Introduction
Esophageal carcinoma is a common malignant tumor of
the digestive tract, mainly comprising squamous cell carcinoma and
adenocarcinoma, that is typically associated with poor patient
prognosis (1,2). In China, esophageal carcinoma is has a
high incidence and is the fourth most common of all malignant
tumors (3). In addition, 90% cases
of esophageal carcinoma are squamous cell carcinoma (4). Although clinical diagnostic methods and
treatments for esophageal squamous cell carcinoma (ESCC) have
improved in recent years, the 5-year survival rate of patients with
ESCC remains ~20% (5,6). Distant metastasis is the major cause of
poor patient prognosis (7). It has
been reported that the invasion and metastasis of tumor cells is a
complex process comprising multiple genes, steps and stages, and
the molecular mechanism that regulates this process is not clear
(8). It is therefore important to
understand the molecular mechanisms underlying the invasion and
metastasis of ESCC and to identify novel genes involved.
MicroRNA (miRNA or miR) is a class of non-coding
small RNA molecules (18–22 nucleotides) that regulate gene
expression at the post-transcriptional level (9). miRNAs are associated with the
regulation of tumor cell proliferation, invasion, metastasis,
apoptosis and autophagy, as well as being important molecular
targets for early diagnosis and treatment of cancer (10). It has been reported that the
expression of a number of miRNAs in ESCC tissues and the peripheral
blood is disordered; this may be of great value in clinical
diagnosis and prognosis (11). For
example, miR-506 expression is significantly elevated in the
peripheral blood of patients with ESCC and is positively correlated
with lymphatic metastasis, TNM staging and tumor size (12). Furthermore, the expression of
miR-1297 in the peripheral blood of patients with ESCC is
significantly reduced and can be used as a diagnostic marker
(13). miR-613 is significantly
downregulated in ESCC tissues and the peripheral blood, as well as
being negatively correlated with lymphatic metastasis and TNM
staging; Kaplan-Meier analysis has been used to demonstrate that
low miR-613 expression is associated with poor patient prognosis
(14). It has also been reported
that miR-10b, miR-29c and miR-205 have good specificity and
sensitivity as predictors of diagnosis or prognosis in ESCC
(15). Together, these studies
suggest that miRNAs may be valuable in the early diagnosis,
clinical treatment and prognosis of ESCC.
miR-455 is a tumor-associated miRNA molecule whose
gene is located at fragile site region on chromosome 9q32 (16,17).
miR-455 is abnormally expressed in a number of tumors, including
breast cancer, non-small cell lung cancer and gastric cancer
(18). Mature miR-455 includes two
subtypes; miR-455-3p and miR-455-5p (19). miR-455-3p is associated with tumor
proliferation, invasion and metastasis; for example, miR-455-3p
inhibits proliferation and induces apoptosis in colon cancer HCT116
cells (20). miR-455-3p promotes the
invasion and metastasis of triple negative breast cancer cells by
targeting EI24 (21). By contrast,
there have been few studies into the effect of miR-455-5p in tumors
and it is thought that its biological function is associated with
the tumor tissue type (22). For
example, miR-455-5p inhibits the proliferation and metastasis of
gastric cancer by downregulating the expression of Rab18, acting as
a tumor-suppressor gene (23). In
oral squamous cell carcinoma, the transforming growth factor
(TGF)-β-SMAD signaling pathway upregulates miR-455-5p expression
and promotes tumor cell proliferation by targeting UBE2B (24). However, the expression and mechanism
of action of miR-455-5p in ESCC remain unclear. Rab31 is a
tumor-associated gene that has been reported in recent years
(25). The expression of Rab31 is
abnormal in a variety of tumor types and Rab31 serves a role in the
proliferation, invasion and metastasis of tumor cells (26,27).
Furthermore, the association between Rab31 and miR-455-5p in ESCC
has not previously been reported, nor has the role of Rab31. The
aim of the present study was to investigate the expression and
underlying mechanisms of miR-455-5p and Rab31 in ESCC at the
cellular and molecular levels.
Materials and methods
Patients
A total of 60 patients (36 males and 24 females)
with ESCC who received radical or palliative resection of tumor
tissues (ESCC group) and tumor-adjacent tissues (control group) at
Tianjin Integrated Traditional Chinese and Western Medicine
Hospital (Tianjin, China) between December 2014 and January 2016
were included in the present study. ESCC was diagnosed and
classified by two pathologists according to the 2003 WHO cancer
classification criteria (28). The
resected tissues were stored at −80°C before use. The age range of
the patients was 36–73 years and the mean age was 51±1.8 years.
None of the patients received adjuvant therapy prior to the
surgery. Patients with lymph node metastasis (29 cases) were
included in the N1 subgroup, while those without lymph node
metastasis (31 cases) were included in the N0 subgroup. According
to the classification of differentiation degrees (29), 11 patients had poor differentiation,
28 patients had moderate differentiation and 21 patients had high
differentiation. All procedures were approved by the Ethics
Committee of Tianjin Nankai Hospital (Tianjin, China). Written
informed consent was obtained from all patients or their
families.
Cells
The ESCC cell line Eca109 was purchased from the
Shanghai Institute of Cell Biology (Chinese Academy of Sciences,
Shanghai, China). Cells were cultured in fresh RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) (both Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in an atmosphere
containing 5% CO2. The medium was replaced every two days and cells
were passaged when they reached 80–90% confluence.
Eca109 cells were divided into the following groups:
Negative control (NC), miR-455-5p mimics
(5′-TAUGTGCCTTTGGACTACATCG-3′) and miR-455-5p inhibitor
(5′-GCAGTCCATGGGCATATACAC-3′). When cells reached 70–90%
confluence, 1.25 µl miR-455-5p mimics or miR-455-5p inhibitor (20
pmol/µl; Gangzhou RiboBio Co., Ltd., Guangzhou, China) and 2 µl
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
were added into two individual vials containing 50 µl Opti Mem
medium (Thermo Fisher Scientific, Inc.), respectively, for 5 min.
The contents of the vials were mixed together and left to stand for
15 min. The mixture was subsequently applied to the cells and
incubated at 37°C for 6 h, following which the medium was replaced
with RPMI-1640 supplemented with 10% FBS. Cells were cultured at
37°C in an atmosphere containing 5% CO2 for 48 h prior to use.
For rescue experiments, Eca109 cells (2×105) in the
miR-NC, miR-455-5p mimics and inhibitor groups were seeded into
24-well plates (1×105/well) containing antibiotic-free RPMI-1640
medium supplemented with 10% FBS. When cells reached 60%
confluence, Eca109 cells in the miR-455-5p mimics and inhibitor
groups were transfected with 0.5 µg pcDNA-3.1-Rab31 or
pcDNA-3.1-shR-Rab31 plasmids (Hanbio Biotechnology Co., Ltd.,
Shanghai, China) using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.), respectively, while cells in the miR-NC group
were transfected with 0.5 µg negative control (NC) plasmid. Cells
were cultured at 37°C in an atmosphere containing 5% CO2 for 6 h,
following which the medium was replaced with fresh RPMI-1640
containing 10% FBS and cultured for 48 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
ESCC and tumor-adjacent tissues (100 mg) were ground
into powder using liquid nitrogen and mixed with 1 ml TRIzol
(Thermo Fisher Scientific, Inc.) for lysis. Total RNA was
subsequently extracted using the phenol chloroform method. The
purity of RNA was determined using A260/A280 ultraviolet
spectrophotometry (Nanodrop ND2000; Thermo Fisher Scientific, Inc.,
Pittsburgh, PA, USA). cDNA was obtained by RT using a PrimeScript
RT Reagent Kit (Takara Bio, Inc., Otsu, Japan) and stored at −20°C.
The qPCR reaction system comprised 10 µl qRT-PCR-Mix (miScript SYBR
Green PCR Kit; Qiagen GmbH, Hilden, Germany), 0.5 µl upstream
primer (5′-TATGTGCCTTTGGACTACATCG-3′), 0.5 µl downstream primer
(universal primer provided with kit; cat. no. 218073; Qiagen GmbH),
2 µl cDNA and 7 µl ddH2O. Reference gene was U6 (forward,
5′-GCGCGTCGTGAAGCGTTC-3′; reverse, 5′-GTGCAGGGTCCGAGGT-3′).
Thermocycling condition were as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles of denaturation at 95°C for
1 min and elongation at 60°C for 30 sec. The 2−ΔΔCq
method (30) was used to calculate
relative expression of target mRNA against U6. Each sample was
tested in triplicate.
Cell Counting Kit (CCK)-8 assay
Sample cells were inoculated in 96-well plates at a
density of 2,000 cells/well. At 0, 24, 48 and 72 h, 20 µl CCK-8
reagent (5 g/l; Beyotime Institute of Biotechnology, Haimen, China)
was added at 37°C for 2 h. The absorbance (490 nm) of each well was
measured and cell proliferation curves were plotted. Each group was
tested in 3 replicate wells.
Transwell assay
Transwell chambers (8 µm diameter and 24 wells;
Corning Inc., Corning, NY, USA) were used to evaluate the migration
of Eca109 cells. Transfected cells were collected by trypsin
digestion and resuspended at a density of 2×105 cells/ml in
RPMI-1640 medium. The cell suspension (200 µl) was added into the
upper chamber. A total of 500 µl RPMI-1640 medium supplemented with
10% FBS was added to the lower chamber. Following 24 h of
incubation at 37°C, cells in the upper chamber were removed using a
cotton swab. The chamber was subsequently fixed using 4%
formaldehyde for 10 min at room temperature, subjected to Giemsa's
staining at room temperature for 1 min and washed three times.
Cells that had migrated to the lower chamber were counted under a
microscope (5 fields; magnification, ×200) to evaluate migration
ability.
Matrigel invasion chambers (BD Biosciences, Franklin
Lakes, NJ, USA) were used to determine the invasion ability of
cells. Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was first
diluted with serum-free RPMI-1640 medium at a ratio of 1:2. In the
upper chamber, 100 µl diluted Matrigel was added and incubated at
37°C for 1 h. A total of 500 µl RPMI-1640 medium supplemented with
10% FBS was added to the lower chamber. Following incubation at
37°C for 72 h, cells in the upper chamber were removed using a
cotton swab. The chamber was subsequently fixed using 4%
formaldehyde for 10 min at room temperature and then subjected to
Giemsa's staining at room temperature for 1 min. Following three
washes, 3 cells which had invaded the lower chamber were counted
under a microscope (5 fields; magnification, ×200).
Western blotting
Cells in each group were trypsinized and collected.
Cold radioimmunoprecipitation assay lysis buffer (600 µl; Beyotime
Institute of Biotechnology) was mixed with the samples for 30 min
on ice, following which samples were centrifuged at 10,000 × g and
4°C for 10 min. The protein concentration was determined using a
bicinchoninic acid protein concentration determination kit
(RTP7102; Realtimes Biotechnology Co., Ltd., Beijing, China).
Protein samples (5 µl) were separated by 10% SDS-PAGE and
electro-transferred to polyvinylidene difluoride membranes on ice.
Membranes were blocked with 5% skimmed milk at room temperature for
1 h. Subsequently, membranes were incubated with rabbit anti-human
Rab31 polyclonal primary antibody (1:1,000; BS70807) and rabbit
anti-human GAPDH primary antibody (1:5,000; AP0063; both Bioworld
Technology, Inc., St. Louis Park, MN, USA) at 4°C overnight.
Membranes were washed with PBS with Tween 20 (PBST) and incubated
with goat anti-mouse horseradish peroxidase-conjugated secondary
antibodies (1:2,000; BS12478; Bioworld Technology, Inc.) at room
temperature for 1 h. Membranes were washed five times with PBST for
5 min and developed using an enhanced chemiluminescence detection
kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for imaging.
Image lab v3.0 software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) was used to acquire and analyze imaging data. The relative
expression of Rab31 protein was calculated with GAPDH as a
control.
Dual luciferase reporter assay
To investigate the regulatory mechanism of Rab31,
TargetScan (http://www.targetscan.org) was used
to predict miRNA molecules that might regulate Rab31. It was
demonstrated that miR-455-5p was a potential regulator of Rab31
expression. Wild-type (WT) and mutant seeding regions of miR-455-5p
in the 3′-UTR of Rab31 were chemically synthesized, following which
SpeI and HindIII restriction sites were added and
cloned into pMIR-REPORT luciferase reporter plasmids (0.5 µg;
Thermo Fisher Scientific, Inc.) with WT or mutant 3′-UTR DNA
sequences. Plasmids were transfected together with miR-NC or
miR-455-5p mimics into Eca109 cells as described earlier. Following
incubation for 24 h, cells were processed using dual luciferase
reporter assay kit according to the manufacturer's protocol
(Promega Corp., Madison, WI, USA). The fluorescence intensity was
measured using a GloMax 20/20 luminometer (Promega Corp.).
Fluorescence values of each group of were measured using Renilla
fluorescence activity as the internal reference.
Statistical analysis
All results were analyzed using SPSS 17.0
statistical software (SPSS, Inc., Chicago, IL, USA). Student's t
test was used for comparisons between two groups. Multiple group
comparisons were analyzed using one-way analysis of variance with
least significant difference and Student-Newman-Keul's post hoc
tests. For heterogeneous data, Tamhane's T2 or Dunnett's T3 post
hoc tests were used. P<0.05 was considered to indicate a
statistically significant difference.
Results
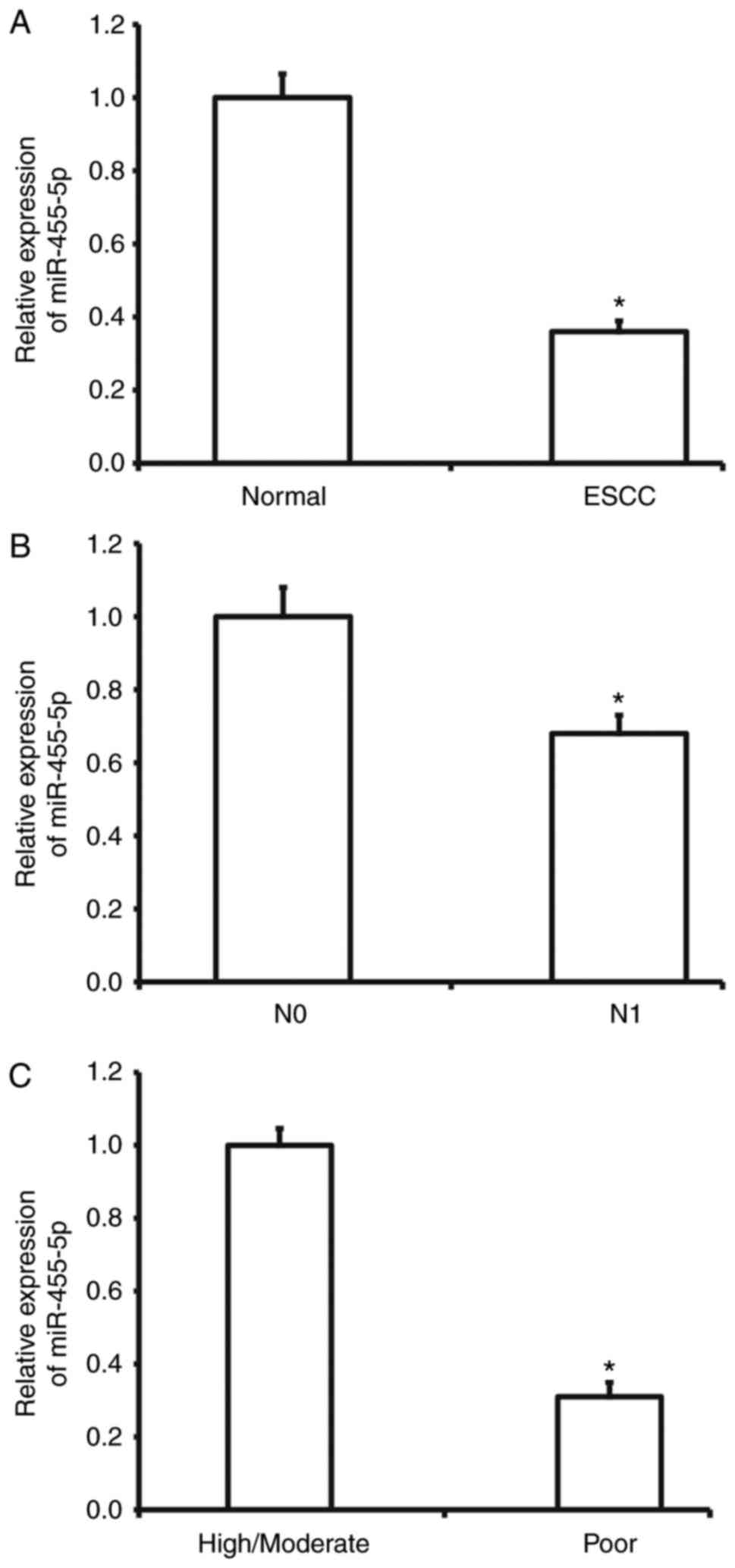
miR-455-5p expression is decreased in
ESCC tissues and is negatively correlated with metastasis and
development of the disease
To measure miR-455-5p expression in ESCC tissues,
RT-qPCR was performed. The results demonstrated that miR-455-5p
expression was significantly decreased in ESCC tissues compared
with tumor-adjacent tissues (P<0.01; Fig. 1A). In addition, miR-455-5p expression
in ESCC tissues from patients with lymphatic metastasis was
significantly reduced compared with tissues from patients without
lymphatic metastasis (P<0.01; Fig.
1B). miR-455-5p was also significantly downregulated in ESCC
tissues from patients with poor differentiation compared with
tissues from patients with high or moderate differentiation
(P<0.05; Fig. 1C). These results
suggest that miR-455-5p expression is decreased in ESCC tissues and
is negatively correlated with metastasis and disease
development.
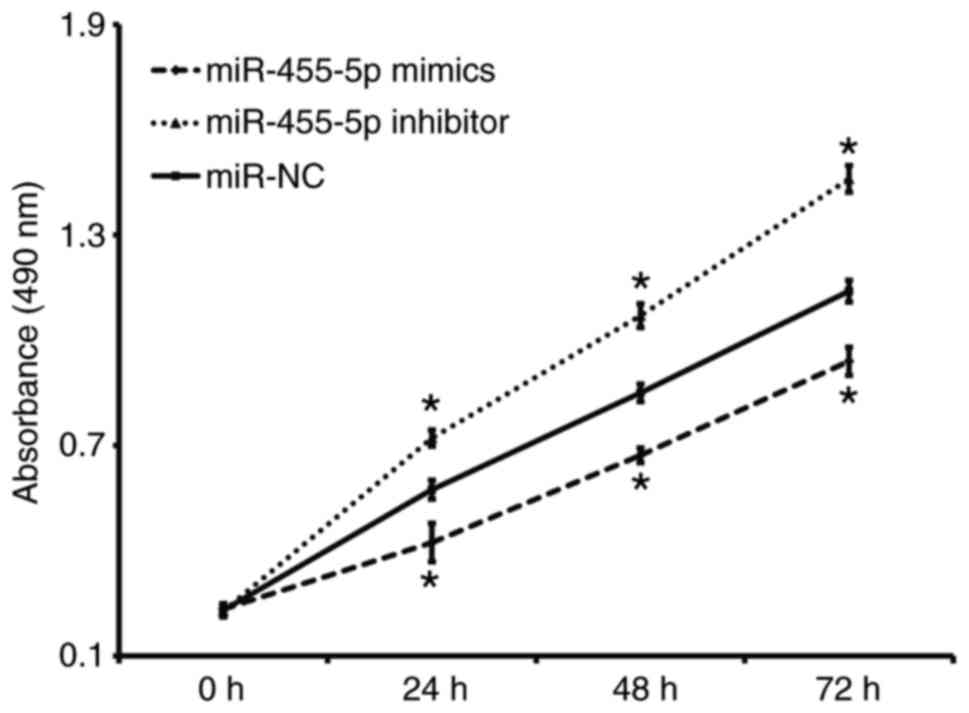
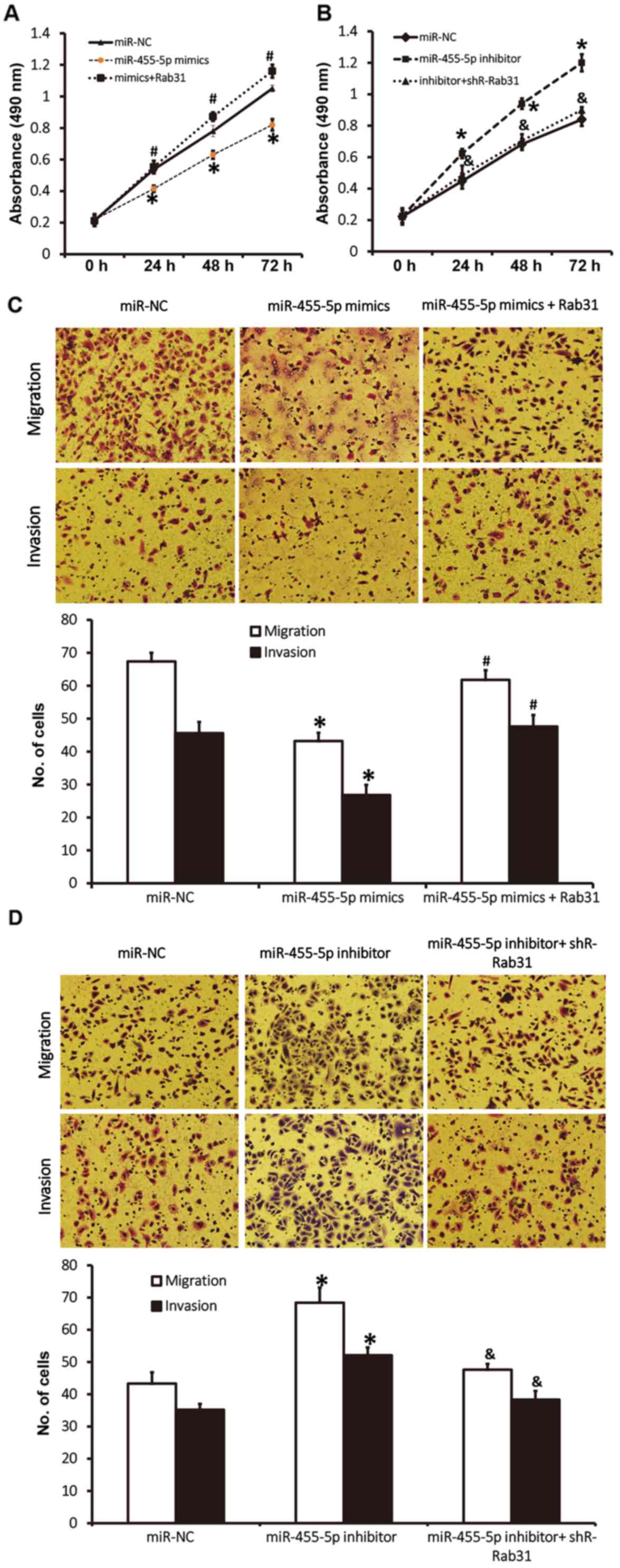
miR-455-5p inhibits the proliferation
of Eca109 cells
A CCK-8 assay was performed to assess proliferation.
The results revealed that the absorbance of Eca109 cells t
transfected with miR-455-5p mimics was significantly reduced
compared with the miR-NC group at all time points (P<0.05),
while the absorbance of Eca109 cells in the miR-455-5p inhibitor
group was significantly increased compared with the miR-NC group
(P<0.05; Fig. 2). These results
indicate that miR-455-5p expression inhibits ESCC Eca109 cell
proliferation.
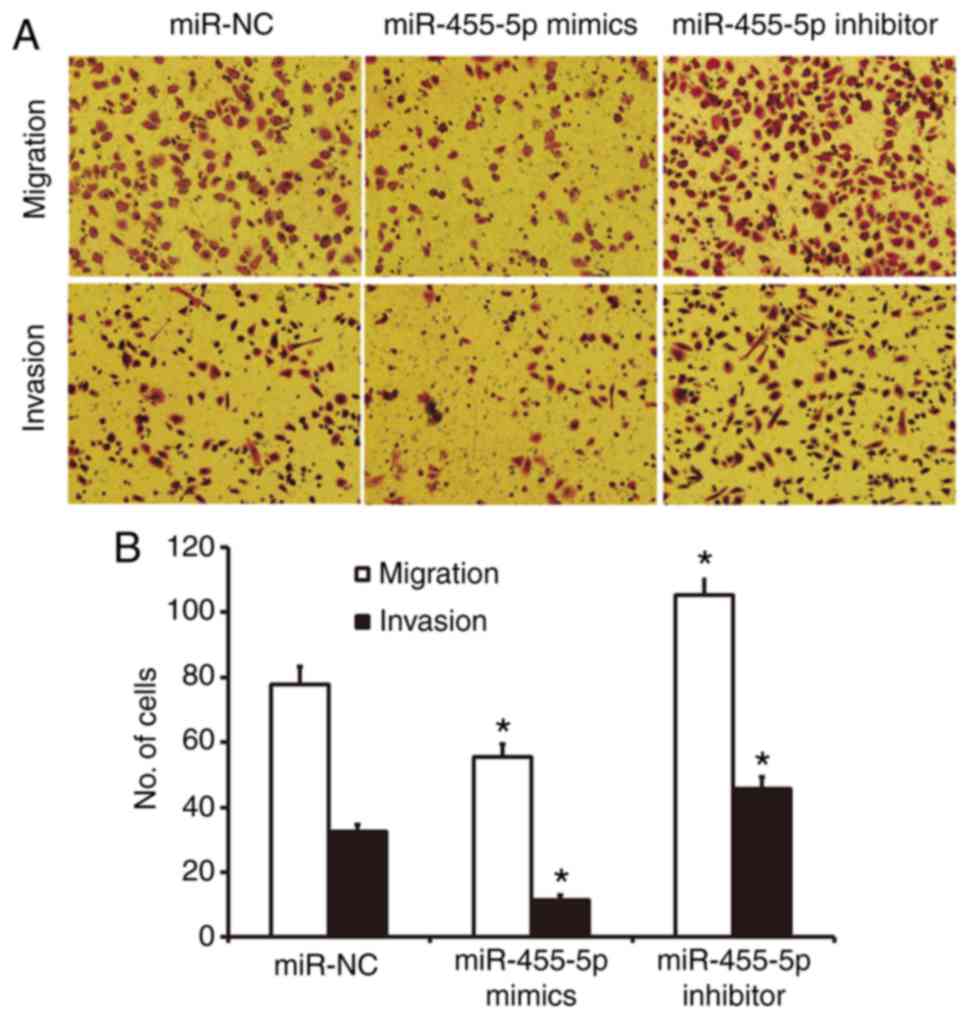
miR-455-5p expression inhibits the
migration and invasion abilities of Eca109 cells
A Transwell assay was used to investigate the
migration and invasion abilities of Eca109 cells. The results
revealed fewer cells in crossed the chamber membrane the miR-455-5p
mimics group compared with the miR-NC group (P<0.05; Fig. 3). In contrast, the number of cells in
the miR-455-5p inhibitor group that crossed the chamber membrane
was significantly increased compared with the miR-NC group
(P<0.05; Fig. 3). In the invasion
assay, the number of cells that crossed the chamber membrane was
lower in the miR-455-5p mimics group compared with the miR-NC group
(P<0.05; Fig. 3). In the
miR-455-5p inhibitor group, there was a significant increase in the
number of invading cells compared with the miR-NC group (P<0.05;
Fig. 3). These results suggest that
miR-455-5p expression inhibits the migration and invasion abilities
of Eca109 cells.
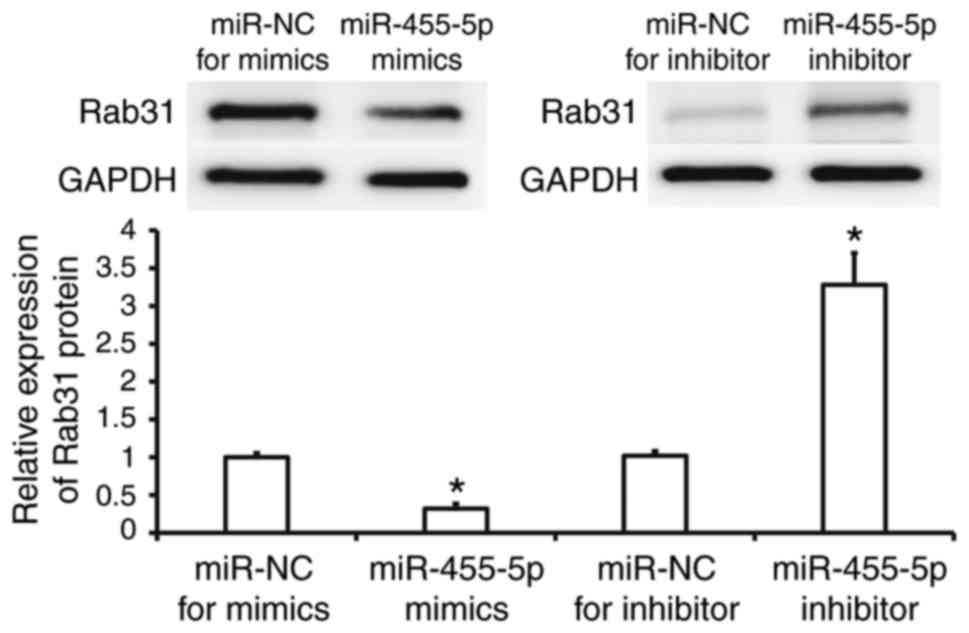
Rab31 expression is regulated by
miR-455-5p in Eca109 cells
Western blotting was performed to assess Rab31
protein expression in Eca109 cells. The results revealed that Rab31
protein expression was significantly reduced in the miR-455-5p
mimics group compared with the miR-NC group (P<0.05), while it
was significantly increased in the miR-455-5p inhibitor group
(P<0.05; Fig. 4). These results
suggest that miR-455-5p regulates the expression of Rab31 protein
in Eca109 cells.
Rab31 promotes the proliferation,
migration and invasion of ESCC Eca109 cells
To assess how the regulation of Rab31 by miR-455-5p
affects the biological functions of Eca109 cells, Rab31 protein was
silenced or overexpressed. The results of a CCK-8 assay revealed
that miR-455-5p-induced Rab31 downregulation significantly
decreased the proliferation of Eca109 cells (P<0.05; Fig. 5A). However, co-transfection with
pcDNA3.1-Rab31 (which reversed Rab31 downregulation) increased the
proliferation of Eca109 cells to a level higher than the miR-NC
group (P<0.05; Fig. 5A). Rab31
overexpression induced by the inhibition of miR-455-5p
significantly promoted the proliferation of Eca109 cells
(P<0.05), whereas Rab31 downregulation induced by
co-transfection with pcDNA3.1-shR-Rab31 reduced the proliferation
of Eca109 cells to a level similar to that observed in the miR-NC
group (Fig. 5B). Transwell assay
data revealed that cotransfection with pcDNA3.1-Rab31 increased the
migration and invasion abilities of Eca109 cells to levels similar
to those observed in the miR-NC group (P<0.05; Fig. 5C), whereas Rab31 downregulation
induced by miR-455-5p overexpression reduced migration and invasion
compared with the miR-NC group (P<0.05; Fig. 5D). Rab31 overexpression induced by
miR-455-5p inhibition resulted in increased migration and invasion
compared with the miR-NC group (P<0.05), whereas co-transfection
with pcDNA3.1-shR-Rab31 (which downregulated Rab31 expression)
reduced the migration and invasion abilities of Eca109 cells to
levels similar to the miR-NC group (Fig.
5D). The results indicate that Rab31 promotes the
proliferation, migration and invasion of ESCC Eca109 cells.
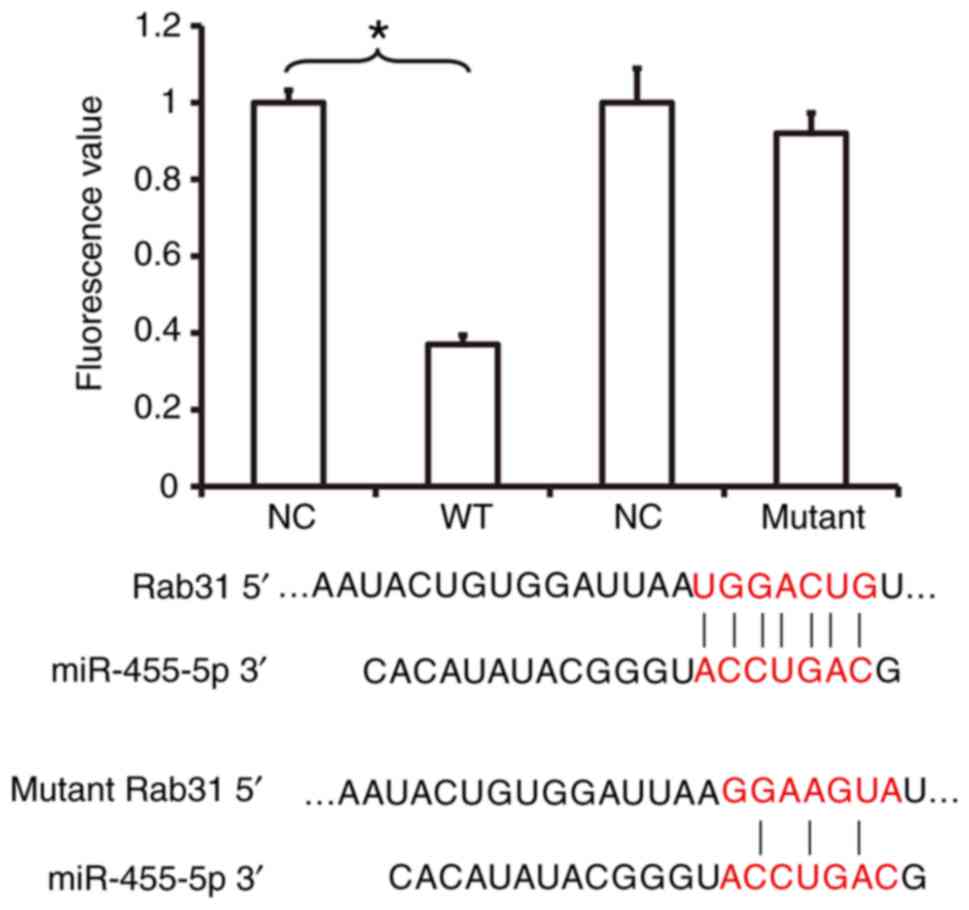
miR-455-5p binds with the 3′-UTR
seeding region of Rab31 mRNA to regulate its expression
To investigate the interaction between miR-455-5p
and the 3′-UTR of Rab31 mRNA, a dual luciferase reporter assay was
performed. The fluorescence value of cells co-transfected with
miR-455-5p mimics and pMIR-REPORT-WT luciferase reporter plasmids
was significantly lower compared with the NC group (P<0.05;
Fig. 6). In contrast, no significant
difference was observed in the fluorescence of cells co-transfected
with miR-455-5p mimics and pMIR-REPORT-mutant luciferase reporter
plasmids compared with the NC group (Fig. 6). These results indicate that
miR-455-5p is able to bind with the 3′-UTR seeding region of Rab31
mRNA to regulate its expression.
Discussion
The initiation and development of ESCC is a complex
process that involves multiple genes, phases and steps. It is
associated with gene mutation, abnormal expression, epigenetic
inheritance and tumor stem cells (31,32). As
an important post-transcriptional regulator, miRNAs serve important
roles in the occurrence and development of tumors (33). It has been reported that the miRNA
expression profile is significantly altered in ESCC tissues and
that various miRNA molecules act as oncogenes or tumor-suppressor
genes in ESCC (34,35). In the present study it was revealed
that miR-455-5p expression is downregulated in ESCC tissues and is
negatively correlated with lymphatic metastasis and
differentiation. In vitro experiments were used to
demonstrate that miR-455-5p inhibits the proliferation, migration
and invasion of ESCC cells. The results of bioinformatics and
molecular biology studies indicate that miR-455-5p inhibits the
proliferation, migration and invasion of ESCC cells by directly
regulating the expression of Rab31. These results suggest that
miR-455-5p downregulation promotes the occurrence and development
of ESCC.
The miR-455 family includes two members, miR-455-5p
and miR-455-3p, both of which participate in the proliferation,
migration and invasion of several types of tumor cells (36). For example, miR-455-5p expression is
downregulated in gastric cancer tissues and cells and miR-455-5p
overexpression inhibits the proliferation and migration of gastric
cancer cells by targeting Rab18, acting as a tumor-suppressor
(23). In addition, miR-455-5p
inhibits the proliferation and promotes the apoptosis of HCT116
colon cancer cells (20). The
biological functions of miR-455 vary with tumor type and it may
serve as an oncogene in some tumors. Li et al (21) reported that miR-455-3p promotes the
proliferation and metastasis of triple-negative breast cancer by
targeting EI24 gene expression. In oral squamous cell carcinoma,
the TGF-β/Smad signaling pathway upregulates miR-455-5p and
promotes the proliferation, migration and invasion of tumor cells
(24). The results of the present
study demonstrate that miR-455-5p is significantly downregulated in
ESCC tissues and is negative correlated with lymphatic metastasis
and differentiation, suggesting that miR-455-5p may be an oncogene
for ESCC. Transfection with miR-455-5p mimics inhibits the
proliferation of Eca109 cells, whereas transfection with miR-455-5p
inhibitor promotes proliferation. Transwell results revealed that
the number of migrated and invading cells in the miR-455-5p mimics
group was significantly lower compared with the NC, whereas
invasion and migration were increased in the miR-455-5p inhibitor
group. This suggests that miR-455-5p inhibits the migration and
invasion of ESCC cells.
miRNA molecules exert their biological functions
mainly by inhibiting the expression of target genes. Bioinformatics
used in the present study suggest that Rab31 is a potential target
gene of miR-455-5p. Western blotting data revealed that Rab31 was
downregulated in the miR-455-5p mimics group, whereas it is
upregulated in the miR-455-5p inhibitor group, suggesting that
miR-455-5p may exert its effect by regulating the expression of
Rab31. Rab31 belongs to the Rab protein family and serves important
regulatory roles in vesicle transport in cells (26). It has been reported that Rab31 is
important for the apoptosis, proliferation and metastasis of tumors
(26). For example, Rab31 inhibits
apoptosis and promotes proliferation in hepatoma carcinoma cells by
regulating the phosphoinositide 3-kinase/protein kinase B/B-cell
lymphoma 2 (Bcl-2)/Bcl-2-associated X protein signaling pathway
(27). Grismayer et al
(37) reported that Rab31 is
associated with the regulation of chemoresistance in breast cancer
cells and affects the prognosis of patients (37). The results of the present study
demonstrate that Rab31 overexpression in cells transfected with
miR-455-5p mimics inhibits ESCC cell proliferation, while Rab31
downregulation in cells transfected with miR-455-5p inhibitor
increases it. Transwell results revealed that Rab31 upregulation in
the miR-455-5p mimics group facilitated the regulatory effect of
miR-455-5p on ESCC cells, while Rab31 downregulation in the
miR-455-5p inhibitor reduced the regulatory effect of miR-455-5p.
Dual luciferase reporter assay results demonstrated that miR-455-5p
directly binds with the 3′-UTR of Rab31 mRNA, suggesting that Rab31
is a direct target gene of miR-455-5p. However, the present study
is not without limitations. The function of miR-455-5p in ESCC was
not investigated in vivo and the mechanism by which Rab31
regulates ESCC development remains to be elucidated. In conclusion,
the present study demonstrates that miR-455-5p inhibits the
proliferation, migration and invasion of ESCC cells by directly
regulating the expression of Rab31. miR-455-5p downregulation is an
important factor that contributes to the occurrence and development
of ESCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and YT collaborated to design the study. YL and
PL were responsible for experiments. YL and YT analyzed the data.
All authors collaborated to interpret results and develop the
manuscript. The final version of the manuscript has been read and
approved by all authors, and each author believes that the
manuscript represents honest work.
Ethics approval and consent to
participate
All procedures performed in the current study were
approved by the Ethics Committee of Tianjin Integrated Traditional
Chinese and Western Medicine Hospital (Tianjin, China). Written
informed consent was obtained from all patients or their
families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lin D, Ma L, Ye T, Pan Y, Shao L, Song Z,
Jiang S, Chen H and Xiang J: Results of neoadjuvant therapy
followed by esophagectomy for patients with locally advanced
thoracic esophageal squamous cell carcinoma. J Thorac Dis.
9:318–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mao Y, Han Y and Shi W: The expression of
aplysia ras homolog I (ARHI) and its inhibitory effect on cell
biological behavior in esophageal squamous cell carcinoma. Onco
Targets Ther. 10:1217–1226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cong L and Hu L: The value of the
combination of hemoglobin, albumin, lymphocyte and platelet in
predicting platinum-based chemoradiotherapy response in male
patients with esophageal squamous cell carcinoma. Int
Immunopharmacol. 46:75–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Z, Li G, Tang L and Li Y: SIX1
overexpression predicts poor prognosis and induces radioresistance
through AKT signaling in esophageal squamous cell carcinoma. Onco
Targets Ther. 10:1071–1079. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qiu B, Li J, Wang B, Wang Z, Liang Y, Cai
P, Chen Z, Liu M, Fu J, Yang H and Liu H: Adjuvant therapy for a
microscopically incomplete resection margin after an esophagectomy
for esophageal squamous cell carcinoma. J Cancer. 8:249–257. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morimoto H, Yano T, Yoda Y, Oono Y,
Ikematsu H, Hayashi R, Ohtsu A and Kaneko K: Clinical impact of
surveillance for head and neck cancer in patients with esophageal
squamous cell carcinoma. World J Gastroenterol. 23:1051–1058. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao Y, Ma K, Yang S, Zhang X, Wang F,
Zhang X, Liu H and Fan Q: MicroRNA-125a-5p enhances the sensitivity
of esophageal squamous cell carcinoma cells to cisplatin by
suppressing the activation of the STAT3 signaling pathway. Int J
Oncol. 53:644–658. 2018.PubMed/NCBI
|
|
8
|
Sugase T, Takahashi T, Serada S, Nakatsuka
R, Fujimoto M, Ohkawara T, Hara H, Nishigaki T, Tanaka K, Miyazaki
Y, et al: Suppressor of cytokine signaling-1 gene therapy induces
potent antitumor effect in patient-derived esophageal squamous cell
carcinoma xenograft mice. Int J Cancer. 140:2608–2621. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue L, Nan J, Dong L, Zhang C, Li H, Na R,
He H and Wang Y: Upregulated miR-483-5p expression as a prognostic
biomarker for esophageal squamous cell carcinoma. Cancer Biomark.
19:193–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie R, Wu SN, Gao CC, Yang XZ, Wang HG,
Zhang JL, Yan W and Ma TH: MicroRNA-30d inhibits the migration and
invasion of human esophageal squamous cell carcinoma cells via the
post-transcriptional regulation of enhancer of zeste homolog 2.
Oncol Rep. 37:1682–1690. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi Q, Wang Y, Mu Y, Wang X and Fan Q:
miR-433-3p inhibits proliferation and invasion of esophageal
squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem.
46:2187–2196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li SP, Su HX, Zhao D and Guan QL: Plasma
miRNA-506 as a prognostic biomarker for esophageal squamous cell
carcinoma. Med Sci Monit. 22:2195–2201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang C, Li Q, Liu F, Chen X, Nesa EU, Guan
S, Liu B, Han L, Tan B, Wang D, et al: Serum miR-1297: A promising
diagnostic biomarker in esophageal squamous cell carcinoma.
Biomarkers. 21:517–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guan S, Wang C, Chen X, Liu B, Tan B, Liu
F, Wang D, Han L, Wang L, Huang X, et al: miR-613: A novel
diagnostic and prognostic biomarker for patients with esophageal
squamous cell carcinoma. Tumour Biol. 37:4383–4391. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu H, Yao Y, Meng F, Qian X, Jiang X, Li
X, Gao Z and Gao L: Predictive value of serum miR-10b, miR-29c, and
miR-205 as promising biomarkers in esophageal squamous cell
carcinoma screening. Medicine (Baltimore). 94:e15582015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen W, Chen L, Zhang Z, Meng F, Huang G,
Sheng P, Zhang Z and Liao W: MicroRNA-455-3p modulates cartilage
development and degeneration through modification of histone H3
acetylation. Biochim Biophys Acta. 1863:2881–2891. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yao S, Tang B, Li G, Fan R and Cao F:
miR-455 inhibits neuronal cell death by targeting TRAF3 in cerebral
ischemic stroke. Neuropsychiatr Dis Treat. 12:3083–3092. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong N, Khwaja SS, Baker CM, Gay HA,
Thorstad WL, Daly MD, Lewis JS Jr and Wang X: Prognostic microRNA
signatures derived from the cancer genome atlas for head and neck
squamous cell carcinomas. Cancer Med. 5:1619–1628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR,
Ding Y, Liang JQ, Li TT and Zhao J: miR-497-5p, miR-195-5p and
miR-455-3p function as tumor suppressors by targeting hTERT in
melanoma A375 cells. Cancer Manag Res. 10:989–1003. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng J, Lin Z, Zhang L and Chen H:
MicroRNA-455-3p inhibits tumor cell proliferation and induces
apoptosis in HCT116 human colon cancer cells. Med Sci Monit.
22:4431–4437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Meng Q, Pan A, Wu X, Cui J, Wang Y
and Li L: MicroRNA-455-3p promotes invasion and migration in triple
negative breast cancer by targeting tumor suppressor EI24.
Oncotarget. 8:19455–19466. 2017.PubMed/NCBI
|
|
22
|
Chai J, Guo D, Ma W, Han D, Dong W, Guo H
and Zhang Y: A feedback loop consisting of
RUNX2/LncRNA-PVT1/miR-455 is involved in the progression of
colorectal cancer. Am J Cancer Res. 8:538–550. 2018.PubMed/NCBI
|
|
23
|
Liu J, Zhang J, Li Y, Wang L, Sui B and
Dai D: miR-455-5p acts as a novel tumor suppressor in gastric
cancer by down-regulating RAB18. Gene. 592:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Up-regulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kotzsch M, Kirchner T, Soelch S, Schäfer
S, Friedrich K, Baretton G, Magdolen V and Luther T: Inverse
association of rab31 and mucin-1 (CA15-3) antigen levels in
estrogen receptor-positive (ER+) breast cancer tissues with
clinicopathological parameters and patients' prognosis. Am J Cancer
Res. 7:1959–1970. 2017.PubMed/NCBI
|
|
26
|
Pan Y, Zhang Y, Chen L, Liu Y, Feng Y and
Yan J: The critical role of Rab31 in cell proliferation and
apoptosis in cancer progression. Mol Neurobiol. 53:4431–4437. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sui Y, Zheng X and Zhao D: Rab31 promoted
hepatocellular carcinoma (HCC) progression via inhibition of cell
apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumour Biol.
36:8661–8670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang J, Wu N, Zheng QF, Yan S, Lv C, Li SL
and Yang Y: Evaluation of the 7th edition of the TNM classification
in patients with resected esophageal squamous cell carcinoma. World
J Gastroentero. 20:18397–18403. 2014. View Article : Google Scholar
|
|
29
|
Ke W, Zeng L, Hu Y, Chen S, Tian M and Hu
Q: Detection of early-stage extrahepatic cholangiocarcinoma in
patients with biliary strictures by soluble B7-H4 in the bile. Am J
Cancer Res. 8:699–707. 2018.PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang W, Hong R, Xue L, Ou Y, Liu X, Zhao
Z, Xiao W, Dong D, Dong L and Fu M: Piccolo mediates EGFR signaling
and acts as a prognostic biomarker in esophageal squamous cell
carcinoma. Oncogene. 36:3890–3902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mei LL, Qiu YT, Zhang B and Shi ZZ:
MicroRNAs in esophageal squamous cell carcinoma: Potential
biomarkers and therapeutic targets. Cancer Biomark. 19:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng X, Chen X, Lu P, Ma W, Yue D, Song L
and Fan Q: miR-202 promotes cell apoptosis in esophageal squamous
cell carcinoma by targeting HSF2. Oncol Res. 25:215–223. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi J, Jin L, Chen J, Feng B, He Z, Chen L
and Song H: miR-375 suppresses invasion and metastasis by direct
targeting of SHOX2 in esophageal squamous cell carcinoma. Acta
Biochim Biophys Sin (Shanghai). 49:159–169. 2017.PubMed/NCBI
|
|
35
|
Ren Y, Chen Y, Liang X, Lu Y, Pan W and
Yang M: miRNA-638 promotes autophagy and malignant phenotypes of
cancer cells via directly suppressing DACT3. Cancer Lett.
390:126–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Wang Y, Sun D, Bu J, Ren F, Liu B,
Zhang S, Xu Z, Pang S and Xu S: miR-455-5p promotes cell growth and
invasion by targeting SOCO3 in non-small cell lung cancer.
Oncotarget. 8:114956–114965. 2017.PubMed/NCBI
|
|
37
|
Grismayer B, Sölch S, Seubert B, Kirchner
T, Schäfer S, Baretton G, Schmitt M, Luther T, Krüger A, Kotzsch M
and Magdolen V: Rab31 expression levels modulate tumor-relevant
characteristics of breast cancer cells. Mol Cancer. 11:622012.
View Article : Google Scholar : PubMed/NCBI
|