Introduction
Oral cavity is a naturally open system, and the
immune molecules in the saliva play important roles in immune
defense and protection (1). The
regulation of the flora in the mouth to keep them balanced, and the
prevention of dental caries decide the oral health. The integrity
of the mucosa should be maintained to avoid direct effects of
harmful substances on the mucosa. The substances in the oral cavity
should be cleared and excreted to keep the mouth relatively clean
and sanitary, e.t.c. (2,3). As an important member of mucosal
immune, polymeric immunoglobulin receptor (pIgR) is a member of
immunoglobulin superfamily, and the only transporting receptor of
polymeric immunoglobulin A (pIgA) and polymeric immunoglobulin M
(pIgM). It is synthesized from mucosa epithelial cells and exocrine
gland epithelial cells, and can clear antigen and harmful
substances in the mucosa, which plays an important role in the
first line of defense in the immune system (4,5).
Therefore, the study on the active ingredient (pIgR) in the saliva
is of great significance and prospect. This study analyzed the
mechanism of agonists in regulating transcriptional level of pIgR
in salivary gland epithelial cells, thus revealing the defense
effect of salivary immune on bacteria in the oral cavity.
Materials and methods
General data
A total of 60 patients with oral bacterial infection
and 70 patients suffering from oral diseases without bacterial
infection were selected randomly from patients that were treated in
Renmin Hospital of Wuhan University (Wuhan, China) from April 2015
to April 2017. Inclusion criteria: patients whose sublingual gland
tissues needed to be removed for the treatment of their diseases.
Exclusion criteria: patients with severe diseases in cardiovascular
or digestive systems, mental diseases, infectious diseases, e.t.c.,
that had an impact on the test results. Patients in the group with
bacterial infection of other organs were aged 19–51 years old with
an average of 36.12±10.32 years old, including 38 males and 22
females, among which 22 patients were mainly infected with oral
streptococci, 10 with Streptococcus salivarius and 28 with
Streptococcus sanguis. Patients in the group without
bacterial infection were aged 18–52 years old with an average of
35.78±11.42 years old, including 44 males and 26 females. The data
(such as age and sex) of the two groups of patients had no
statistical differences (p>0.05), and they were comparable. The
study was approved by the Ethics Committee of Renmin Hospital of
Wuhan University and written informed consents were signed by the
patients.
Reagents
DMEM/F12 culture medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA); fetal bovine serum (FBS) (Lanzhou
Bailing, Lanzhou, China); TRIzol (Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA); reverse transcription kit
(Takara Biotechnology Co., Ltd., Dalian, China); SYBR®
Premix Ex Taq™ II kit (Takara Biotechnology Co., Ltd.); rabbit
anti-human pIgR primary antibody and goat anti-rabbit secondary
antibody (Abcam, Cambridge, MA, USA); cellulose acetate membrane
(EMD Millipore, Burlington, MA, USA); developing liquid and fixing
liquid (Beijing Transgen Biotech Co., Ltd., Beijing, China),
internal reference glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), primer of pIgR (Shanghai Sangon, Shanghai, China), and
Cell Counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc.,
Shanghai, China).
Methods
Isolation of salivary gland epithelial
cells (6)
The removed salivary glands were washed with sterile
phosphate-buffered saline (PBS) for 2–3 times. They were cut into
small pieces (1–2 mm3) with a scissor, and incubated in
the culture plates. Dulbecco's modified Eagle's medium/F12
(DMEM/F12) [containing penicillin, streptomycin, epidermal growth
factor (EGF), insulin and hydrocortisone] with 3% FBS was added.
Then they were placed in the incubator containing 5% carbon dioxide
(CO2) for culturing. Three days later, the culture
solution was replaced. Thereafter, the culture solution was
replaced once every 3 days. The cells were passaged when 70–80%
cells grew and fused.
Fluorescent quantitative polymerase
chain reaction (FQ-PCR) analysis on transcriptional level of
pIgR
Ribonucleic acid (RNA) of the cell specimen was
extracted, and the concentration was measured. RNA (1 µg) was taken
to carry out reverse transcription reaction with reverse
transcription enzyme-reagent kits, thus obtaining complementary DNA
(cDNA). The concentration of cDNA was adjusted, and Bio-Rad CFX 96
PCR (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to
measure the relative expression volume of messenger RNA (mRNA) of
different groups according to the illustration of SYBR®
Premix Ex Taq™ II kits. The sequences of the corresponding primers
are shown in Table I, and the
setting of PCR program is demonstrated in Table II.
 | Table I.Primer sequences of GAPDH, alkaline
phosphatase (ALP) and osteocalcin (OCN). |
Table I.
Primer sequences of GAPDH, alkaline
phosphatase (ALP) and osteocalcin (OCN).
| Name of the gene | Sequence of
primers |
|---|
| β-actin | Forward:
5′-GCTTGGAATGAGACTGCTGA-3′ |
|
| Reverse:
5′-CTGGCCATATCCACCAGAGT-3′ |
| pIgR | Forward:
5′-TCAGGTGCTTTGCTAGATG-3′ |
|
| Reverse:
5′-TTTGGGTGTAAGAATGGTAA-3′ |
 | Table II.Program of PCR. |
Table II.
Program of PCR.
| Step | Temperature | Time | Circulation |
|---|
| 1 | 94°C | 15 min | 1 |
| 2 | 94°C | 10 sec | 40 |
|
| 50°C | 30 sec |
|
|
| 72°C | 15 sec |
|
|
| Read |
|
| 3 | 72°C | 10 min | 1 |
Detection of protein expression of
pIgR with western blotting (7,8)
Salivary gland cells were digested with pancreatin
and transferred to the radio-immunoprecipitation assay (RIPA)
protein lysate. The resulted solution was lysed for 30 min on ice,
during which vortex mixing was conducted for 3 times. Then it was
centrifuged for 10 min at 4°C in a rate of 12,000 × g. The
supernatant was filled in a 1.5 ml Eppendorf (EP) tube. Part of the
supernate was taken to detect the concentration of protein by
bicinchoninic acid (BCA) method. Then part of the supernatant was
taken to ensure that the content of protein in each specimen
waiting for test was 100 µg. Reduced loading buffer (5X) was added,
and boiled with boiled water for 10 min. The afore-mentioned sample
solution was added slowly to the prepared gel wells for sample
loading with microsyringes. Sodium dodecyl sulfate polyacrylamide
gel electrophoresis (SDS-PAGE; 15%) was conducted under the voltage
of 80 V. Wet transfer was conducted for 0.5 h under the voltage of
40 V after the end of electrophoresis. The target protein in the
gel was transferred to nitrocellulose (NC) membrane. Then the
membrane was rinsed with eluent for at least 3 times (10 min/time),
and rabbit anti-human pIgR polyclonal antibody (1:600; cat no.
ab96196; Abcam) was added to the membrane. The membrane was blocked
overnight with skimmed milk powder at 4°C. The primary antibody was
incubated for 2 h, and the goat anti-rabbit secondary polyclonal
antibody (1:1,000; cat no. ab6721; Abcam) was incubated for 1 h at
room temperature. Fluorogenic substrate was added, and pressed in
the dark room for the formation of images.
Detection of protein expression in
pIgR with immunofluorescence (9)
The slides filled with cells in the culture plates
were rinsed with PBS for 3 times (3 min/time). The slides were
fixed with 4% paraformaldehyde for 15 min, and rinsed with PBS for
3 times (3 min/time). Triton X-100 (0.5%) (prepared with PBS) was
added, and allowed to stand at room temperature for 20 min. The
glass slides were rinsed with PBS for 3 times (3 min/time); then
absorbent paper was used to absorb PBS. Normal goat serum was
dripped on the glass slides, and they were blocked at room
temperature for 30 min. A sufficient amount of diluted primary
antibody was dripped to each slide. Then the slides were incubated
in the wet box overnight at 4°C. Fluorescent secondary antibody was
added. The slides were rinsed with PBS containing Tween-20 (PBST)
for 3 times (30 min/time), and incubated in the wet box at room
temperature for 1 h. They were rinsed with PBST, and cut into
slices for 3 times (3 min/time). Diamidino-phenyl-indole (DAPI) was
added, and the slides were incubated away from light for 5 min.
Staining was conducted for the specimen, and the slides were rinsed
with PBST to remove excessive DAPI. The slides were sealed with
slide-sealing solution containing anti-fluorescence quenching
agent; then the slides were placed under the fluorescence
microscope to observe and collect the images.
Detection of survival rate of cells
with CCK-8
Cell suspension (100 µl) was prepared in 96-well
plates. All the blank group, observation group and control group
had 3 repeated wells. The culture plates were cultured in the
incubator until the density of the cells reached 50%. Agonist
solution (10 µl) was added to the observation group, and the
equivalent volume of culture medium was added to the control group
for incubation. Twenty-four hours later, 10 µl of CCK-8 solution
was added to each well, and they were placed in the incubator for 2
h. The optical density at 450 nm was detected with microplate
reader. Survival rate of cells = (OD in the observation group - OD
in the blank group) / (OD in the control group - OD in the blank
group).
Statistical methods
Statistical treatment was carried out with
Statistical Product and Service Solutions (SPSS, Inc., Chicago, IL,
USA) 17.0 software, and the test data were expressed as (mean ±
SD). t-test was used for comparison. P<0.05 suggested that the
comparison had a statistical difference.
Results
Isolation of salivary gland epithelial
cells
The tissue blocks of salivary gland epithelial cells
adhered to the wall for growing. They lined up closely and showed a
typical shape of ‘paving stones’. It could be seen that small round
and polygonal cells inlaid each other (Fig. 1).
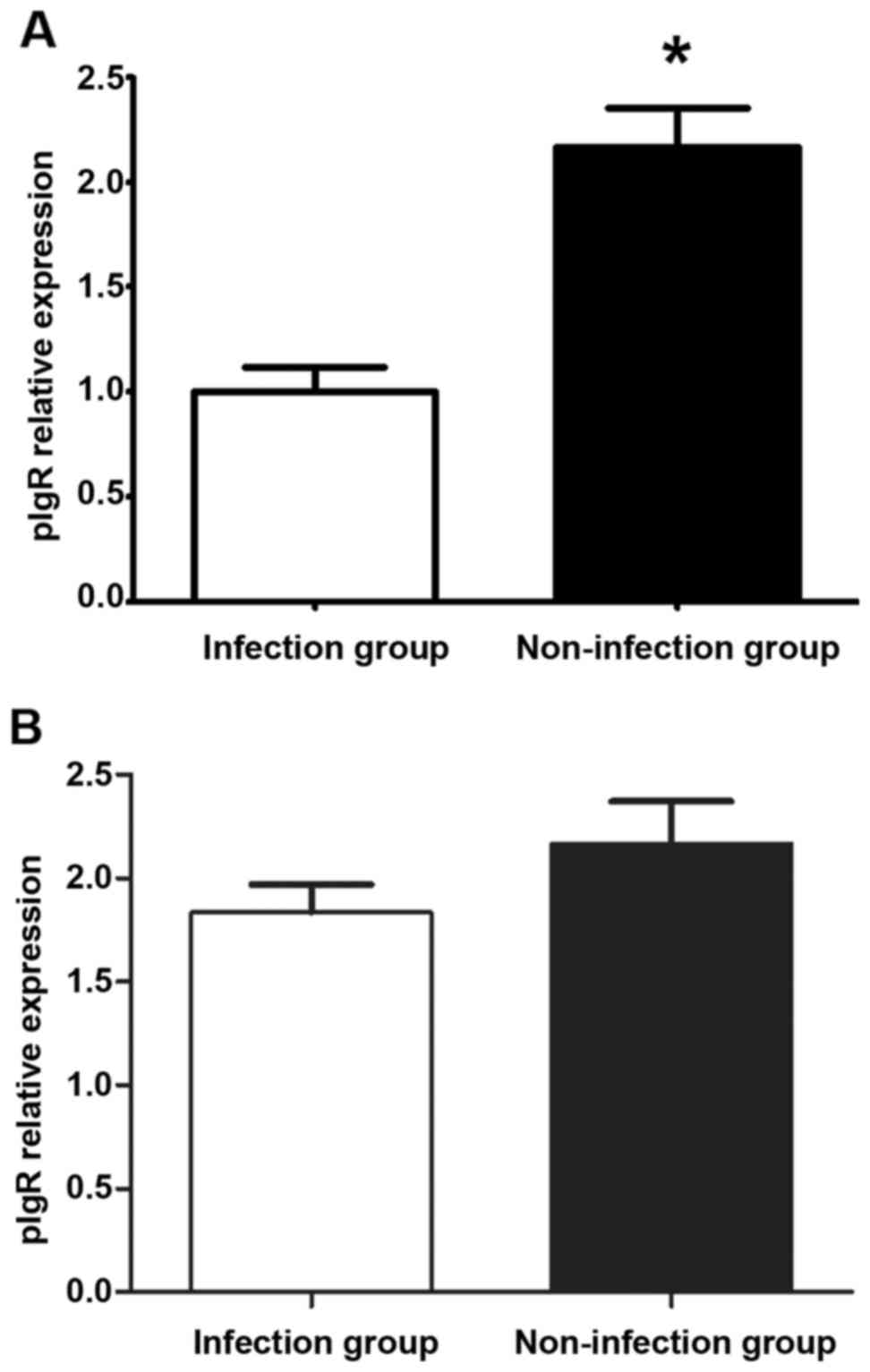
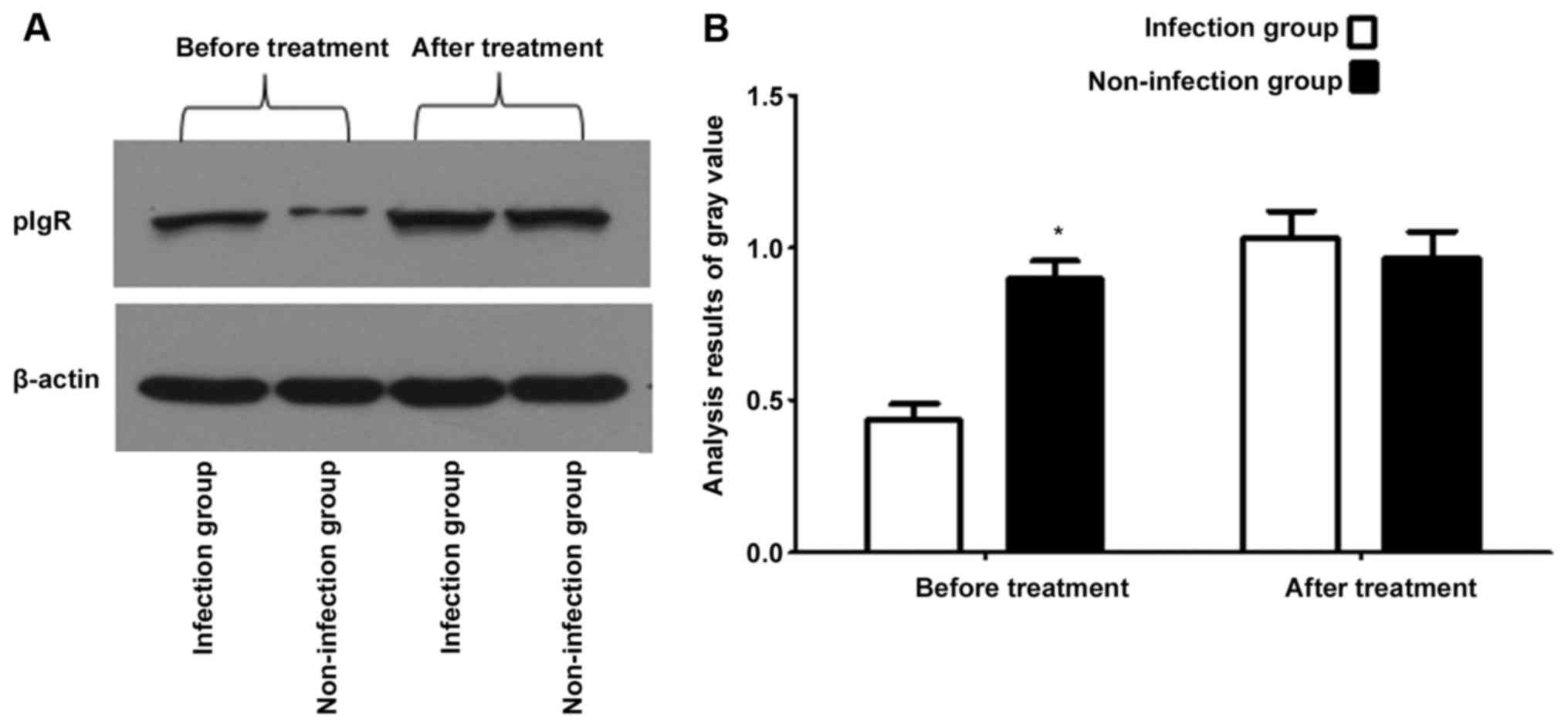
Comparison of transcriptional level of
pIgR in the infection group with that in the non-infection group
before and after treatment
Total RNA of salivary gland epithelial cells with
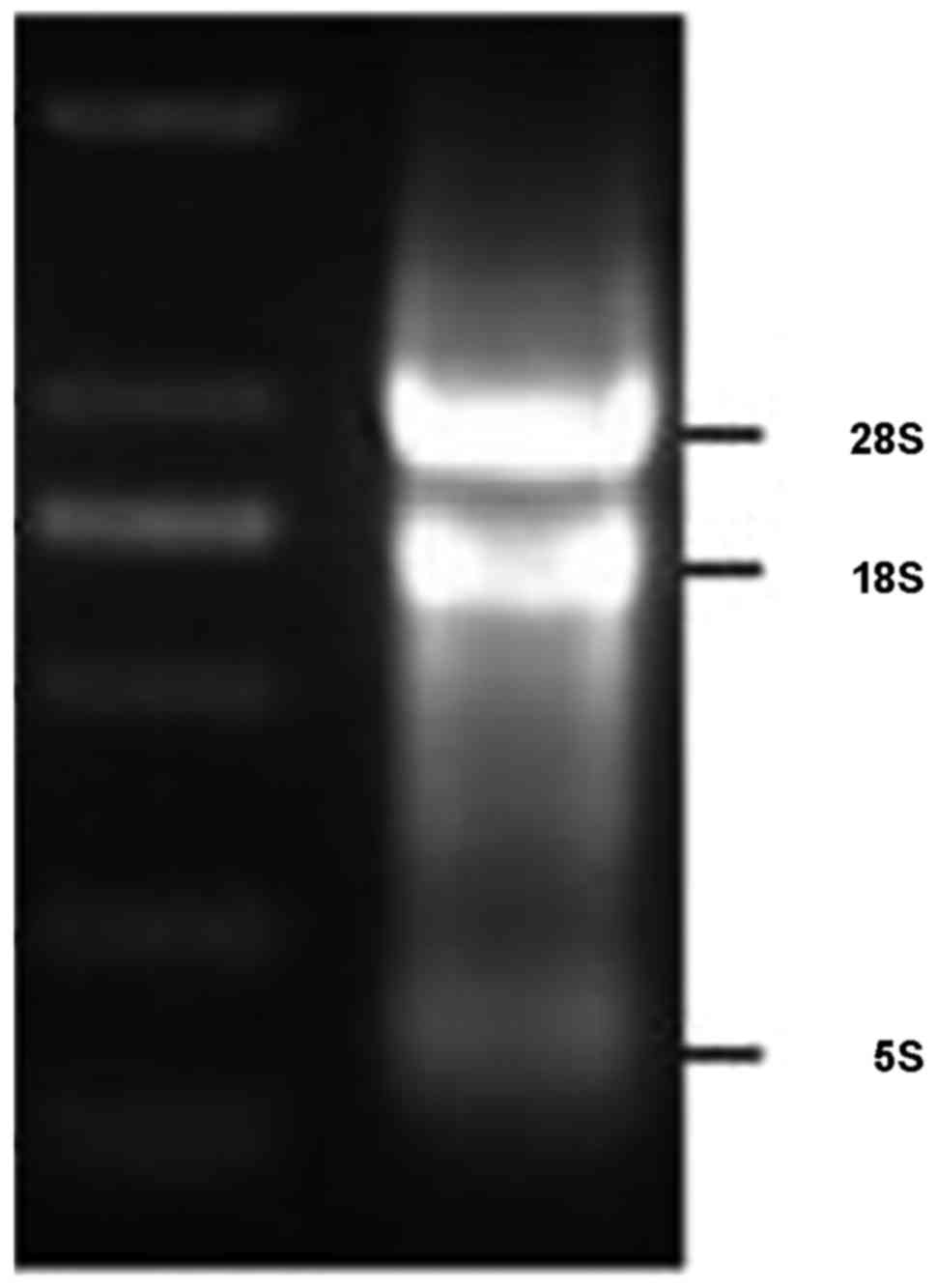
the purity (A260/A280) of 2.02 was extracted (Fig. 2). The results of FQ-PRC and western
blotting methods showed that the transcriptional level of pIgR in
the bacterial infection group was lower than that in the
non-infection group before treatment (p<0.05), while the
transcriptional level of pIgR in the bacterial infection group was
increased after treatment, and the difference with that in the
non-infection group showed no statistical significance (p>0.05)
(Figs. 3 and 4).
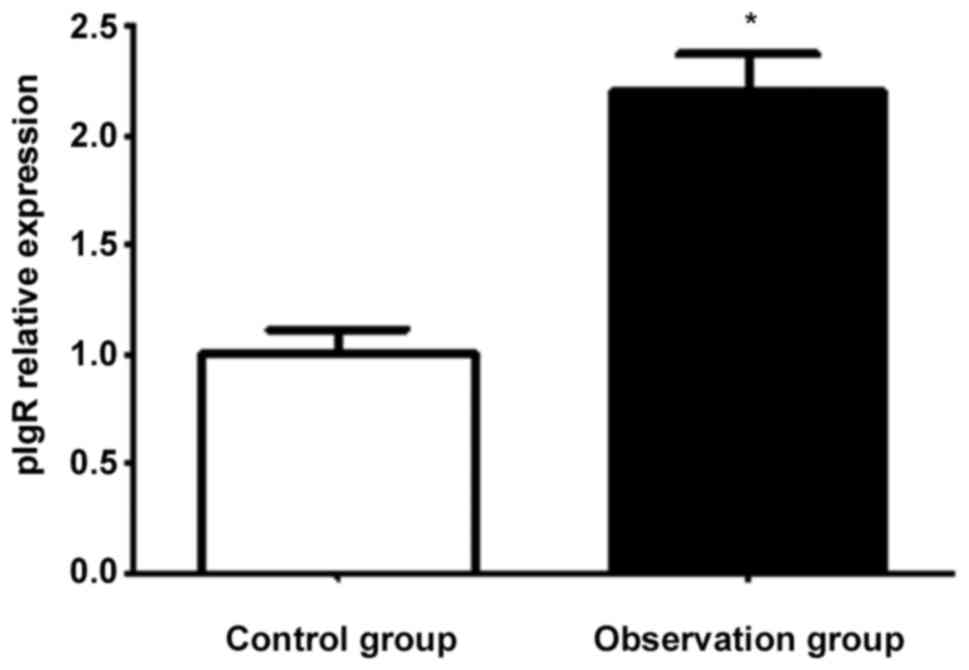
Impacts of agonists on transcription
of pIgR in salivary gland epithelial cells
FQ-PCR results showed that the transcriptional level
of pIgR in the observation group was higher than that in the
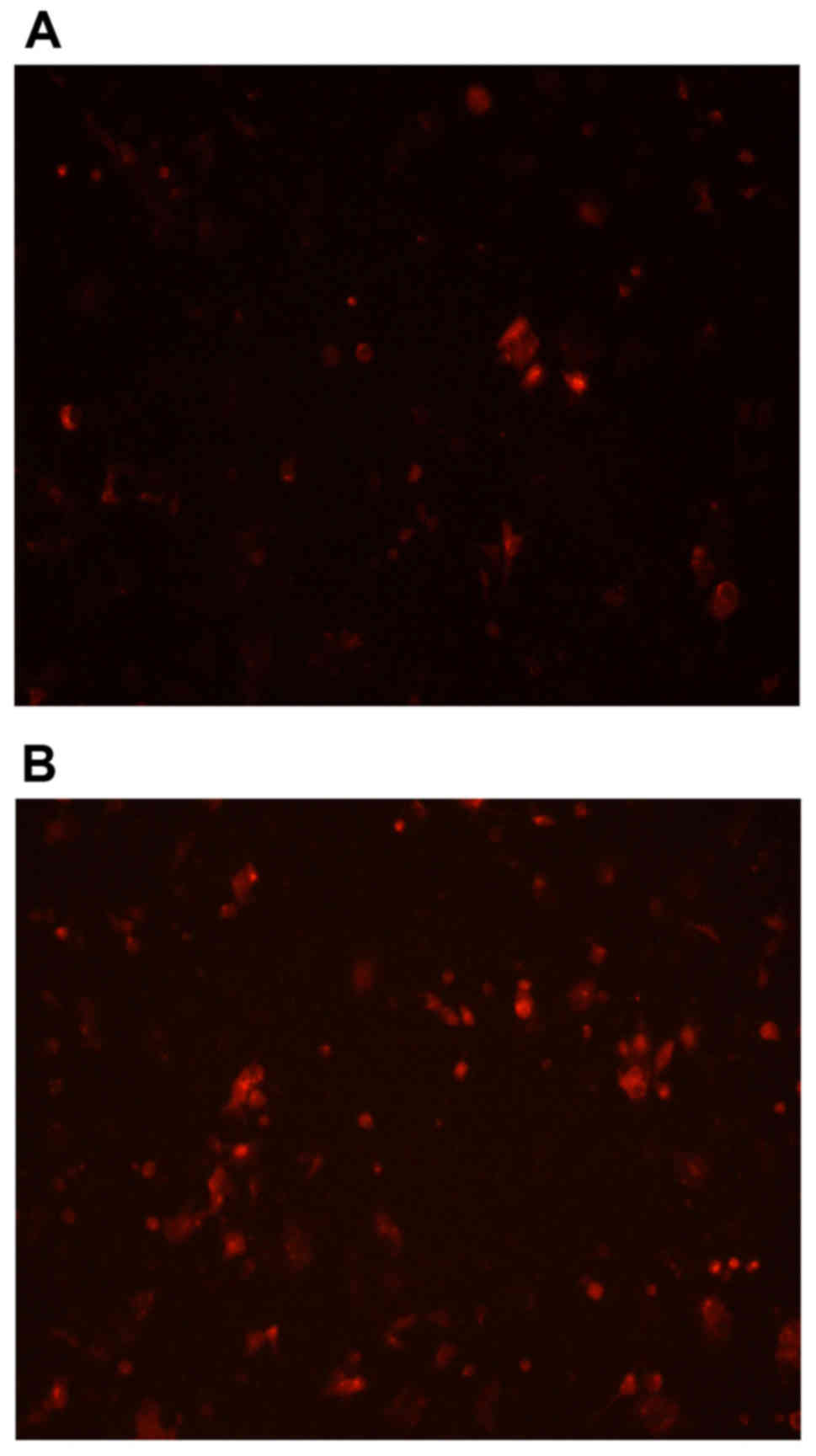
control group (p<0.05) after acting with agonists (Fig. 5). The immunofluorescence results
indicated that the protein expressed by transcription in the
observation group was higher than that in the control group
(Fig. 6).
Detection of toxicity of agonists on
the cells with CCK-8
The average OD of blank wells was 0.212, and that in
the control group and the observation group was 0.4114 and 0.402,
respectively. It was calculated that the survival rate of cells in
the observation group was 95.9%, while that in the control group
was 100%. The difference had no statistical significance
(p>0.05).
Discussion
There is a large number of microbial communities in
the oral cavity, and in a sense, the health of the oral cavity is a
reflection of the ecological balance of the bacteria. Once the
balance is broken, the beneficial bacteria will be reduced, which
will result in all kinds of oral inflammation and diseases
(10,11). Oral streptococcus that often lives in
the oral cavity plays an important role in the oral
micro-ecological system, among which Streptococcus mutans
are the main pathogenic bacteria of caries, Streptococcus
sanguis is the pioneer in dental plaque, and Streptococcus
buccalis and Streptococcus salivarius are the early
colonized bacteria of oral mucosa (12). Colonization and dissemination of oral
microorganisms are regulated and defended by the salivary immune
system (13). Saliva is an important
substance in oral immunity, and it contains lysozyme. There are
lymphocytes and plasmacytes in the mesenchyme of salivary glands.
IgA secreted by plasmacytes binds to the protein secretory
components secreted by glandular cells, thus forming secreted IgA,
which is released into the oral cavity with the saliva, and has an
immunogenic effect (14). In recent
years, the discussion on the effects of Streptococcus
salivarius and Streptococcus mutans from the aspect of
immunity has been a hotspot of research. A study of Carrasco-Yepez
et al (15) found that the
effects of Streptococcus salivarius and Streptococcus
mutans are not specific. They are a manifestation of
broad-spectrum non-specific effects. Kortum et al (16) held the opinion that alkaline
phosphatase, lactate dehydrogenase and other relevant components in
saliva immunity are highly correlated with the incidence of dental
caries.
pIgR mediates the transcription of polymeric
immunoglobulin in the epithelial cells, and participates in the
formation of secretory immunoglobulin A (S-IgA) (17). pIgR has high affinity with dimer IgA
(dIgA) with J chain near the external side of the base of the
lamina propria, and forms a complex of ligand and receptor. Then
the complex is actively transported to the apical end of the cell
from the external side of the base through intracellular transport.
The extracellular segment of pIgR binds covalently to dIgA at the
apical end of the cell, and cleaves between transmembrane region
and extracellular segment, thus secreting S-IgA. When viruses or
foreign antigens penetrate the mucosal surface, and enter the
lamina propria, pIgR also binds to immune complexes formed by IgA
and antigen (Ag) to expel pathogens such as viruses and bacteria
out of the body through transcytosis (18). A study conducted by DeSantis et
al (19) showed that the impacts
of pIgR on the structure, expression distribution, function and
expression regulation are significantly associated with fish
mucosal immunity. Qin et al (20) believed that polyimmunoglobulin
receptors and secretion components are involved in multiple
molecular mechanisms, and they play a very important role in
mucosal immunity. The results of this study also found that the
transcriptional expression of pIgR in salivary gland epithelial
cells had a great relationship with disease-treating streptococcus
in the oral cavity. The transcriptional level of pIgR in the
bacterial infection group was lower than that in the non-infection
group before treatment, while the transcriptional level of pIgR in
the bacterial infection group was increased after treatment, and
the level was equivalent to that in the non-infection group, which
showed that oral bacterial infection can reduce the content of pIgR
in oral cavity, which in turn leads to the decrease of oral
salivary immunity. Agonist is a substance that enhances the
transcriptional level of pIgR. This study also proved that
transcriptional level of pIgR in the observation group was higher
than that in the control group after acting with agonists
(p<0.05), which suggested that promoters can affect the oral
salivary immune function by improving the transcriptional level of
pIgR, thus preventing and treating pathogens such as bacteria and
virus in the mouth. Moreover, the use of agonists has no influence
on normal proliferation of salivary gland epithelial cells, and it
has relatively high safety.
In conclusion, agonists can promote the rise of the
transcriptional level of pIgR in salivary gland epithelial cells,
and the increase in pIgR is closely related to the cure of oral
bacterial infection. Therefore, agonists can improve the oral
immune function by regulating the transcription of pIgR.
Acknowledgements
Not applicable.
Funding
This study was sponsored by the National Natural
Science Fund (81600860).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LH and CS assisted with PCR and western blotting. RP
and ZL were responsible for immunofluorescence. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Renmin Hospital of Wuhan University (Wuhan, China) and
written informed consents were signed by the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rombout JH, Yang G and Kiron V: Adaptive
immune responses at mucosal surfaces of teleost fish. Fish
Shellfish Immunol. 40:634–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
López MC: Chronic alcohol consumption
regulates the expression of poly immunoglobulin receptor (pIgR) and
secretory IgA in the gut. Toxicol Appl Pharmacol. 333:84–91. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ward-Caviness CK, Neas LM, Blach C, Haynes
CS, LaRocque-Abramson K, Grass E, Dowdy ZE, Devlin RB, Diaz-Sanchez
D, Cascio WE, et al: A genome-wide trans-ethnic interaction study
links the PIGR-FCAMR locus to coronary atherosclerosis via
interactions between genetic variants and residential exposure to
traffic. PLoS One. 12:e01738802017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li D, Wang FJ, Yu L, Yao WR, Cui YF and
Yang GB: Expression of pIgR in the tracheal mucosa of
SHIV/SIV-infected rhesus macaques. Zool Res. 38:44–48.
2017.PubMed/NCBI
|
|
5
|
Yue X, Ai J, Xu Y, Chen Y, Huang M, Yang
X, Hu B, Zhang H, He C, Yang X, et al: Polymeric immunoglobulin
receptor promotes tumor growth in hepatocellular carcinoma.
Hepatology. 65:1948–1962. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang S, Liu S, Qu B, Dong Y and Zhang S:
Identification of sea bass pIgR shows its interaction with
vitellogenin inducing antibody-like activities in HEK 293T cells.
Fish Shellfish Immunol. 63:394–404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Armitage CW, O'Meara CP and Beagley KW:
Chlamydial infection enhances expression of the polymeric
immunoglobulin receptor (pIgR) and transcytosis of IgA. Am J Reprod
Immunol. 77:e126112017. View Article : Google Scholar
|
|
8
|
Qi X, Li X and Sun X: Reduced expression
of polymeric immunoglobulin receptor (pIgR) in nasopharyngeal
carcinoma and its correlation with prognosis. Tumour Biol.
37:11099–11104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang F, Liu D, Wang L, Li T, Chang Q, An
L and Yang G: Characterization of IgM-binding protein: A pIgR-like
molecule expressed by intestinal epithelial cells in the common
carp (Cyprinus carpio L.). Vet Immunol Immunopathol. 167:30–35.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamamoto Y, To M, Hayashi T, Shimizu T,
Kamata Y, Saruta J, Takahashi T and Tsukinoki K: Intake of
indigestible carbohydrates influences IgA response and polymeric Ig
receptor expression in the rat submandibular gland. Br J Nutr.
113:1895–1902. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Simpfendorfer KR, Strugnell RA, Brodnicki
TC and Wijburg OL: Increased autoimmune diabetes in pIgR-deficient
NOD mice is due to a ‘Hitchhiking’ interval that refines the
genetic effect of Idd5.4. PLoS One. 10:e01219792015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sankineni S, Cho Y, Hosseinian N and
Kolliputi N: Does pIgR down-regulation in COPD cause reprogramming
of bronchial epithelium? Lung. 193:1–2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niu H, Wang K and Wang Y: Polymeric
immunoglobulin receptor expression is predictive of poor prognosis
in glioma patients. Int J Clin Exp Med. 7:2185–2190.
2014.PubMed/NCBI
|
|
14
|
Iovino F, Molema G and Bijlsma JJ:
Streptococcus pneumoniae Interacts with pIgR expressed by the brain
microvascular endothelium but does not co-localize with PAF
receptor. PLoS One. 9:e979142014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carrasco-Yepez M, Campos-Rodriguez R,
Lopez-Reyes I, Bonilla-Lemus P, Rodriguez-Cortes AY, de Oca
Contis-Montes A, Jarillo-Luna A, Miliar-Garcia A and
Rojas-Hernandez S: Intranasal coadministration of Cholera toxin
with amoeba lysates modulates the secretion of IgA and IgG
antibodies, production of cytokines and expression of pIgR in the
nasal cavity of mice in the model of Naegleria fowleri
meningoencephalitis. Exp Parasitol. 145 Suppl:S84–S92. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kortum AN, Rodriguez-Nunez I, Yang J, Shim
J, Runft D, O'Driscoll ML, Haire RN, Cannon JP, Turner PM, Litman
RT, et al: Differential expression and ligand binding indicate
alternative functions for zebrafish polymeric immunoglobulin
receptor (pIgR) and a family of pIgR-like (PIGRL) proteins.
Immunogenetics. 66:267–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trevisi P, Gandolfi G, Priori D, Messori
S, Colombo M, Mazzoni M, Lallès JP and Bosi P: Age-related
expression of the polymeric immunoglobulin receptor (pIgR) in the
gastric mucosa of young pigs. PLoS One. 8:e814732013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu T, Xie W, Ma Y, Zhou S, Zhang L, Chen
J, Cai M, Sun R, Zhang P, Yu S, et al: Leptin/OB-R pathway promotes
IL-4 secretion from B lymphocytes and induces salivary gland
epithelial cell apoptosis in Sjögren's syndrome. Oncotarget.
8:63417–63429. 2017.PubMed/NCBI
|
|
19
|
DeSantis KA, Stabell AR, Spitzer DC,
O'Keefe KJ, Nelson DA and Larsen M: RARα and RARγ reciprocally
control K5+ progenitor cell expansion in developing
salivary glands. Organogenesis. 13:125–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin R, Steel A and Fazel N: Oral mucosa
biology and salivary biomarkers. Clin Dermatol. 35:477–483. 2017.
View Article : Google Scholar : PubMed/NCBI
|