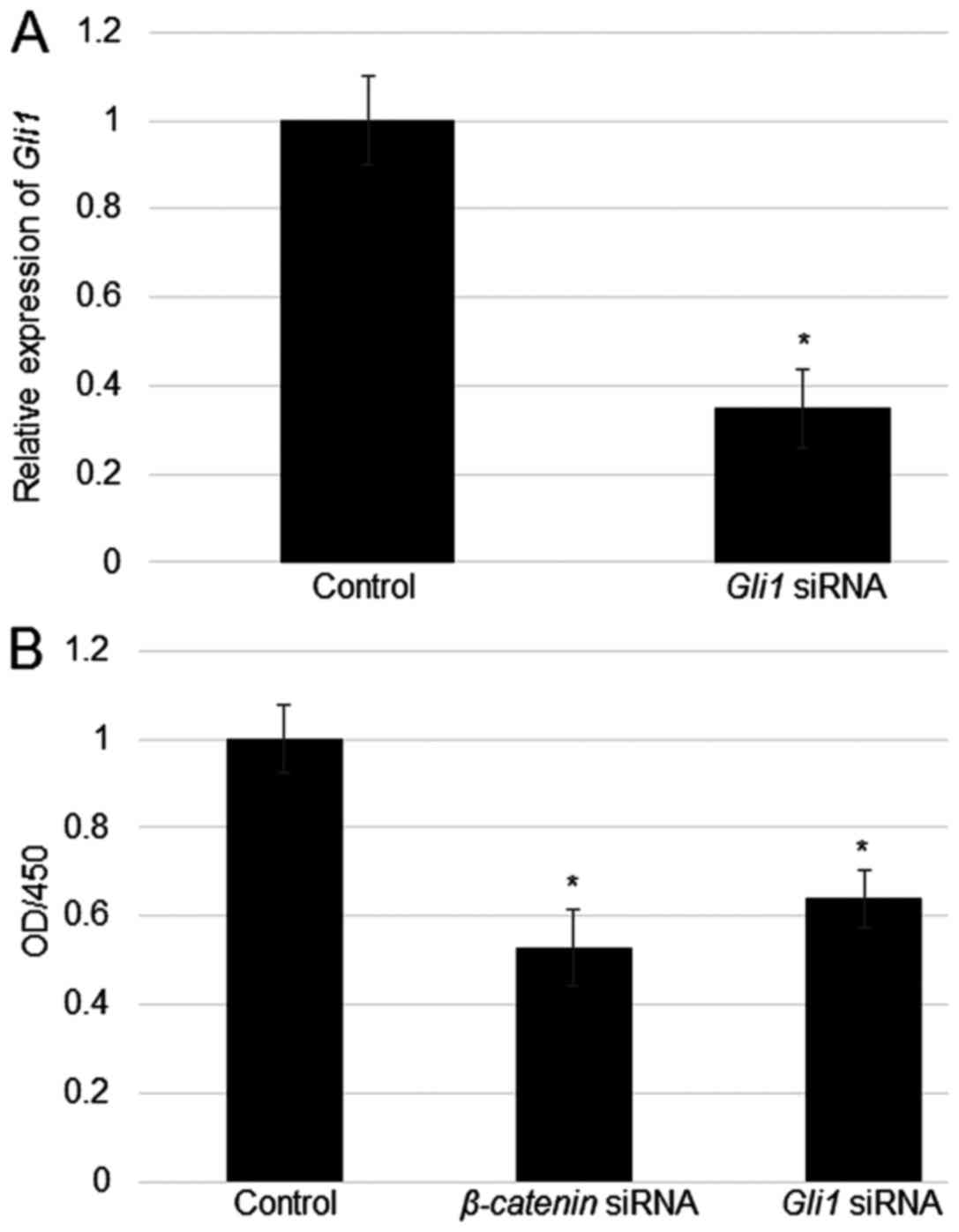
Effects of β-catenin and Gli1
suppression on cell proliferation in fibroblasts
Given that the Wnt and Hedgehog signalling pathways
have been demonstrated to be involved in cell proliferation, we
examined the effects of β-catenin and Gli1
suppression on fibroblast proliferation. Before evaluating cell
proliferation ability, fibroblasts were transfected with
Gli1 siRNA and control siRNA, after which Gli1
expression levels were measured by RT-qPCR. Gli1 was
significantly downregulated in Gli1-siRNA transfected cells
relative to those of control cells (Fig.
5A). In addition, cell proliferation rates were determined in
β-catenin and Gli1 siRNA-transfected cells relative
to those of the control group. Cell density measurements evidently
showed that suppression of β-catenin and Gli1
inhibited cell proliferation (Fig.
5B).
Discussion
The skin is the tissue layer that protects the body
from external damage. Fibroblasts are a cell type that play a major
role in wound repair. Extensive studies have been conducted to
elucidate the regulatory mechanisms underlying fibroblast cell
migration and proliferation (20–23). TCF
and β-catenin are master transcriptional regulators of Wnt
signalling, and nuclear translocation of β-catenin leads to the
formation of a TCF/β-catenin transcription factor complex (2–4).
GSK3β-β-catenin signalling was reported to regulate cell migration
in human fibroblasts via feedback regulation of basic fibroblast
signalling (20). Another study
demonstrated that key regulators of Hedgehog signalling, including
Smo and Gli1, are controlled by β-catenin to modulate
fibroblast cell migration (22). In
addition, connections between Wnt and hedgehog signalling was
reported (27). In colon cancer,
GSK3β and CK1α phosphorylate full-length Gli3 (28), leading to the degradation of
C-terminal peptides of Gli3 to produce truncated form, Gli3R. Gli3R
further inhibits Gli1 activity (29). A negative regulator (kinase) of both
Wnt/β-catenin and Hedgehog/Gli signaling pathways, Sufu interacts
with β-catenin and Gli1 to modulate their nuclear-cytoplasmic
distributions (30,31). However, the relationship between
β-catenin and Gli as well as Wnt and hedgehog signalling remains
unclear in fibroblasts. Our previous transcriptome study revealed
that Wnt3a stimulation in fibroblasts induced the expression of
Hedgehog signalling genes, such as Smo, PTCH, and Gli
(21). To further confirm the
relationship between Wnt and Hedgehog signalling, fibroblast cells
were subjected to Wnt3a treatment, and expression patterns of key
hedgehog signalling genes were analysed. The results of RT-qPCR and
western blot analyses demonstrated that Wnt3a-induced Wnt
signalling activation upregulated the expression of Smo, PTCH,
Gli1, Gli2, and Gli3 (Fig.
1).
A previous study demonstrated that the TCF/β-catenin
complex activates various downstream genes by binding to specific
sequences in the promoters of the target genes (7). Interestingly, promoter sequence
analysis revealed that the putative TCF/LEF binding motifs
(7) to which the TCF/β-catenin
complex binds were located within 1.5 kb of the Smo and
Gli1 promoters. We next performed ChIP and yeast-one hybrid
assays to determine whether the TCF/β-catenin complex directly
binds to the Smo and Gli1 promoters. Our findings
indicated that the TCF4/β-catenin complex directly binds to two and
one TCF/LEF motifs in the Smo and Gli1 promoters,
respectively (Fig. 2B).
Given that the TCF4/β-catenin complex binds to the
Smo and Gli1 promoters, the expression levels of
Smo and Gli1 were examined in the cells
overexpressing TCF4 or β-catenin. TCF4 or
β-catenin was highly expressed in the cells transfected with
pCDNA3.1-TCF4 or pCDNA3.1-β-catenin (Fig. 3). In addition, the results indicated
that TCF4 or β-catenin overexpression activated
Smo and Gli1 transcription. Moreover, Smo and
Gli1 expression levels were examined in β-catenin
siRNA-transfected cells. siRNA transfection evidently suppressed
β-catenin levels. In addition, Smo and Gli1
levels were lower in β-catenin siRNA-transfected cells
compared to those of control cells (Fig.
4), which suggested that β-catenin is located upstream of
Smo and Gli1. To confirm whether Wnt3a
stimuli-mediated induction of Smo and Gli1 are
mediate by β-catenin, control siRNA- and β-catenin
siRNA-transfected cells were treated with Wnt3a. The results
indicated that Wnt3a induced β-catenin, Smo, and Gli1
expression in both the control siRNA- and β-catenin
siRNA-transfected cells (Fig. 4).
However, the fold inductions in β-catenin siRNA-transfected
cells were lower than those in control cells, suggesting that
β-catenin mediates Wnt3a-induced activation of Hedgehog
signalling.
We further analysed the biological function of
β-catenin as a key regulator of Hedgehog signalling genes. The Wnt
signalling pathway is known to regulate cell proliferation
(2). Thus, β-catenin and
Gli1 expression was suppressed via siRNA transfection of
human fibroblasts, and cell proliferation rates were determined.
The results suggested that β-catenin and Gli1
participate in the same signalling pathway and act as positive
regulators of cell proliferation.
Wnt and Hedgehog signalling play crucial roles in
diverse processes involved in mammalian development. In the present
study, our findings revealed that the Wnt and Hedgehog signalling
pathways directly share a common molecular mechanism, which is the
binding of the TCF4/β-catenin complex to Smo and Gli1
promoters in human fibroblasts. Furthermore, β-catenin and
Gli1 were demonstrated to positively regulate cell migration
and proliferation in human fibroblasts (2; Fig. 5B), thereby demonstrating the strong
link between gene regulation and biological function. Previous
study showed the post-transcription regulation of Gli family
proteins by Wnt signalling regulators GSK3β and CK1α (26–28), but
transcriptional regulation between two pathways has not been
observed. In this study, we identified a transcriptional regulation
between β-catenin and Smo or Gli1, which extended
knowledge of Wnt and hedgehog relationship, which could be
important for understanding regulatory networks not only in
fibroblasts, but also in cancer cells.
Acknowledgements
The authors would like to thank Mr. Rui Wen
(Department of Basic Medical Science, Wenzhou Medical University,
Zhejiang, China) for their assistance.
Funding
The present study was supported by Wenzhou City
Public Welfare Technology Projects (grant no. Y20170033).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YPW, PPL and QW performed the experiments. YPW, MQZ
and LXP analyzed and interpreted the data. YPW, MQZ and LXP were
the major contributors in writing the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Wenzhou Medical University (Wenzhou, Zhejiang, China)
and written informed consent was obtained from all of the patients
involved.
Patient consent for publication
The patients provided written informed consent for
the publication of any associated data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, D'Amico M, Pestell R and Ben-Ze'ev A: The cyclin D1
gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad
Sci USA. 96:5522–5527. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moon RT, Kohn AD, De Ferrari GV and Kaykas
A: WNT and beta-catenin signalling: Diseases and therapies. Nat Rev
Genet. 5:691–701. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ,
Keller C and Rando TA: Increased Wnt signaling during aging alters
muscle stem cell fate and increases fibrosis. Science. 317:807–810.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gordon MD and Nusse R: Wnt signaling:
Multiple pathways, multiple receptors, and multiple transcription
factors. J Biol Chem. 281:22429–22433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aberle H, Bauer A, Stappert J, Kispert A
and Kemler R: beta-catenin is a target for the ubiquitin-proteasome
pathway. EMBO J. 16:3797–3804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schuijers J, Mokry M, Hatzis P, Cuppen E
and Clevers H: Wnt-induced transcriptional activation is
exclusively mediated by TCF/LEF. EMBO J. 33:146–156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bushman W: Hedgehog signaling in
development and cancer. Prostate Cancer. 107–118. 2007. View Article : Google Scholar
|
|
9
|
Taipale J and Beachy PA: The Hedgehog and
Wnt signalling pathways in cancer. Nature. 411:349–354. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Beauchamp EM, Ringer L, Bulut G, Sajwan
KP, Hall MD, Lee YC, Peaceman D, Ozdemirli M, Rodriguez O,
Macdonald TJ, et al: Arsenic trioxide inhibits human cancer cell
growth and tumor development in mice by blocking Hedgehog/GLI
pathway. J Clin Invest. 121:148–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang K, Pan L, Che X, Cui D and Li C:
Sonic Hedgehog/GLI1 signaling pathway inhibition
restricts cell migration and invasion in human gliomas. Neurol Res.
32:975–980. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huangfu D and Anderson KV: Signaling from
Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways
from Drosophila to vertebrates. Development. 133:3–14. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rohatgi R, Milenkovic L and Scott MP:
Patched1 regulates hedgehog signaling at the primary cilium.
Science. 317:372–376. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan R, Peng X, Yuan X, Huang D, Chen J, Lu
Q, Lv N and Luo S: Suppression of growth and migration by blocking
the Hedgehog signaling pathway in gastric cancer cells. Cell Oncol
(Dordr). 36:421–435. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inaguma S, Kasai K and Ikeda H: GLI1
facilitates the migration and invasion of pancreatic cancer cells
through MUC5AC-mediated attenuation of E-cadherin. Oncogene.
30:714–723. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tanimoto Y, Veistinen L, Alakurtti K,
Takatalo M and Rice DP: Prevention of premature fusion of calvarial
suture in GLI-Kruppel family member 3 (Gli3)-deficient mice by
removing one allele of Runt-related transcription factor 2 (Runx2).
J Biol Chem. 287:21429–21438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brem H and Tomic-Canic M: Cellular and
molecular basis of wound healing in diabetes. J Clin Invest.
117:1219–1222. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martin P: Wound healing-aiming for perfect
skin regeneration. Science. 276:75–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goldin A, Beckman JA, Schmidt AM and
Creager MA: Advanced glycation end products: Sparking the
development of diabetic vascular injury. Circulation. 114:597–605.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Zhu Y, Sun C, Wang T, Shen Y, Cai
W, Sun J, Chi L, Wang H, Song N, et al: Feedback activation of
basic fibroblast growth factor signaling via the Wnt/β-catenin
pathway in skin fibroblasts. Front Pharmacol. 8:322017.PubMed/NCBI
|
|
21
|
Wang Y, Zheng X, Wang Q, Zheng M and Pang
L: Activation of Wnt signaling reduces high-glucose mediated
damages on skin fibroblast cells. Iran J Basic Med Sci. 20:944–950.
2017.PubMed/NCBI
|
|
22
|
Zhu ZX, Sun CC, Zhu Ting Y, Wang Y, Wang
T, Chi LS, Cai WH, Zheng JY, Zhou X, Cong WT, et al: Hedgehog
signaling contributes to basic fibroblast growth factor-regulated
fibroblast migration. Exp Cell Res. 355:83–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xuan YH, Huang BB, Tian HS, Chi LS, Duan
YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al: High-glucose
inhibits human fibroblast cell migration in wound healing via
repression of bFGF-regulating JNK phosphorylation. PLoS One.
9:e1081822014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Wong K, Walsh K, Gao B and Zang M:
Retinoic acid receptor β stimulates hepatic induction of fibroblast
growth factor 21 to promote fatty acid oxidation and control
whole-body energy homeostasis in mice. J Biol Chem.
288:10490–10504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zittermann SI and Issekutz AC: Basic
fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte
recruitment to inflammation by enhancing endothelial adhesion
molecule expression. Am J Pathol. 168:835–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song L, Li ZY, Liu WP and Zhao MR:
Crosstalk between Wnt/β-catenin and Hedgehog/Gli signaling pathways
in colon cancer and implications for therapy. Cancer Biol Ther.
16:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tempé D, Casas M, Karaz S,
Blanchet-Tournier MF and Concordet JP: Multisite protein kinase A
and glycogen synthase kinase 3beta phosphorylation leads to Gli3
ubiquitination by SCFbetaTrCP. Mol Cell Biol. 26:4316–4326. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang B and Li Y: Evidence for the direct
involvement of {beta}TrCP in Gli3 protein processing. Proc Natl
Acad Sci USA. 103:33–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dunaeva M, Michelson P, Kogerman P and
Toftgard R: Characterization of the physical interaction of Gli
proteins with SUFU proteins. J Biol Chem. 278:5116–5122. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tukachinsky H, Lopez LV and Salic A: A
mechanism for vertebrate Hedgehog signaling: Recruitment to cilia
and dissociation of SuFu-Gli protein complexes. J Cell Biol.
191:415–428. 2010. View Article : Google Scholar : PubMed/NCBI
|