Introduction
ATP-binding cassette sub-family E member 1 (ABCE1)
is a member of the ATP-binding cassette superfamily (1). ABCE1 acts as an RNase L inhibitor or
host protein (HP) 68 and has been reported to participate in HIV-1
capsid assembly (2). After the ABCE1
gene was silenced in the human small cell lung cancer cell line
NCI-H446 using RNAi technology, in vitro cell biology
experiments, including cell adhesion, wound healing, migration, and
invasion experiments, were performed. These assays demonstrated
that the migration and invasiveness of small cell lung cancer cells
were significantly inhibited (3).
ABCE1 was confirmed to be one of the key factors that promotes the
development and metastasis of lung cancer following the inoculation
of nude mice with the lung adenocarcinoma (AC) cell line LTEP-a-2,
which has upregulated ABCE1 expression (4).
Chemokine (C-C motif) ligand 7, which was also known
as monocyte chemotactic factor-3 for a long period of time, can
induce the majority of immune inflammatory cells, especially
monocytes (5). CCL7 plays an
important role in various pathologies, including cancers,
auto-immune diseases and chronic inflammation (6). Monocytes have strong chemotactic
ability towards tumor-associated macrophages (TAMs), and several
chemokines, including CCL7, interact with cancer-associated
fibroblasts (CAFs), which can influence the tumor microenvironment
and promote tumor angiogenesis and infiltration by TAMs (7).
The relationship between ABCE1 and chemokine CCL7 in
lung cancer has never been reported. This study attempted to
determine the relationship between ABCE1 and chemokine CCL7 in lung
cancer using a PCR microarray and immunohistochemistry to provide a
new basis for the roles of ABCE1 and CCL7 in the pathogenesis of
lung cancer.
Materials and methods
Cell culture and lentiviral packaging
vector transfection
The lung cancer cell line H1299 was purchased from
the Shanghai Chinese Academy of Science. The H1299 cells were
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum under the following conditions: 37°C, 5% CO2, and
an aseptic environment. The culture medium was changed every 1 or 2
days. After they reached confluence, the cells were digested with
0.25% trypsin for subculture, cryopreservation and lentivirus
transfection.
The lentiviral packaging vector that overexpressed
ABCE1 was purchased from JiKai Gene Chemical Technology (Shanghai,
China). The elements sequence incorporated into the GV358 vector
was Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin. The restriction enzyme
site was located in AgeI. Recombinant clones were screened by
puromycin, and the constructs expressed green fluorescent protein
(GFP) reporter genes. The experimental cells were divided into an
overexpression group and an empty vector group and were seeded in
six-well plates. After the cells reached 30% confluence, culture
medium with enhanced transfection reagent and polybrene (5 µg/ml)
were added to the wells for the transfection experiments; the
amount of virus added was calculated based on the pre-experimental
values of the multiplicity of infection (MOI). A fluorescence
microscope was used to observe the transfection efficiency, which
was 80% or greater.
RT2 Profiler™ PCR
Array
Total RNA in the cells was extracted using a TaKaRa
RNA extraction kit (TaKaRa Bio Inc., Dalian, China) and was stored
at −80°C. An RT2 First Strand Kit (Qiagen GmbH, Hilden,
Germany) was used to synthesize cDNA, and a comparative study was
performed using an RT2 Profiler™ PCR Array Human Tumor
Metastasis (PAHS-028Z) chip and RT-SYBR-Green Master Mix. qPCR was
performed using an ABI7500 PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). A number of
housekeeping genes served as internal controls for sample
normalization, and the 2−ΔΔCq values were compared
(8).
Western blotting
After the H1299 cells were transfected with the
lentiviral vector, total protein was extracted. The BCA method was
used to determine the protein concentration, and 30 µg of total
protein was loaded onto a 10% SDS-PAGE gel for protein
electrophoresis and then transferred onto a PVDF film, which was
incubated with an ABCE1 rabbit anti-human monoclonal antibody
(1:2,000 dilution; Abcam, Cambridge, MA, USA), a CCL7 rabbit
anti-human polyclonal antibody (1:1,000 dilution; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and a GAPDH mouse anti-human
polyclonal antibody (1:1,000 dilution; Abcam) overnight at 4°C.
After the membranes were incubated with the indicated secondary
antibodies (goat anti-rabbit monoclonal antibody and goat
anti-mouse monoclonal antibody, 1:1,000 dilution) for 2 h at room
temperature, the bands were visualized with enhanced
chemiluminescence (ECL kit; Thermo Fisher Scientific, Inc.) with
dark room exposure and development. The gray values of the protein
bands, which represented the relative expression levels of the
proteins, were determined.
Patient selection and tissue
specimens
Cancer tissues and adjacent normal tissues (NTs)
(located more than 2 cm from the edge of the tumor) surgically
resected from 30 patients with non-small cell lung cancer (NSCLC)
in the Department of Thoracic Surgery, Fourth Affiliated Hospital
of China Medical University (Shenyang, China), from 2014 to 2016
were embedded in paraffin. These patients, including 13 males and
17 females with an average age of 62.4 years (range, 52 to 78
years), did not receive preoperative radiotherapy; the group
included 12 cases of squamous cell carcinoma (SCC) and 18 cases of
AC.
Immunohistochemical analysis
The tissue specimens were sliced, baked for 2 h,
soaked in xylene for deparaffinization, subjected to benzene
removal using 100% ethanol, and then subjected to gradient ethanol
hydration before they were rinsed with 0.01 mol/l
phosphate-buffered saline (PBS). The slices were then incubated in
0.01 M citric acid buffer (pH=6.0) for 20 min at 97°C for antigen
retrieval. After endogenous peroxidase was blocked and the slides
were incubated with non-immune animal serum at room temperature,
each section was incubated with 50 µl of the appropriate primary
antibody (ABCE1 1:500; CCL7 1:250) in a humidified chamber
overnight at 4°C. After the sections were rinsed with PBS, a
biotinylated secondary antibody was added to them, and the slides
were incubated for 10 min at room temperature. Streptavidin
peroxidase solution was then added to the slides, which were
incubated for another 10 min. Approximately 100 µl of
diaminobenzidine (DAB) liquid was added to each of the tissue
sections, all of which were observed under a microscope for 10 min
before the reaction was terminated. Hematoxylin solution was then
added to the slides as a counterstain for 5 min to visualize the
nuclei.
Cells with a brownish-yellow membrane and cytoplasm
were considered positively stained. Ten continuous high-power
fields (×400) in each slice were observed under a light microscope
and given scores of 0, 1, 2 or 3 points according to the color
intensity; the average score was then recorded. Fields with a
positive cell rate of <5%, 5–25%, 26–50%, 51–75%, or >75%
were given scores of 0, 1, 2, 3 or 4 points, respectively. Both
scores were multiplied, and the final score was categorized as
follows: A score of 0–2 points was considered negative (−), a score
of 3–4 points was considered weakly positive (+), a score of 5–8
points was considered moderately positive (++), and a score of 9–12
points was considered strongly positive (+++). In addition, (++)
and (+++) were considered high expression, and (−) and (+) were
considered low expression.
Quantitative PCR (qPCR)
PCR was performed in a 96-well plate. Each well
contained 20 µl of the reaction system, which included 10 µl of
SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd., Dalian,
China) and a total of 1.6 µl of upstream and downstream primers.
The primers were as follows: CCL7 primer:
5′-GACAAGAAAACCCAAACTCCAAAG-3′ and 5′-TCAAAACCCACCAAAATCCA-3′;
ABCE1 primer: 5′-CAGCCTTTGTTGTGGAACATGA-3′ and
5′-ATTCGTGGCCTATAGTTGTTTGGA-3′; and GAPDH primer:
5′-CACAAGAAGGTGGTGAAGCAG-3′ and 5′-AAAGGTGGAGGAGTGGGTGT-3′. PCR
analysis was performed using an ABI7500 PCR reaction system.
Statistical analysis
The results of the qPCR Array: The GAPDH gene
was used as the internal control gene; the relative expression of
the genes was calculated as ΔCq=Cqtarget
gene-Cqinternal control, and the difference
between groups was calculated as ΔΔCq=ΔCqexperimental
group-ΔCqcontrol group. The relationship between
the experimental and control groups was expressed as
2−ΔΔCq (8).
Statistical analysis software provided by Qiagen
GmbH was used for statistical analysis and mapping. P<0.05 was
considered to indicate a statistically significant difference.
Western blotting: Black bands on the PVDF film,
which indicated positive results, were scanned by a gel imaging
system for quantitative analysis based on the gray values. The
protein band of GAPDH was used as the control. SPSS21.0 statistical
analysis software (SPSS, Inc., Chicago, IL, USA) was used for the
analysis. Measurement data are presented as the mean ± standard
deviation and were analyzed using the paired sample t-test and
one-way ANOVA with Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Immunohistochemistry and qPCR: SPSS 21.0 statistical
analysis software (SPSS, Inc.) was employed. Measurement data are
presented as mean ± standard deviation and were analyzed using the
paired sample t-test. P<0.05 was considered to indicate a
statistically significant difference, and correlations were
evaluated using Pearson correlation analysis.
Results
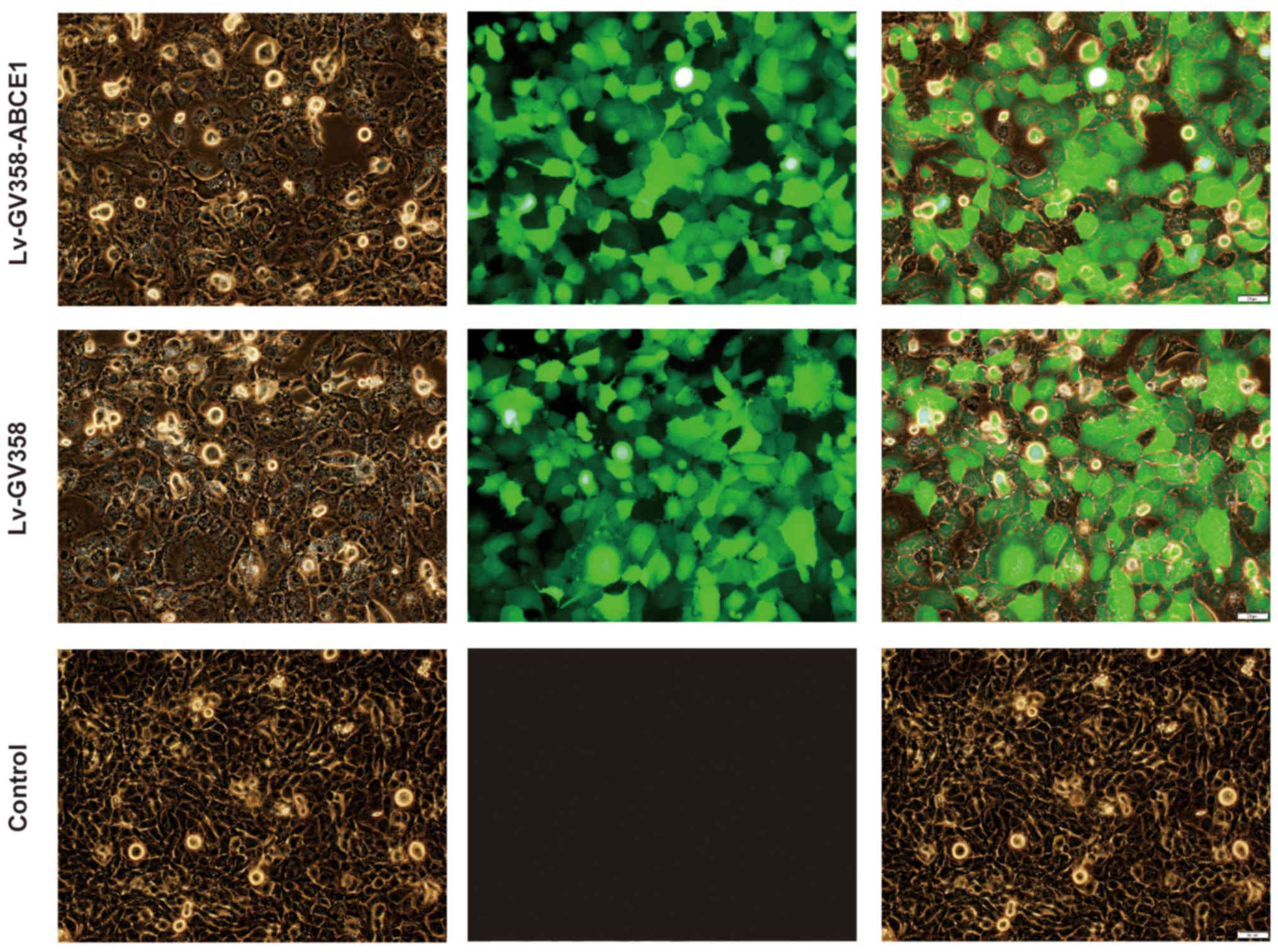
Transfection of the NSCLC cell line
H1299 using a lentiviral vector with ABCE1 overexpression
The H1299 cells were transfected with Eni.S and
polybrene. As the LV-GV358-ABCE1 and LV-GV358 constructs contained
GFP, the transfected cells were observed to have visible green
fluorescence in the cytoplasm when viewed under a fluorescence
microscope. The transfection efficiency was 80% or greater. No
fluorescence was observed in the untreated control cells (Fig. 1).
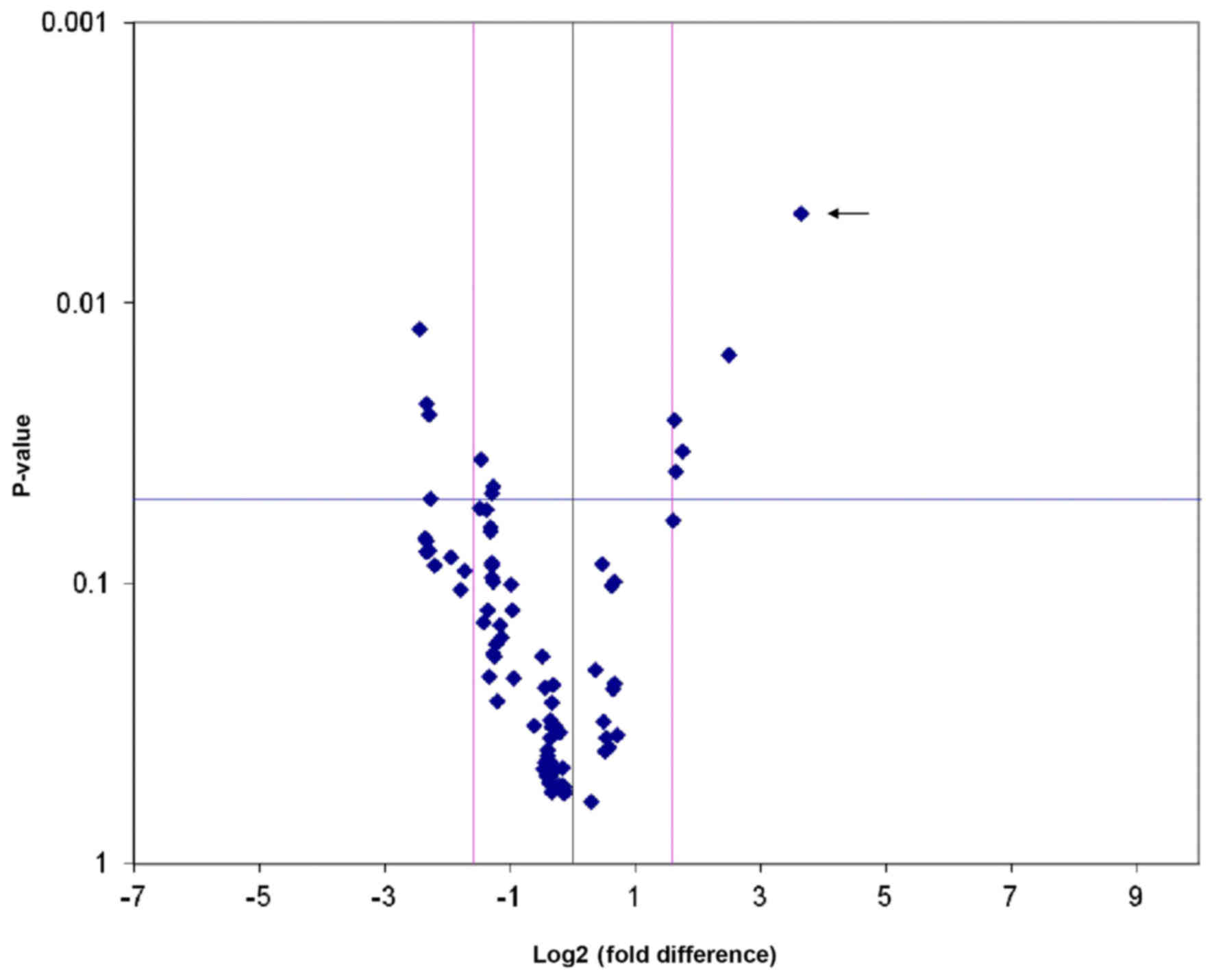
Screening of ABCE1-related genes by a
PCR Array chip
Total mRNA in the LV-GV358-ABCE1- and
LV-GV358-transfected H1299 cells was extracted and then reverse
transcribed to obtain cDNA fragments. Using the cDNA as the
template, we used an RT2 Profiler™ PCR Array Human Tumor
Metastasis chip for qPCR. The experiments were performed in
triplicate, and in all, nine tumor metastasis-related genes were
screened by statistical analysis. The difference in CCL7 expression
was the most significant (Fig. 2;
Table I), and the CCL7 gene was
selected as the primary research target from the group of
ABCE1-related metastasis genes.
 | Table I.Genes differentially expressed between
ABCE1 overexpression and empty vector groups. |
Table I.
Genes differentially expressed between
ABCE1 overexpression and empty vector groups.
| Gene | Description | Fold change | P-value |
|---|
| CCL7 | Chemokine (C-C motif)
ligand 7 | 12.49 | 0.0048 |
| TIMP3 | TIMP metallopeptidase
inhibitor 3 | 5.63 | 0.0154 |
| CXCR2 | Chemokine (C-X-C
motif) receptor 2 | 3.37 | 0.0338 |
| ETV4 | Ets variant 4 | 3.16 | 0.0401 |
| TNFSF10 | TNF superfamily
member 10 | 3.11 | 0.0262 |
| SERPINE1 | Serpin peptidase
inhibitor, clade E, member 1 | −5.41 | 0.0124 |
| CXCL12 | Chemokine (C-X-C
motif) ligand 12 | −5.02 | 0.0228 |
| MMP11 | Matrix
metallopeptidase 11 | −4.87 | 0.0250 |
| ITGA7 | Integrin, α7 | −4.76 | 0.0498 |
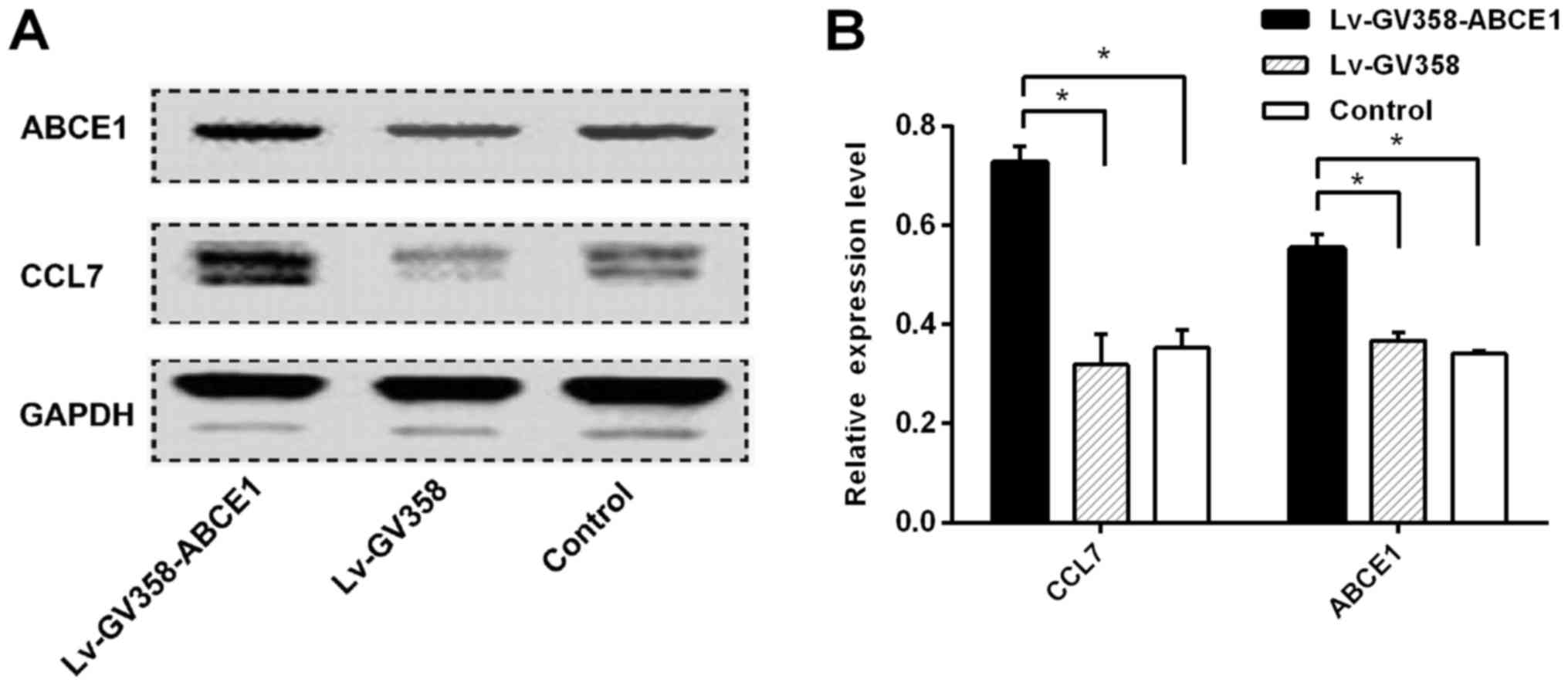
Western blotting
The expression of CCL7 in LV-GV358-ABCE1-transfected
H1299 cells (0.73±0.019) was significantly higher than that in
LV-GV358-transfected H1299 cells (0.32±0.019) and normal H1299
cells (0.35±0.021), P<0.01, and the expression of ABCE1 in
LV-GV358-ABCE1-transfected H1299 cells (0.56±0.016) was
significantly higher than that in LV-GV358-transfected H1299 cells
(0.37±0.016) and normal H1299 cells (0.34±0.003), P<0.01
(Fig. 3).
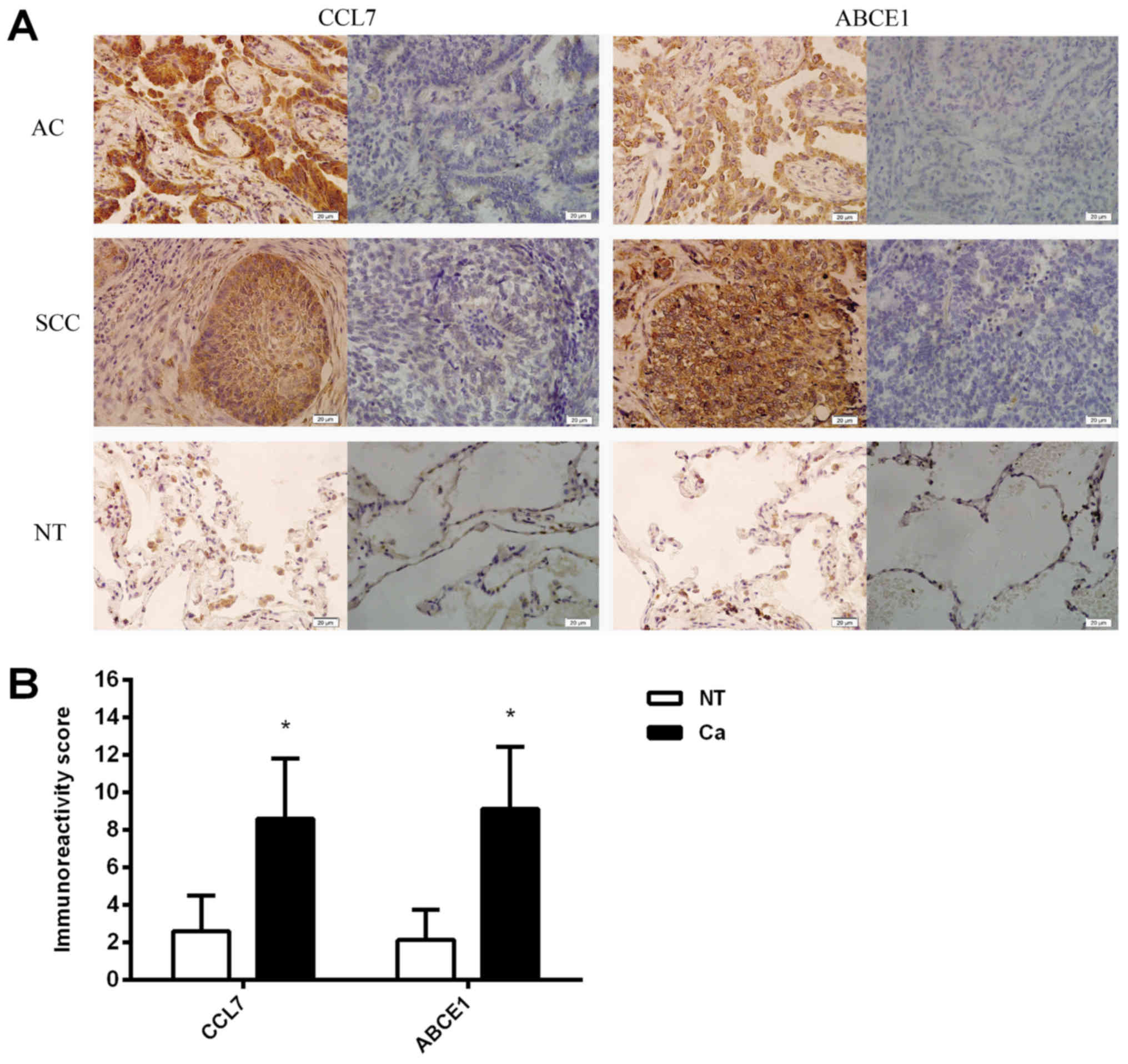
Immunohistochemistry
The expression of the CCL7 and ABCE1 proteins was
mainly localized in the cytoplasm, but CCL7 was also expressed in
some fibroblasts and capillary endothelial cells. The expression
level of CCL7 in lung cancer tissues (8.6±0.58) was higher than
that in adjacent NTs (2.6±0.35), P<0.01, and the expression
level of ABCE1 in lung cancer tissues (9.13±0.6) was higher than
that in adjacent NTs (2.13±0.29), P<0.01 (Fig. 4). The rate of positive CCL7
expression in lung cancer tissues was 70%, and the rate of positive
ABCE1 expression in lung cancer tissues was 87%.
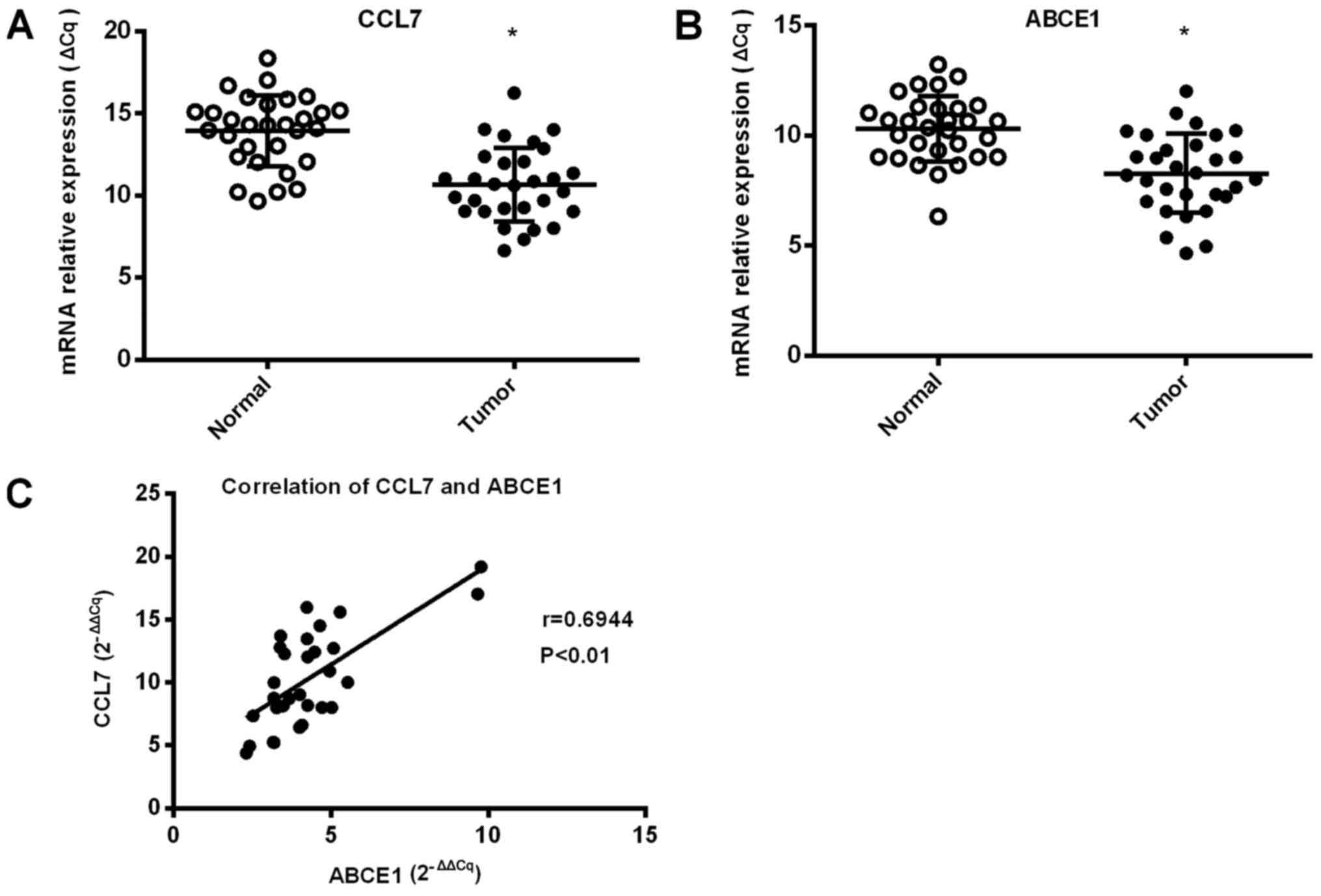
qPCR
The relative expression levels (ΔCq values) of CCL7
mRNA in NSCLC tissues and adjacent tissues were 10.66±0.41 and
13.93±0.39, respectively, P<0.01, and the ΔCq values of ABCE1
mRNA in NSCLC tissues and adjacent tissues were 8.29±0.33 and
10.31±0.27, respectively, P<0.01. The mRNA expression of CCL7
was positively correlated with that of ABCE1 in NSCLC, with a
Pearson correlation coefficient of r=0.6944, P<0.01 (Fig. 5).
Discussion
Due to its malignancy and threat to human health,
lung cancer is a hot topic in the field of cancer research. The
incidence and mortality of lung cancer are currently increasing
annually, the age at onset is decreasing, and the disease is
widespread and occurs worldwide (8).
In recent years, with the development of molecular biology, more
cancer genes and pathogenic mechanisms have been identified;
however, the survival and disease remission rates of lung cancer
are still low. Lymph node and organ metastases are important
factors in determining the degree of malignancy in lung cancer;
thus, further investigations into the mechanism of lung cancer
metastasis are essential.
As a specific inhibitor of RNase and a key enzyme in
the interferon-dependent 2–5A/RNase L pathway, ABCE1 plays an
important physiological role in the regulation of the stability of
cell RNA (9). ABCE1 also plays an
important role in the initiation, extension and termination of
eukaryotic protein translation, as well as in ribosome recycling
(10,11). Ren et al (12) found that ABCE1 was highly expressed
in human lung AC and metastatic lymph nodes and was associated with
clinical stage. Gao et al (13) observed that the expression levels of
ABCE1 was correlated with histopathological type, but not with age,
gender, the grade of tumor differentiation. In AC, the expression
level of ABCE1 protein were higher than that in the squamous
carcinoma. Recently, a series of studies revealed that ABCE1 may be
a new interaction protein for β-actin and that the binding of ABCE1
to β-actin requires the Fe-S cluster domain (14,15).
These results show that ABCE1 is highly expressed in many malignant
tumor cells, indicating that this protein is closely related to the
proliferation, invasion and metastasis of lung cancer.
Chemokine CCL7, which was initially identified as a
cytokine in mononuclear cells, acts on a variety of target cells,
including neutrophils, eosinophils, basophils, natural killer
cells, T lymphocytes and other inflammatory cells, as well as
dendritic cells and mononuclear cells, particularly mononuclear
cells (16). Further research has
shown that CCL7 has functions in many diseases. For example,
Tsuneyama et al (17) found
that CCL7 and mononuclear cell infiltration were present in the
portal area of the liver in more than 80% of patients with primary
biliary cirrhosis, suggesting that elevated CCL7 expression is
associated with biliary cirrhosis. The study by Edman et al
(18) found that CCL2 and CCL7
selectively enhanced the differentiation of Nurr1+ precursors into
dopaminergic (DA) neurons. Gonzalez et al (19) confirmed that CCL7 plays a dual role
in renal tubulointerstitial fibrosis by altering the extracellular
matrix, an effect that is detrimental at the early stage but
beneficial at the later stage.
The role of chemokine CCL7 in tumor growth and
metastasis is very complicated, as studies have shown that CCL7 not
only promotes tumor metastasis but also inhibits the growth of some
malignant tumors (20,21). As CCL7 can play a chemotactic role in
many leukocyte subsets, which identify and kill tumor cells, some
researchers conducted anti-tumor experiments using mast cells
transfected with CCL7. Interestingly, the tumor cells did not die,
but the surrounding tumor tissue was infiltrated with TAMs,
eosinophils, neutrophils, granulocytes and lymphocytes (22,23). In
addition, a large number of dendritic cells accumulated around the
peripheral vasculature of the tumor. Wetzel et al found that
transfection with a virus containing CCL7 inhibited the growth of
cervical cancer cells in humans (24). In contrast, other studies found that
CCL7 promoted the brain metastasis of breast cancer cells and was
conducive to the growth of cancer cells in the brain, while reduced
CCL7 expression inhibited the metastasis of breast cancer cells to
the brain (25). Cho et al
found that high CCL7 expression is associated with the metastasis
of colorectal cancer to the liver (26), and Rajaram et al (27) found that CCL7 plays an important role
in promoting the migration and proliferation of tumor cells during
the process of mutual transformation involving tumor cells and
stromal cells in breast cancer. CCL7 and its receptor, CCR2,
promote the brain metastasis of renal cell carcinoma (28). In addition, CCL7 plays an important
role in the infiltration and invasion of cancer cells in oral SCC
(29).
In the present study, screening using an
RT2 Profiler™ PCR Array chip showed that the change in
the mRNA expression of chemokine CCL7 was significant in NSCLC cell
lines that exhibited upregulation of ABCE1, showing that the
expression of these two genes is strongly correlated during the
processes of NSCLC invasion and metastasis. Western blotting was
performed to verify the high expression of CCL7 protein in NSCLC
cells that overexpressed ABCE1. The expression of CCL7 and ABCE1 in
NSCLC tissues was significantly higher than that in adjacent
tissues, as confirmed by immunohistochemistry and qPCR, and a
positive correlation between the two genes was observed. These
results indicate that CCL7 and ABCE1 are closely associated with
the development and metastasis of NSCLC. ABCE1 may change the tumor
microenvironment through the chemokine CCL7 pathway; this is a new
direction for future research.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 30973502).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW and DT conceived and designed the study. DT and
XY had full access to all the data in the study, and took
responsibility for the integrity of the data and the accuracy of
the data analysis. ZW, YT, QY, ZT and HL extracted the data. ZW, DT
and XY analyzed the data. ZW, DT and XY interpreted the data. ZW,
DT and XY wrote the first draft of the manuscript. All authors
critically revised the manuscript and approved the final
version.
Ethics approval and consent to
participate
Ethical approval was given by the Fourth Affiliated
Hospital of China Medical University Ethics Committee. All
procedures performed in studies involving human participants were
in accordance with the ethical standards of the institutional
and/or national research committee and with the 1964 Declaration of
Helsinki and its later amendments or comparable ethical standards.
All patients provided written informed consent prior to their
inclusion in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bisbal C, Martinand C, Silhol M, Lebleu B
and Salehzada T: Cloning and characterization of a RNAse L
inhibitor. A new component of the interferon-regulated 2–5A
pathway. J Biol Chem. 270:13308–13317. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lingappa JR, Dooher JE, Newman MA, Kiser
PK and Klein KC: Basic residues in the nucleocapsid domain of Gag
are required for interaction of HIV-1 gag with ABCE1 (HP68), a
cellular protein important for HIV-1 capsid assembly. J Biol Chem.
281:3773–3784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang B, Gao Y, Tian D and Zheng M: A
small interfering ABCE1-targeting RNA inhibits the proliferation
and invasiveness of small cell lung cancer. Int J Mol Med.
25:687–693. 2010.PubMed/NCBI
|
|
4
|
Tian Y, Tian X, Han X, Chen Y, Song CY,
Jiang WJ and Tian DL: ABCE1 plays an essential role in lung cancer
progression and metastasis. Tumour Biol. 37:8375–8382. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zlotnik A and Yoshie O: Chemokines: A new
classification system and their role in immunity. Immunity.
12:121–127. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Menten P, Wuyts A and Van Damme J:
Monocyte chemotactic protein-3. Eur Cytokine Netw. 12:554–560.
2001.PubMed/NCBI
|
|
7
|
Mishra P, Banerjee D and Ben-Baruch A:
Chemokines at the crossroads of tumor-fibroblast interactions that
promote malignancy. J Leukoc Biol. 89:31–39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–1083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hassel BA, Zhou A, Sotomayor C, Maran A
and Silverman RH: A dominant negative mutant of 2–5A-dependent
RNase suppresses antiproliferative and antiviral effects of
interferon. EMBO J. 12:3297–3304. 1993.PubMed/NCBI
|
|
10
|
Chen ZQ, Dong J, Ishimura A, Daar I,
Hinnebusch AG and Dean M: The essential vertebrate ABCE1 protein
interacts with eukaryotic initiation factors. J Biol Chem.
281:7452–7457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barthelme D, Dinkelaker S, Albers SV,
Londei P, Ermler U and Tampé R: Ribosome recycling depends on a
mechanistic link between the FeS cluster domain and a
conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci
USA. 108:3228–3233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren Y, Li Y and Tian D: Role of the ABCE1
gene in human lung adenocarcinoma. Oncol Rep. 27:965–970. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao Y, Xu HH, Wang R, Fang H, Xue YD, Liu
JW and Tian DL: Expression of a new tumor metastasis-related gene
ABCE1 in non-small cell lung cancer and its significance. J Chin
Med Univ. 40:911–914. 2011.
|
|
14
|
Han X, Tian Y and Tian D: Tumor metastatic
promoter ABCE1 interacts with the cytoskeleton protein actin and
increases cell motility. Oncol Rep. 35:3623–3629. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Q, Han X and Tian DL: Deficiency of
functional iron-sulfur domains in ABCE1 inhibits the proliferation
and migration of lung adenocarcinomas by regulating the biogenesis
of beta-actin in vitro. Cell Physiol Biochem. 44:554–566. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ali S, Robertson H, Wain JH, Isaacs JD,
Malik G and Kirby JA: A non-glycosaminoglycan-binding variant of CC
chemokine ligand 7 (monocyte chemoattractant protein-3) antagonizes
chemokine-mediated inflammation. J Immunol. 175:1257–1266. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsuneyama K, Harada K, Yasoshima M,
Hiramatsu K, Mackay CR, Mackay IR, Gershwin ME and Nakanuma Y:
Monocyte chemotactic protein-1, −2, and −3 are distinctively
expressed in portal tracts and granulomata in primary biliary
cirrhosis: Implications for pathogenesis. J Pathol. 193:102–109.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edman LC, Mira H and Arenas E: The
beta-chemokines CCL2 and CCL7 are two novel differentiation factors
for midbrain dopaminergic precursors and neurons. Exp Cell Res.
314:2123–2130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonzalez J, Mouttalib S, Delage C, Calise
D, Maoret JJ, Pradère JP, Klein J, Buffin-Meyer B, Van der Veen B,
Charo IF, et al: Dual effect of chemokine CCL7/MCP-3 in the
development of renal tubulointerstitial fibrosis. Biochem Biophys
Res Commun. 438:257–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hwang TL, Lee LY, Wang CC, Liang Y, Huang
SF and Wu CM: CCL7 and CCL21 overexpression in gastric cancer is
associated with lymph node metastasis and poor prognosis. World J
Gastroenterol. 18:1249–1256. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dempe S, Lavie M, Struyf S, Bhat R,
Verbeke H, Paschek S, Berghmans N, Geibig R, Rommelaere J, Van
Damme J and Dinsart C: Antitumoral activity of parvovirus-mediated
IL-2 and MCP-3/CCL7 delivery into human pancreatic cancer:
Implication of leucocyte recruitment. Cancer Immunol Immunother.
61:2113–2123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fioretti F, Fradelizi D, Stoppacciaro A,
Ramponi S, Ruco L, Minty A, Sozzani S, Garlanda C, Vecchi A and
Mantovani A: Reduced tumorigenicity and augmented leukocyte
infiltration after monocyte chemotactic protein-3 (MCP-3) gene
transfer: Perivascular accumulation of dendritic cells in
peritumoral tissue and neutrophil recruitment within the tumor. J
Immunol. 161:342–346. 1998.PubMed/NCBI
|
|
23
|
Luster AD: Antichemokine immunotherapy for
allergic diseases. Curr Opin Allergy Clin Immunol. 1:561–567. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wetzel K, Menten P, Opdënakker G, Van
Damme J, Gröne HJ, Giese N, Vecchi A, Sozzani S, Cornelis JJ,
Rommelaere J and Dinsart C: Transduction of human MCP-3 by a
parvoviral vector induces leukocyte infiltration and reduces growth
of human cervical carcinoma cell xenografts. J Gene Med. 3:326–337.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu K, Fukuda K, Xing F, Zhang Y, Sharma S,
Liu Y, Chan MD, Zhou X, Qasem SA, Pochampally R, et al: Roles of
the cyclooxygenase 2 matrix metalloproteinase 1 pathway in brain
metastasis of breast cancer. J Biol Chem. 290:9842–9854. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cho YB, Lee WY, Choi SJ, Kim J, Hong HK,
Kim SH, Choi YL, Kim HC, Yun SH, Chun HK and Lee KU: CC chemokine
ligand 7 expression in liver metastasis of colorectal cancer. Oncol
Rep. 28:689–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rajaram M, Li J, Egeblad M and Powers RS:
System-wide analysis reveals a complex network of tumor-fibroblast
interactions involved in tumorigenicity. PLoS Genet.
9:e10037892013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wyler L, Napoli CU, Ingold B, Sulser T,
Heikenwälder M, Schraml P and Moch H: Brain metastasis in renal
cancer patients: Metastatic pattern, tumour-associated macrophages
and chemokine/chemoreceptor expression. Br J Cancer. 110:686–694.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bae JY, Kim EK, Yang DH, Zhang X, Park YJ,
Lee DY, Che CM and Kim J: Reciprocal interaction between
carcinoma-associated fibroblasts and squamous carcinoma cells
through interleukin-1α induces cancer progression. Neoplasia.
16:928–938. 2014. View Article : Google Scholar : PubMed/NCBI
|