Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide and >80% cases are
non-small cell lung cancer (NSCLC) (1). Despite consistent advances in the
diagnosis and molecular targeted therapy techniques for effective
control of lung cancer, the clinical outcomes remain unsatisfactory
(2). The number of mortalities due
to lung cancer is estimated to be 154,050 in 2018, accounting for
25.27% of all cancer-associated mortalities (1), and the overall 5-year survival rate of
patients with lung cancer remains poor at <20% (3). These poor statistics are primarily due
to the fact that the majority of patients present with advanced
disease stage at the time of diagnosis, at which point, the optimal
time to perform surgery has passed (4). Therefore, improving the rate of early
diagnosis is particularly important in increasing the opportunities
for surgical intervention of patients with NSCLC, and
identification of promising diagnostic markers for early detection
is urgent.
Circular RNA (circRNA), which widely exists in
mammalian cells, has received increasing focus in endogenous
noncoding RNA research (5). One
important characteristic of circRNA is the high level of stability
that provides great possibilities as a biological marker (5,6).
Additionally, emerging studies have suggested the aberrant
expression of circRNAs in various diseases, including esophageal
squamous cell carcinoma, gastric cancer and pancreatic ductal
adenocarcinoma (7,8). It has been suggested that circRNAs act
as microRNA (miRNA) sponges, regulating gene expression, and
serving important roles in tumorigenesis and tumor progression
(7,8). The aforementioned evidence indicates
the potential value of circRNAs as novel biomarkers and therapeutic
targets for cancer diagnosis and treatment. At present, the roles
of circRNAs in the progression of NSCLC remain unclear (9,10). The
majority of valuable circRNAs, including hsa_circ_0000190 in
gastric cancer (11), are potential
biomarkers with an increased sensitivity and specificity compared
with classic biomarkers for the diagnosis of malignancies and
includes carcinoembryonic antigen and CA19-9. circRNAs have not
been comprehensively investigated in the field of heterogeneous
NSCLC (12).
In the present study the circRNA, hsa_circ_0033155
was investigated, which was demonstrated to be downregulated in
NSCLC tissues according to our previous circRNA microarray analysis
(unpublished data). To the best of our knowledge, the present study
revealed for the first time that the expression level of
hsa_circ_0033155 was significantly downregulated in NSCLC tissues.
Subsequently, the association between hsa_circ_0033155 expression
and clinicopathological parameters of NSCLC was analyzed. In order
to study the role of hsa_circ_0033155 in NSCLC progression, the
biological functions of hsa_circ_0033155 were investigated. In
addition, the levels of phosphatase and tensin homolog deleted on
chromosome 10 (PTEN), a modulator of cell survival, which has been
identified as a tumor suppressor in various tumor types (13), were evaluated.
Materials and methods
Specimens
All clinical samples were collected from the
respiratory department of Shanghai Jiaotong University Affiliated
Sixth People's Hospital (Shanghai, China) between April 2013 and
June 2016. NSCLC tissues and their matched adjacent non-tumorous
tissues 5 cm from the edge of the tumor were obtained from 40
surgical patients, (males, 23; females, 17; age range, 36–57
years). All tissue specimens were immediately preserved in
RNA-fixer reagent (Bioteke Corporation, Beijing, China) following
resection and stored at −80°C prior to use in subsequent
experiments. No patients received radiotherapy, chemotherapy or
targeted therapy prior to surgery. Tumor size was calculated using
the widest diameter determined on computerized tomography (CT)
images. Magnetic resonance imaging (MRI) and CT findings were used
to diagnose lymphatic metastasis and to evaluate tumor
differentiation, respectively. Tumor histological grading and
staging were performed according to the World Health Organization
classification criteria and the Tumor Node Metastasis system of the
International Union Against Cancer (7th edition). The present study
was approved by the Human Research Ethics Committee of Shanghai
Jiaotong University and written informed consent was obtained from
all patients.
Cell culture
NSCLC cell lines HCC827 and H1975 were purchased
from American Type Culture Collection (Manassas, VA, USA) and
normal human bronchus epithelium cell line BEAS-2B and NSCLC cell
lines PC9 and H1650 were from Shanghai Suer Biological Technology
Co., Ltd. (Shanghai, China). The cell lines were cultured in
RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin (all Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). All cells were incubated at 37°C with 5%
CO2.
Transient transfection
PC9 and H1650 were cultured in the above mentioned
medium, seeded in 6-well plates at a density of 4.0×105
cells/well and incubated at 37°C in 5% CO2. The
following day, cells were transfected with the overexpression
vector (14 µg) for hsa_circ_0033155 (pLCDH-ciR-hsa_circ_0033155;
Geneseed Biotech Co., Ltd., Guangzhou, China) using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. Negative control (NC) groups were
established by transfection with the control vector (pLCDH-ciR;
Geneseed Biotech Co., Ltd.). Cells were cultured for 24 h following
transfection. For selection of the successfully transfected clones,
cells were seeded in 6-well plates (4,000 cells/well) and treated
with 400 µg/ml G418 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
at 37°C for 10 days. Medium containing G418 was replaced every 3
days. Positive clones were verified using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Total RNA extraction and RT-qPCR
Total RNA from paired NSCLC and adjacent
non-tumorous tissues as well as cell lines were extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Then, ReverTra Ace
(Toyobo Life Science, Osaka, Japan) was applied to reverse
transcribe total RNA into single-stranded cDNA following the
manufacturer's protocol. qPCR was performed using the SYBR Premix
Ex Taq™ II kit (Takara Biotechnology Co., Ltd., Dalian, China) in a
final volume of 10 µl containing 0.5 µl cDNA, 0.5 µl each primer
and 5 µl SYBR Green. Primers were synthesized by Sangon Biotech
Co., Ltd., (Shanghai, China) and their sequences were as follows:
GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′; hsa_circ_0033155, forward
5′-GGGGTCAGGAAAGAAACTGC-3′ and reverse 5′-TGTCGTCTTCTTGCATCTGG-3′;
PTEN, forward 5′-TGGATTCGACTTAGACTTGACCT-3′ and reverse
5′-GGTGGGTTATGGTCTTCAAAAGG-3′. Thermal cycling was as follows: 94°C
for 5 min, then 42 cycles at 94°C for 5 sec and 60°C for 1 min.
Data were analyzed using the 2−ΔΔCq method (14), whereby a higher ΔCq values indicates
a lower expression of hsa_circ_0033155.
Cell proliferation assay
Cells in NC and hsa_circ_0033155 groups were seeded
in 96-well plates at a density of 3.5×103 cells/well and
incubated for 1–7 days, under the culture conditions stated above.
A total of 5 mg/ml MTT (Sigma-Aldrich; Merck KGaA) was subsequently
added to the wells and cultured at 37°C for 3 h, then the
supernatant was discarded and dimethyl sulfoxide (Sigma-Aldrich;
Merck KGaA) was added. Subsequently, the absorbance was detected at
492 nm to determine the proliferation rate. Cell proliferation was
evaluated using the following equation: Proliferation
rate=A(sample)/A(control); where A indicates the absorption
measured at 492 nm and the control describes the sample analyzed at
day 0.
Colony formation assay
Cells in NC and hsa_circ_0033155 groups were seeded
in 6-well plates (6.0×102 cells/well) and incubated at
37°C for 14 days. The medium was replaced every 2 days during the
culture. Subsequently, the plates were washed with PBS and the
colonies were fixed with 100% methanol at room temperature (RT) for
15 min, then stained with 1% crystal violet at RT for 30 min.
Finally, stained colonies with >50 cells were counted under a
light microscope at ×200 magnification.
Migration assay
The migration assay was performed in cells from NC
and hsa_circ_0033155 groups using 24-well Transwell inserts (8-µm
pore size; EMD Millipore, Billerica, MA, USA) according to the
manufacturer's protocol. A total of 1.0×104 cells were
suspended in serum-free RPMI-1640 and seeded in the upper chamber,
whereas RPMI-1640 containing 10% FBS was added to the lower
chamber. Following incubation at 37°C for 24 h, cells in the upper
surface were removed and cells remaining on the bottom surface were
fixed with 100% methanol at RT for 15 min, followed by staining
with 1% crystal violet at RT for 30 min. Finally, stained cells
were photographed under a light microscope at ×200 magnification
and counted in at least five randomly selected fields.
Western blot analysis
NC and hsa_circ_0033155 cell lysates were prepared
using cell lysis buffer (Sigma-Aldrich; Merck KGaA) and then
centrifuged (13,800 × g; 5 min; 4°C). Total protein concentration
was detected using Bradford protein assay kit from Hangzhou
MultiSciences (Lianke) Biotech Co., Ltd. (Hangzhou, China)
following the manufacturer's protocol. Protein (20 µg) was
separated on 12% SDS-PAGE gels and subsequently transferred to
polyvinylidene fluoride membranes. The membranes were incubated
with the corresponding monoclonal antibody at 4°C overnight
followed by incubation with a secondary antibody at RT for 1 h.
GAPDH was used as the loading control. Primary antibodies against
PTEN (#9188; 1:1,000), GAPDH (#5174; 1:1,000) and horseradish
peroxidase-conjugated secondary antibody (#7074; 1:2,000) were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Bands were visualized with an enhanced chemiluminescence kit from
Pierce Biotechnology, Inc. (Thermo Fisher Scientific, Inc.).
Statistical analysis
SPSS 16.0 (IBM Corp., Chicago, IL, USA) and GraphPad
Prism 5.0 (GraphPad Software, La Jolla, CA, USA) were used for data
processing. Data are presented as the mean ± standard deviation,
and each experiment was performed at least three times. Paired and
unpaired Student's t-tests were used to analyze significant
differences between two groups in tissue samples and cultured
cells, respectively. Comparisons of data among multiple groups were
performed by one-way analysis of variance followed by Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Hsa_circ_0033155 is downregulated in
NSCLC tissues and cell lines
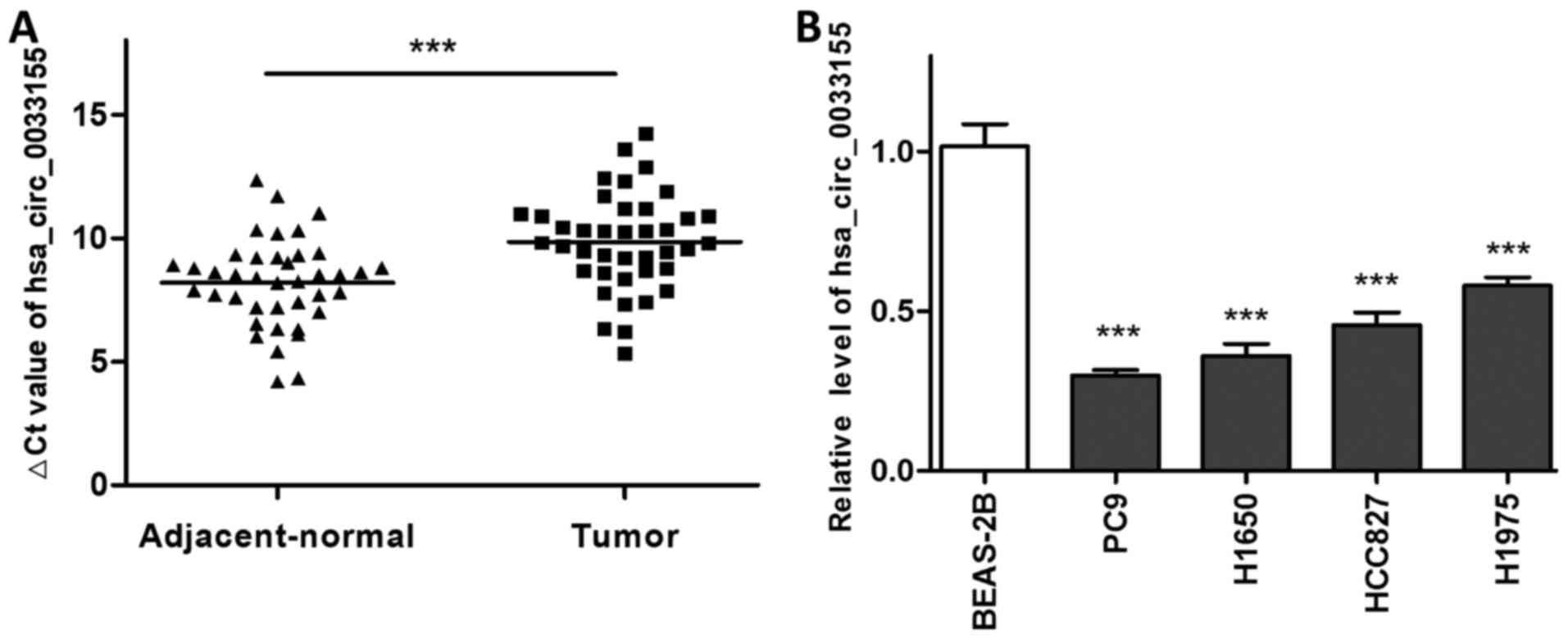
The expression level of hsa_circ_0033155 was
initially detected in NSCLC and the matched adjacent non-tumorous
tissues using RT-qPCR. As presented in Fig. 1A, the ΔCq values in the NSCLC tissues
were significantly higher compared with those in the adjacent
non-tumorous tissues, suggesting that the expression of
hsa_circ_0033155 in the tumor samples was downregulated. To
validate this, the expression level of hsa_circ_0033155 was then
measured in four NSCLC cell lines. qPCR results indicated that the
levels of circRNA in PC9, H1650, HCC827 and H1975 cells were
significantly lower compared with that in BEAS-2B, a normal human
bronchus epithelium cell line (Fig.
1B).
Clinical diagnostic value of
hsa_circ_0033155 in NSCLC
Subsequently, the association between
hsa_circ_0033155 expression levels and clinicopathological
characteristics of patients with NSCLC was evaluated. As presented
in Table I, the aberrant expression
of hsa_circ_0033155 in NSCLC tissues was significantly correlated
with lymphatic metastasis (P=0.0237), whereas no significant
association was detected between hsa_circ_0033155 expression and
other clinicopathological characteristics.
 | Table I.Associations between hsa_circ_0033155
expression levels and clinical pathological characteristics of
patients with NSCLC. |
Table I.
Associations between hsa_circ_0033155
expression levels and clinical pathological characteristics of
patients with NSCLC.
| Characteristic | Cases, n (%) | ΔCq (mean ± SD) | P-value |
|---|
| Age, years |
|
|
|
| ≤60 | 24 (60.0) | 9.86±1.47 | 0.973 |
|
>60 | 16 (40.0) | 9.83±2.56 |
|
| Gender |
|
|
|
| Male | 22 (55.0) | 9.51±2.22 | 0.229 |
|
Female | 18 (45.0) | 10.26±1.51 |
|
| Tumor size, cm |
|
|
|
| ≤5 | 21 (52.5) | 9.48±1.72 | 0.213 |
|
>5 | 19 (47.5) | 10.25±2.14 |
|
| Lymphatic
metastasis |
|
|
|
| Yes | 12 (30.0) | 10.90±2.27 | 0.024 |
| No | 28 (70.0) | 9.40±1.63 |
|
| TNM stage |
|
|
|
| ≤II | 18 (45.0) | 9.83±1.78 | 0.971 |
|
>II | 22 (55.0) | 9.86±2.11 |
|
| Tumor
differentiation |
|
|
|
| Well | 13 (32.5) | 10.01±1.85 | 0.722 |
|
Moderate | 16 (40.0) | 9.91±1.95 |
|
| Poor | 11 (27.5) | 9.57±2.20 |
|
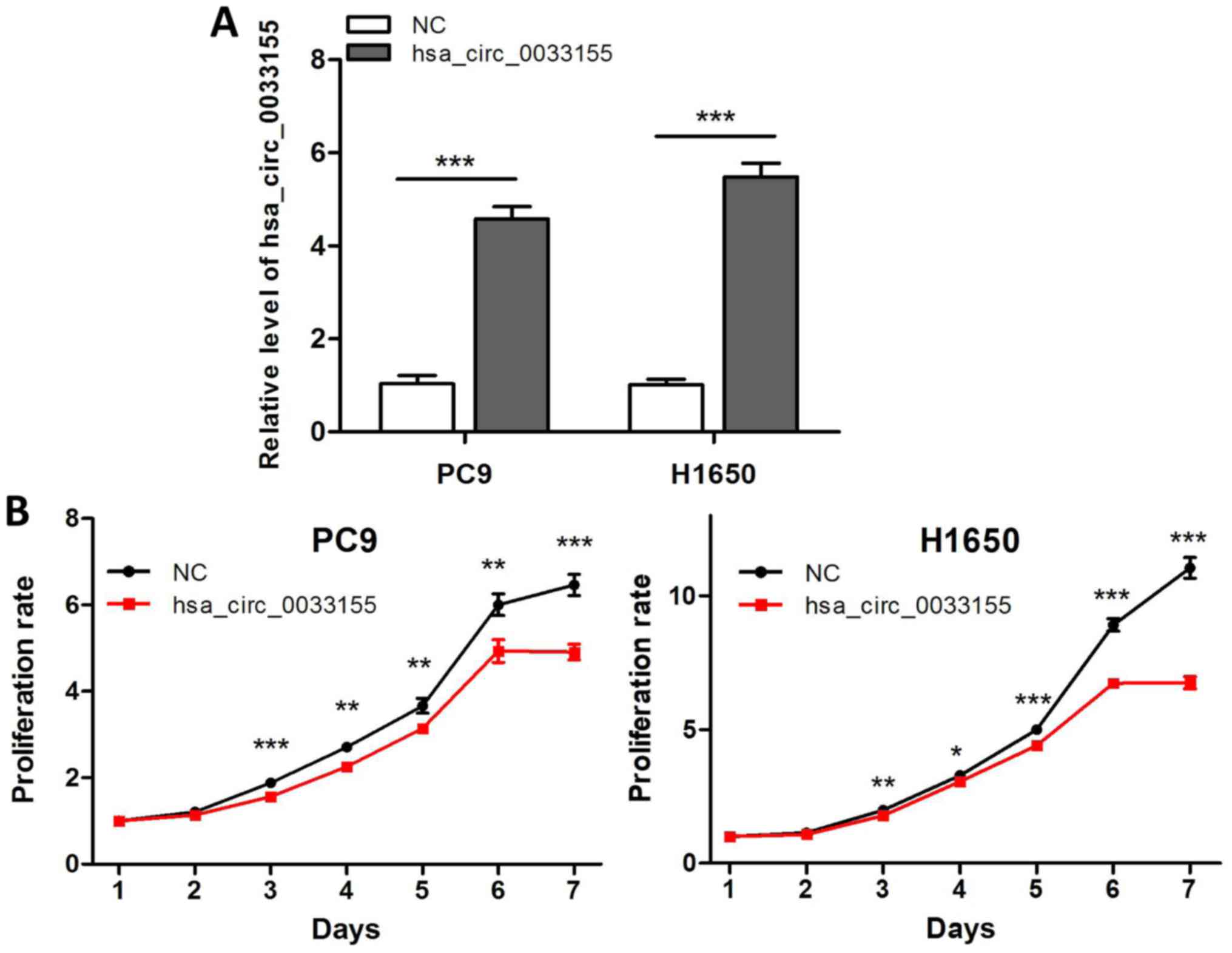
Overexpression of hsa_circ_0033155
decreases cell proliferation
The aforementioned results suggested that
downregulated hsa_circ_0033155 may serve a role in NSCLC
progression. Therefore, the effects of hsa_circ_0033155 on tumor
cell phenotypes were investigated. As relatively lower levels of
hsa_circ_0033155 were observed in PC9 and H1650 cells compared with
in HCC827 and H1975 cells (Fig. 1B),
the former two cell lines were selected to overexpress
hsa_circ_0033155. The results of the qPCR assay in Fig. 2A suggested that hsa_circ_0033155 was
successfully transfected into the two cell lines. The MTT assay
indicated that the overexpression of hsa_circ_0033155 exhibited a
significant decrease in cell proliferation following incubation for
>2 days (Fig. 2B).
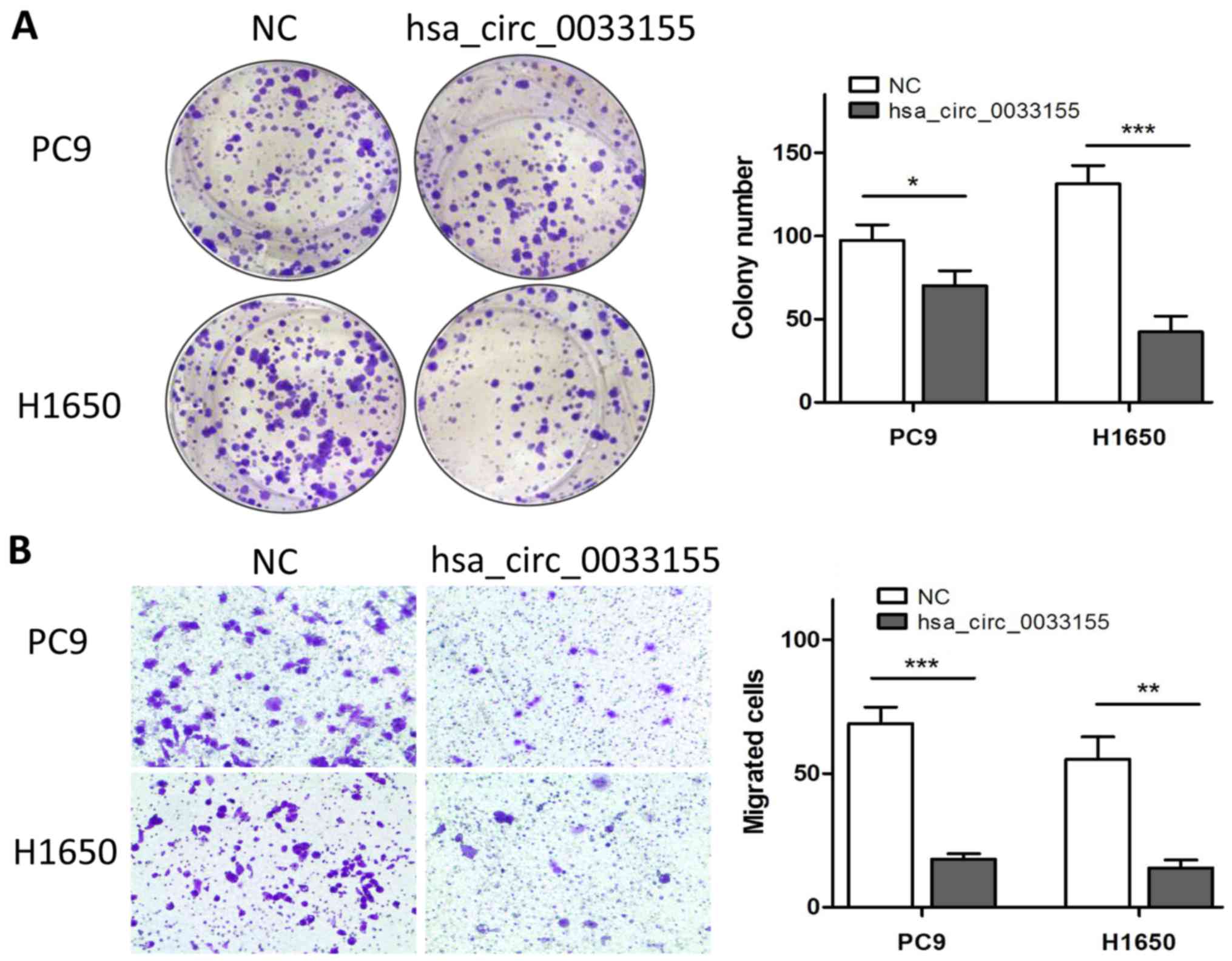
Overexpression of hsa_circ_0033155
inhibits colony formation and migration
The effects of hsa_circ_0033155 on colony formation
were then determined following incubation for 2 weeks. As presented
in Fig. 3A, the overexpression of
hsa_circ_0033155 significantly inhibited the colony formation
ability of the two cell lines. The effect of hsa_circ_0033155 on
H1650 cells was greater than that on PC9 cells. The effects of
hsa_circ_0033155 on the migration ability of cells were also
assessed. The number of cells that migrated was significantly
inhibited following overexpression of hsa_circ_0033155 (Fig. 3B).
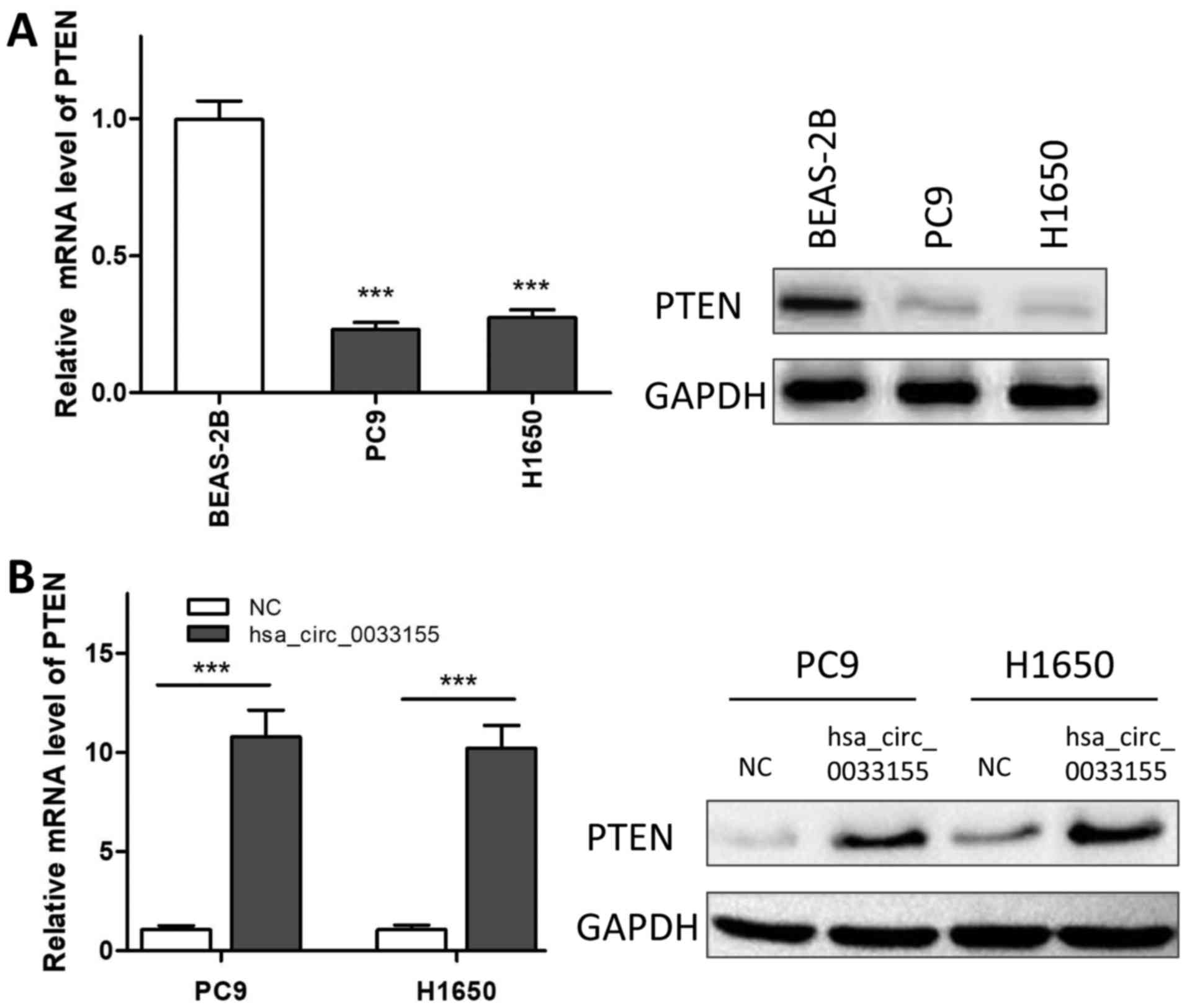
Overexpression of hsa_circ_0033155
upregulates the level of PTEN
As presented in Fig.
4, the mRNA expression of PTEN in PC9 and H1650 cells was
significantly decreased compared with that in BEAS-2B cells, which
was consistent with the western blotting results (Fig. 4A). The expression level of PTEN was
then analyzed in hsa_circ_0033155-overexpressed cells. The results
revealed that overexpression of hsa_circ_0033155 significantly
increased the expression of PTEN at the mRNA and protein levels
(Fig. 4B).
Discussion
circRNAs are a type of novel untranslated RNA
molecule that lacks 5′-3′ UTRs and a poly A tail, covalently
forming closed loops, which were previously considered to be
functionless products of errors in splicing (7). Notably, it is the unique circular
structure that protects them from the degradation of exonuclease
RNase R and confers these RNAs excellent stability (15). In addition, the expression level of
circRNAs, including circular ANRIL, is higher compared with
that of linear RNAs (16,17). In addition, their expression level
and function are independent of linear RNA isomers (16,17). The
aforementioned advantages indicate that, circRNAs may serve as
ideal diagnostic biomarkers for cancer, and be superior to other
non-coding RNAs such as long non-coding RNAs and miRNAs (18,19).
In the present study, the expression of
hsa_circ_0033155 was detected in 40 pairs of NSCLC tissues and the
adjacent non-tumorous tissues with qPCR for the first time, to the
best of our knowledge. Furthermore, the level of hsa_circ_0033155
was validated in NSCLC cell lines. The results demonstrated that
the levels of hsa_circ_0033155 were significantly downregulated in
both NSCLC tissues and NSCLC cell lines compared with their
corresponding controls. The dysregulated expression was
significantly associated with lymphatic metastasis, suggesting the
potential value of hsa_circ_0033155 as a biomarker for NSCLC
diagnosis. Coincidentally, circRNA_100876 was found to be
associated with lymphatic metastasis in NSCLC (20). Therefore, circRNAs including
hsa_circ_0033155 from NSCLC biopsy samples may be combined with
previously reported circ-ITCH and circRNA_100876, which were
correlated with tumor staging, to increase positive diagnosis rate
(21).
An increasing number of studies have revealed that
the aberrant expression of circRNAs is associated with tumor
progression. circRNA_100290 is markedly upregulated in oral
squamous cell carcinoma tissue, and the knockdown of circRNA_100290
significantly inhibits the proliferative ability of cells (22). In lung cancer, interference of
upregulated hsa_circ_0000064 markedly blocked cell cycle
progression, promoted cell apoptosis, and decreased migration and
invasive activities (23).
Therefore, hsa_circ_0033155 was overexpressed in NSCLC cells with
lower levels of the circRNA to detect whether the biological
functions were attenuated. The results indicated that
overexpression of hsa_circ_0033155 significantly decreased cell
proliferation and colony formation. Furthermore, the migration
ability of cells, an important determinant of malignancy
progression and metastasis, was also significantly inhibited. These
results indicated that hsa_circ_0033155 may serve a
cancer-suppressive role in NSCLC progression.
PTEN, a modulator of cell survival and cell cycle
progression, has been identified as a tumor suppressor that is
downregulated and mutated in various cancers, including
hepatocarcinoma, glioblastoma, ovarian and prostate cancer
(13,24,25).
Therefore, aberrant hsa_circ_0033155 may be associated with the
regulation of PTEN, thus regulating cell biological functions. It
was also demonstrated that the level of PTEN in NSCLC cell lines
was markedly decreased, and that the overexpression of
hsa_circ_0033155 significantly enhanced the level of PTEN,
revealing the regulatory role of hsa_circ_0033155 on PTEN
expression. Emerging studies have demonstrated that circRNA
functions as miRNA sponges, removing the inhibitory effect of miRNA
on its target genes, and further regulating the expression of
target genes, including PTEN (26,27). The
miRNA that is regulated by hsa_circ_0033155, subsequently
regulating the level of PTEN remains to be investigated.
In conclusion, downregulated hsa_circ_0033155 is
associated with lymphatic metastasis in NSCLC and overexpression of
hsa_circ_0033155 significantly decreased the proliferation, colony
formation and migration abilities of NSCLC cells. Additionally,
PTEN may serve an important role in these regulations. Overall,
hsa_circ_0033155 may serve as a prospective biomarker and a
promising target for NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by
National Natural Science Foundation of China (grant no.
81673014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request for non-commercial purposes, without breaching participant
confidentiality.
Authors' contributions
GW and XF worked on the conception and design of the
study, and analyzed and interpreted the data. XG and HS collected
and assembled data. All authors contributed to preparation of the
manuscript and approved the final version.
Ethics approval and consent to
participate
The present study was approved by the Human Research
Ethics Committee of Shanghai Jiaotong University (Shanghai, China)
and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan CS, Gilligan D and Pacey S: Treatment
approaches for EGFR-inhibitor-resistant patients with
non-small-cell lung cancer. Lancet Oncol. 16:e447–e459. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institute: Cancer
statistics: SEER fact sheets, lung and bronchus. http://seer.cancer.gov/statfacts/seer.cancer.gov/statfacts/html/lungb.htmlFebruary
5–2018
|
|
4
|
Hrinczenko Borys B and Subramonia-Iyer S:
Method of biopsy and sample quality for genetic mutation testing in
advanced non-small cell lung cancer in a community hospital. J Clin
Oncol. 30 34_suppl:S2712012. View Article : Google Scholar
|
|
5
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han YN, Xia SQ, Zhang YY, Zheng JH and Li
W: Circular RNAs: A novel type of biomarker and genetic tools in
cancer. Oncotarget. 8:64551–64563. 2017.PubMed/NCBI
|
|
8
|
Han C, Seebacher NA, Hornicek FJ, Kan Q
and Duan Z: Regulation of microRNAs function by circular RNAs in
human cancer. Oncotarget. 8:64622–64637. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian F, Yu CT, Ye WD and Wang Q:
Cinnamaldehyde induces cell apoptosis mediated by a novel circular
RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem Biophys
Res Commun. 493:1260–1266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao J, Li L, Wang Q, Han H, Zhan Q and Xu
M: circRNA expression profile in early-stage lung adenocarcinoma
patient. Cell Physiol Biochem. 44:2138–2146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng J, Xiang Y, Xia S, Liu H, Wang J,
Ozguc FM, Lei L, Kong R, Diao L, He C and Han L: CircView: A
visualization and exploration tool for circular RNAs. Brief
Bioinform. Jun 30–2017.(Epub ahead of print). doi:
10.1093/bib/bbx070. View Article : Google Scholar
|
|
17
|
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R
and Li H: The emerging functions and roles of circular RNAs in
cancer. Cancer Lett. 414:301–309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of circRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: circRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX,
Yan PW and Wen J: Emerging roles of circular RNA hsa_circ_0000064
in the proliferation and metastasis of lung cancer. Biomed
Pharmacother. 96:892–898. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim C, Lee CK, Chon HJ, Kim JH, Park HS,
Heo SJ, Kim HJ, Kim TS, Kwon WS, Chung HC and Rha SY: PTEN loss and
level of HER2 amplification is associated with trastuzumab
resistance and prognosis in HER2-positive gastric cancer.
Oncotarget. 8:113494–113501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Xu E, Ren C, Yang HJ, Zhang Y,
Sun W, Kong X, Zhang W, Chen M, Huang E and Chen X: Genetic
ablation of Rbm38 promotes lymphomagenesis in the context of mutant
p53 by downregulating PTEN. Cancer Res. 78:1511–1521. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hsu YL, Hung JY, Chang WA, Jian SF, Lin
YS, Pan YC, Wu CY and Kuo PL: Hypoxic lung-cancer-derived
extracellular vesicle MicroRNA-103a increases the oncogenic effects
of macrophages by targeting PTEN. Mol Ther. 26:568–581. 2018.
View Article : Google Scholar : PubMed/NCBI
|