Introduction
Diarrhea is a common clinical symptom and is the
third leading cause of infectious disease-associated mortalities
worldwide, mainly affecting children (1). Approximately 1.87 million children
succumb to diarrhea annually worldwide (2), and children with an age of <5 years
in developing countries are reported to experience an average of
three diarrheal episodes per year (3). The most common cause of diarrhea is an
infection of the gastrointestinal tract due to viruses (4), bacteria (5), or parasites (6). Enterotoxigenic Escherichia coli
(ETEC) is considered to be the most common cause of bacterial
diarrhea, also known as traveler's diarrhea (7). The major serotypes of ETEC are
O6, O27, O148, O159,
O149 and O101 (8,9).
O101 is commonly associated with diarrhea and poses a
significant threat worldwide (10).
Thus, the present study attempted to establish a rat model of
Escherichia (E.) coli O101-induced
diarrhea.
Gut microbiota, the complex microbial communities
harbored in the digestive tracts of animals, serve a major role in
the host's metabolism (11,12), nutrient absorption or production
(13), and immune system (14), greatly contributing to the overall
health status of the host (15,16).
Accumulating evidence indicated that gut microbiota is closely
associated with the incurrence and development of a variety of
diseases, including obesity (17,18),
diabetes (19) and diarrhea
(20). Previous studies have
suggested that diarrhea can cause changes in intestinal microbiota
(21,22), and altered intestinal microbial
composition and function may result in an increased risk of
bacterial diarrhea (20). However,
the specific changes in intestinal microbiota in individuals
suffering from E. coli-associated diarrhea are poorly
understood. Therefore, it would be of great interest to identify
the systemic microbiome alterations and the specific microorganisms
involved in E. coli-associated diarrhea.
The next-generation sequencing technique facilitates
the investigation of the taxonomic composition of intestinal
microbiota and provides a new perspective for studying E.
coli-induced diarrhea (23). In
the present study, the effects of E. coli O101 on
the intestinal tissues of rats were investigated, the fecal
microbiota from diarrhea rats was compared with that in the control
rats, and the characteristic bacterial diversity and compositions
were identified. In addition, the current study provided an insight
into the pathology of E. coli O101 and provided
evidence for identifying bacteria for the diagnosis and treatment
of diarrhea.
Materials and methods
Animals and ethics statement
Specific pathogen-free male Sprague-Dawley rats
(n=22; 190–210 g; 6 weeks old) were obtained from Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). The
animals were maintained at a temperature of 22°C and a 12-h
light/dark cycle environment for at least one week prior to use in
the experiments. The animals were fed the same batch of standard
laboratory diet to minimize the variation of environmental factors.
The present study was approved by the Institutional Animal Care and
Use Committee of the Academy of Military Medical Sciences (Beijing,
China). All animal care and experimental procedures were conducted
according to the Chinese Laboratory Animals' Welfare and Ethics
guidelines (24).
E. coli O101 treatment
The E. coli O101 strain was
purchased from the China Institute of Veterinary Drug Control
(Beijing, China) and used to establish a diarrhea model in the
rats. A total of 22 rats were divided into two groups, including
the diarrhea (n=11) and control (n=11) groups. The diarrhea group
received intraperitoneal (ip) injections with E. coli
O101 (1×1011 colony-forming units/kg) for
three consecutive days. The normal group received ip injection with
an equivalent volume of sterile physiological saline for three
consecutive days. The animals were sacrifices after 3 days.
Fecal sample collection
The fecal score was recorded two times per day using
a four-grade system, with a score of 0 indicating firm, dry and
normal consistency of feces, 1 indicating pasty feces, 2 indicating
thick and fluid feces, and 3 indicating watery feces (25). Diarrhea was defined as the daily sum
score of ≥2. The diarrhea incidence and diarrhea index (diarrhea
index=rate of loose stools per day * the degree of diarrhea) were
used to assess the establishment of an E. coli
O101-induced diarrhea rat model (26). Fresh fecal samples of rats were
collected individually on the third day, immediately frozen in
liquid nitrogen and stored at −80°C for further analysis.
Histopathological analysis
Partial intestinal tissues were dissected, fixed in
4% paraformaldehyde for 24 h, dehydrated and embedded in paraffin.
Next, 4-µm sections were cut and stained with hematoxylin and
eosin. Histopathological changes were observed and scored under an
Olympus microscope (Olympus Corporation, Tokyo, Japan). The
criteria for grading the intestinal histopathological changes were
as follows (27): Score 0, no
evident pathological changes; score 1–3, mild injury characterized
by slight edema and a decrease in the number of mucous epithelial
cells; score 4–5, moderate injury characterized by inflammatory
cell infiltration, congestion, cell apoptosis and necrosis; score
6–10, severe injury characterized by massive inflammatory cell
infiltration, severe hemorrhage and congestion, evident edema,
coagulation necrosis and focal necrosis.
DNA extraction and pyrosequencing
Microbial DNA was extracted from fecal samples using
the E.Z.N.A.® Soil DNA kit (Omega Bio-tek, Inc.,
Norcross, GA, USA) according to the manufacturer's protocol. The
V3-V4 region of the bacterial 16S ribosomal RNA (rRNA) was
amplified by polymerase chain reaction (PCR), conducted under the
following conditions: 95°C for 3 min, followed by 25 cycles at 95°C
for 30 sec, 55°C for 30 sec and 72°C for 45 sec, a final extension
at 72°C for 10 min, and maintained at 10°C. The primers used in PCR
were as follows: 338F, 5′-ACTCCTACGGGAGGCAGCAG-3′, and 806R,
5′-GGACTACHVGGGTWTCTAAT-3′. The PCR reactions were performed in
triplicate in a mixture with a total volume of 20 µl, which
contained 0.4 µl FastPfu Polymerase, 4 µl 5X FastPfu buffer (both
Beijing Transgen Biotech Co., Ltd., Beijing, China), 2 µl of 2.5 mM
dNTPs (Vazyme, Piscataway, NJ, USA), 0.8 µl of each primer (5 µM)
and 10 ng template DNA. PCR products were purified on agarose gels
using an AxyPrep DNA Gel Extraction kit (Axygen; Corning
Incorporated, Corning, NY, USA) according to the manufacturer's
protocol. Equimolar concentrations of purified PCR products were
pooled and paired-end sequenced (2×300 bp) on an Illumina MiSeq
platform (Illumina, Inc., San Diego, CA, USA) according to the
manufacturer's recommendations.
Sequencing analysis
Raw Fastq files were demultiplexed and
quality-filtered using the QIIME bioinformatics pipeline (version
1.17; http://qiime.org/). The criteria used were as
follows: i) 300 bp reads were truncated at any site receiving an
average quality score of <20 over a 50 bp sliding window,
discarding any truncated reads that were <50 bp; ii) exact
barcode matching, mismatch of 2 nucleotides in primer matching and
reads containing ambiguous characters were removed; and iii) only
sequences that overlapped by >10 bp were assembled according to
their overlap sequence. Reads were discarded if they could not be
assembled.
Bioinformatics analysis
The operational taxonomic unit (OTU) is a classified
operation unit that is set up for a specific unit (such as strain,
species, genus and grouping) for the convenience of analysis in
phylogenetic or population genetics research (28). In the analysis of microbial
diversity, OTU is divided for all sequences based on different
similarity levels (29). Thus, an
OTU is defined by a similarity of >97% (taxonomic rank) between
sequences, and each OTU represents a species (30). In the present study, the OTUs were
clustered with a similarity cutoff value of 97% using UPARSE
software (version 7.1; http://drive5.com/uparse/) with a novel ‘greedy’
algorithm that performs chimera filtering and OTU clustering
simultaneously, as previously described (31). The taxonomy of each 16S rRNA gene
sequence was analyzed using the Ribosomal Database Program (RDP)
classifier (http://rdp.cme.msu.edu/) against the
SILVA (SSU 115) 16S rRNA database (https://www.arb-silva.de/), with a confidence
threshold of 70%. Subsequently, the sequences were classified
taxonomically to different levels (including phylum, class, order,
family, genus and species) using the RDP classifier. The
α-diversity indices, including Ace, Chao1, Shannon and Simpson,
were then calculated using QIIME from rarefied samples for richness
and diversity indices of the bacterial community. OTUs that reached
97% similarity were used for diversity (Shannon and Simpson),
richness (Chao1 and Ace), Good's coverage, rarefaction curve and
Shannon-Wiener curve analyses (32).
The community structure was compared using principal component
analysis (PCA) based on the weighted UniFrac distance. A
hierarchical cluster, rank-abundance and heatmap were constructed
and analyzed using R software package (http://www.r-project.org) (33).
Statistical analysis
Data are represented as the mean ± standard
deviation. Statistical analyses were performed with Student's
t-test using GraphPad Prism software (version 6.0; GraphPad
Software Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
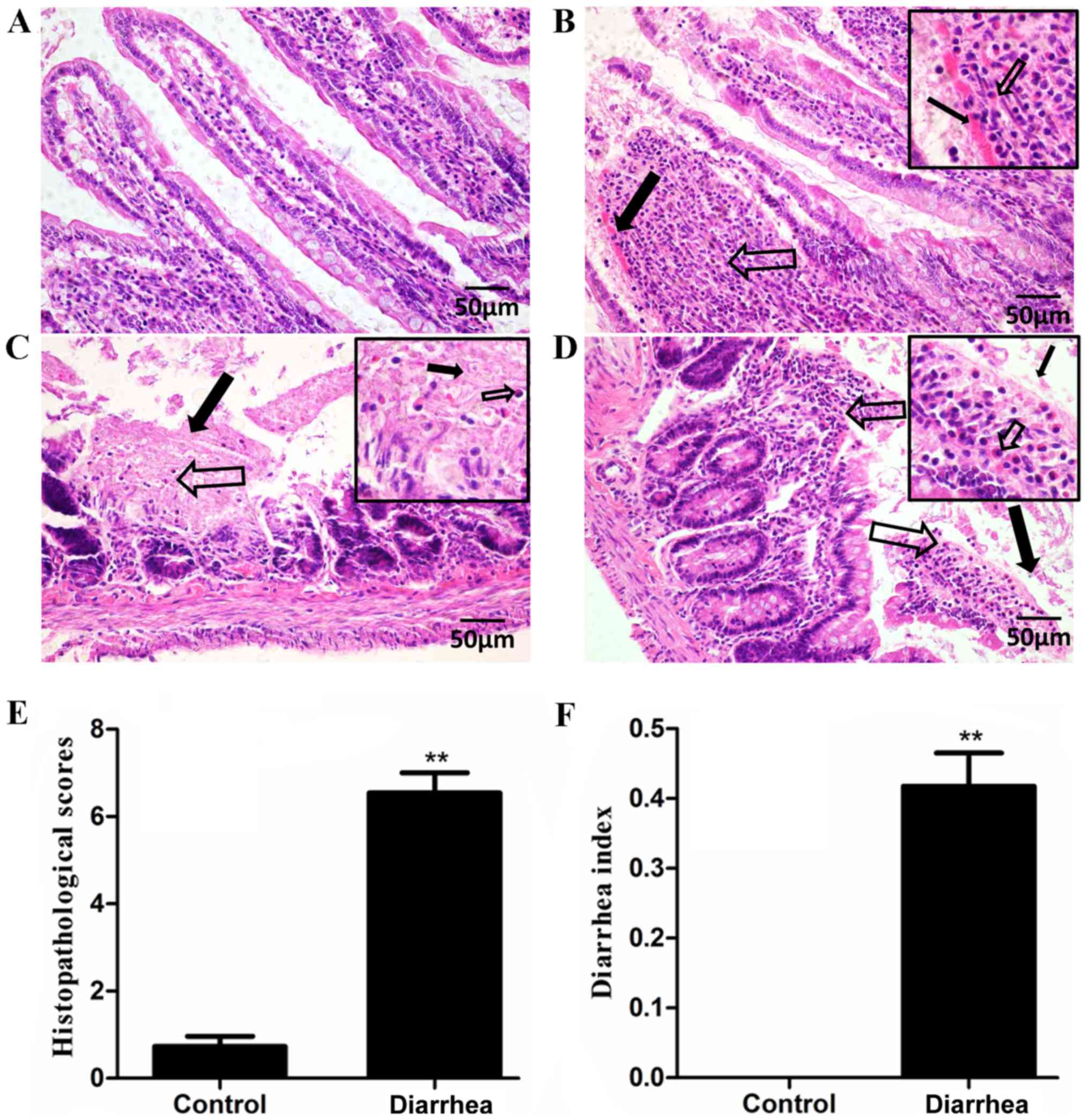
Following an intraperitoneal injection of E.
coli O101, the histopathological changes in the
intestinal tissues of the rats that received bacteria injection
were investigated. The intestinal tissues of healthy rats exhibited
a normal mucosal structure and intact epithelium (Fig. 1A). By contrast, the jejunal tissues
obtained from rats injected with E. coli O101
demonstrated congestion and inflammatory cellular infiltrates in
the mucosal lamina propria (Fig.
1B). The mucosal lamina propria did not exhibit a normal tissue
structure, but coagulation necrosis and focal necrosis with
inflammatory cell infiltration were observed (Fig. 1C). In addition, a disrupted surface
epithelium and inflammatory cellular infiltrate in the villous
lamina propria were observed in the intestinal mucosa (Fig. 1D). The severity scores for the
intestinal lesions are listed in Fig.
1E, which indicates that the histological scores of the
diarrhea group were significantly increased as compared with those
of the control group (P<0.01). In addition, the incidence rate
of liquid stools in rats was 100%, while the control group did not
produce any liquid stools. In addition, the diarrhea rats had a
higher diarrhea index as compared with that of the control group
(P<0.01) (Fig. 1F).
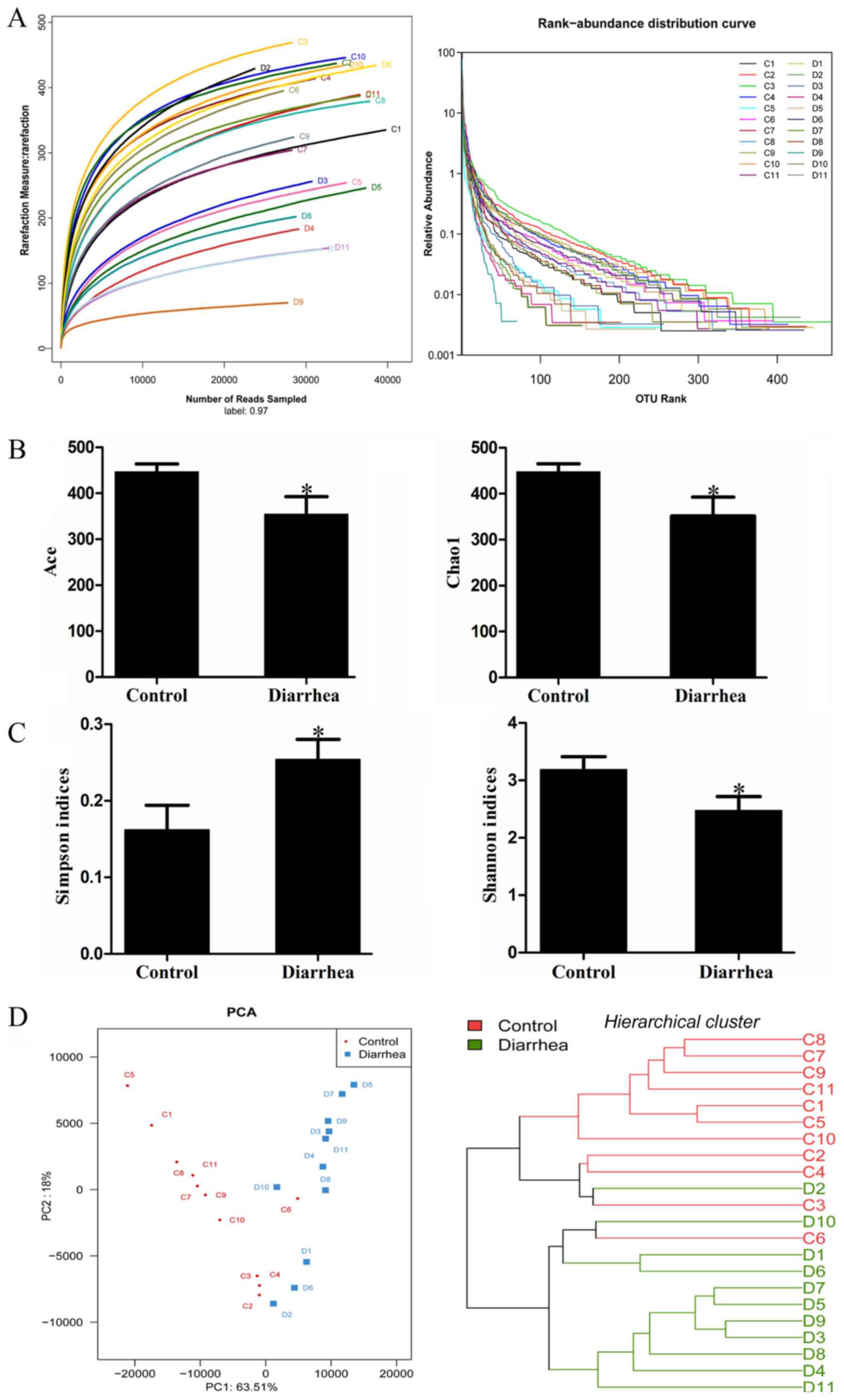
To characterize the bacterial diversity and
abundance in the fecal microbiota of the control and diarrhea rats,
high throughput sequencing was performed on the V3-V4 hypervariable
region of bacterial 16S rRNA gene using the Illumina MiSeq system.
Following denoising and filtering steps, a total of 712,814 valid
reads were obtained from the 22 samples, with a mean of 32,400
reads/sample. A dataset consisting of 32,810±1,315 reads from the
control group (n=11) and 31,990±1,372 from the diarrhea group
(n=11), with an average length of 442 bp, was used in the final
analysis. The Good's coverage of all samples was 0.9977±0.0007%,
indicating that the 16S rRNA sequences represented the majority of
the bacteria in the samples. Based on a sequence similarity of
>97%, an average of 376 and 267 OTUs were defined for the
control and diarrhea groups, respectively. The rarefaction curves
indicated that the diversity of the bacteria was addressed, while
the rank abundance curves presented the abundance and evenness of
the two groups (Fig. 2A). The levels
of the indicators of community abundance (Ace and Chao1) in the
diarrhea group were significantly decreased as compared with those
in the control group (P<0.05) (Fig.
2B). Furthermore, the Simpson and Shannon indices indicated
that the diversity of the microbial community of the control group
was higher than that of the diarrhea group (Fig. 2C).
The differences and similarity between the microbial
communities in the diarrhea and control groups were revealed by
determination of the weighted Unifrac PCA and hierarchical cluster
analysis, respectively (Fig. 2D).
Weighted UniFrac PCA demonstrated a high degree of variation
between individual rats. Nevertheless, the first principal
component (PC1), which explained 63.51% of the variance in the
data, distinctly separated the diarrhea group from the control
group. Further hierarchical cluster analysis revealed that the
control and diarrhea groups were distinguished into two major
clusters, indicating robust differences in the microbial
communities of the two groups.
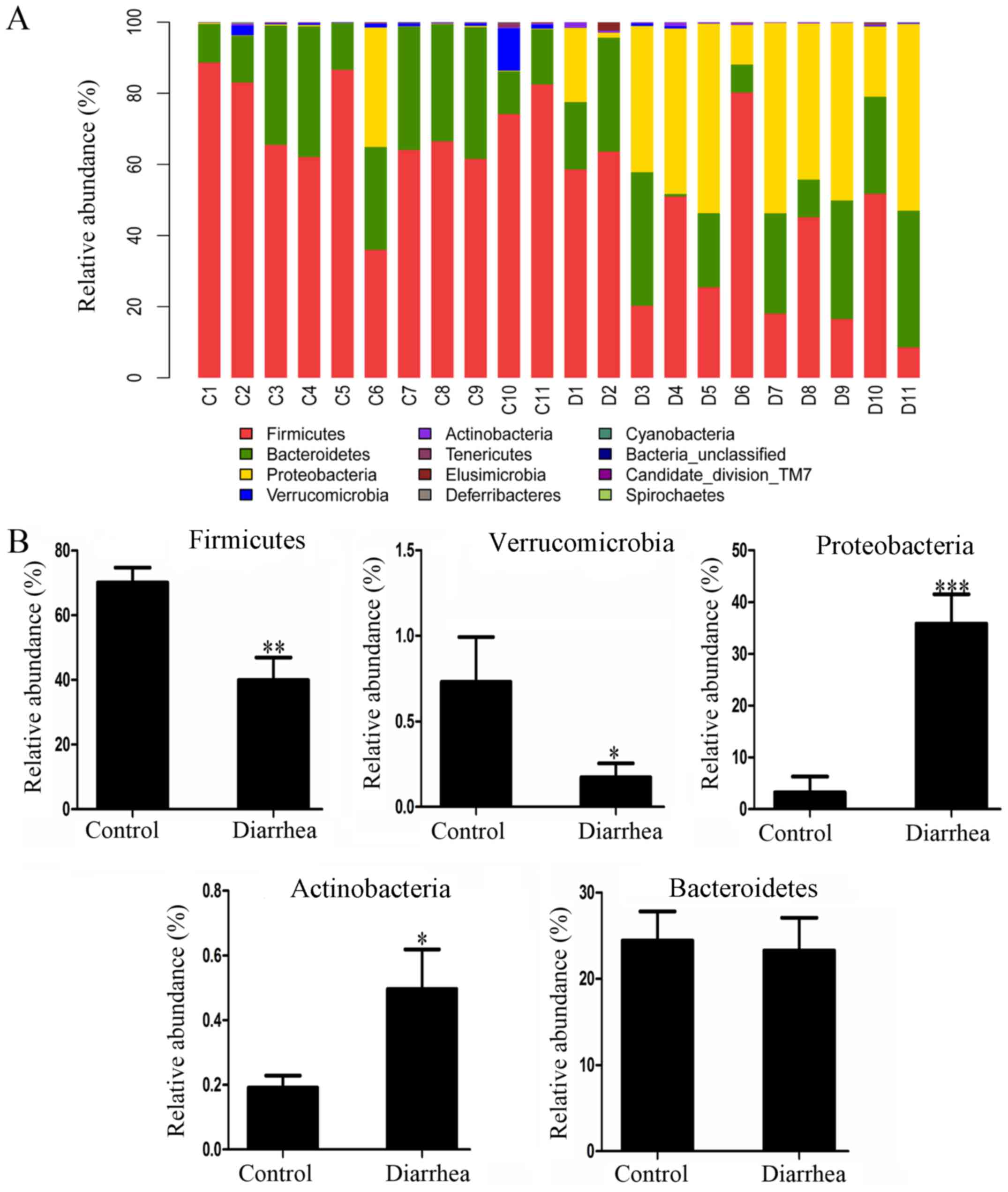
The phylum-level distribution patterns of the
control and diarrhea groups are shown in Fig. 3A. In the control group, the major
bacterial communities included Firmicutes (70.05±4.59%),
Bacteroidetes (24.41±3.39%), Proteobacteria (3.25±3.02%),
Verrucomicrobia (0.73±0.26%) and Actinobacteria (0.19±0.04%). In
the diarrhea group, the order of the major bacterial communities
was Firmicutes (39.91±7.02%), Proteobacteria (35.82±5.70%),
Bacteroidetes (23.27±3.79%), Actinobacteria (0.50±0.12%) and
Verrucomicrobia (0.17±0.08%). Thus, the results suggested that the
most abundant communities in the two groups are the three most
populated bacterial phyla, namely Firmicutes, Bacteroidetes and
Proteobacteria, followed by the low abundance phyla of
Verrucomicrobia, Actinobacteria, Candidate division TM7,
Cyanobacteria, Deferribacteres, Elusimicrobia and Spirochaetes. As
shown in Fig. 3B, statistical
analysis revealed that the relative abundance of Firmicutes and
Verrucomicrobia in the diarrhea group was significantly lower in
comparison with that in the control group (P<0.05). By contrast,
the relative abundance of Proteobacteria and Actinobacteria in the
diarrhea group was significantly higher compared with that in the
control group (P<0.05). Bacteroidetes, one of the most dominant
phyla, was not significantly altered in the diarrhea group.
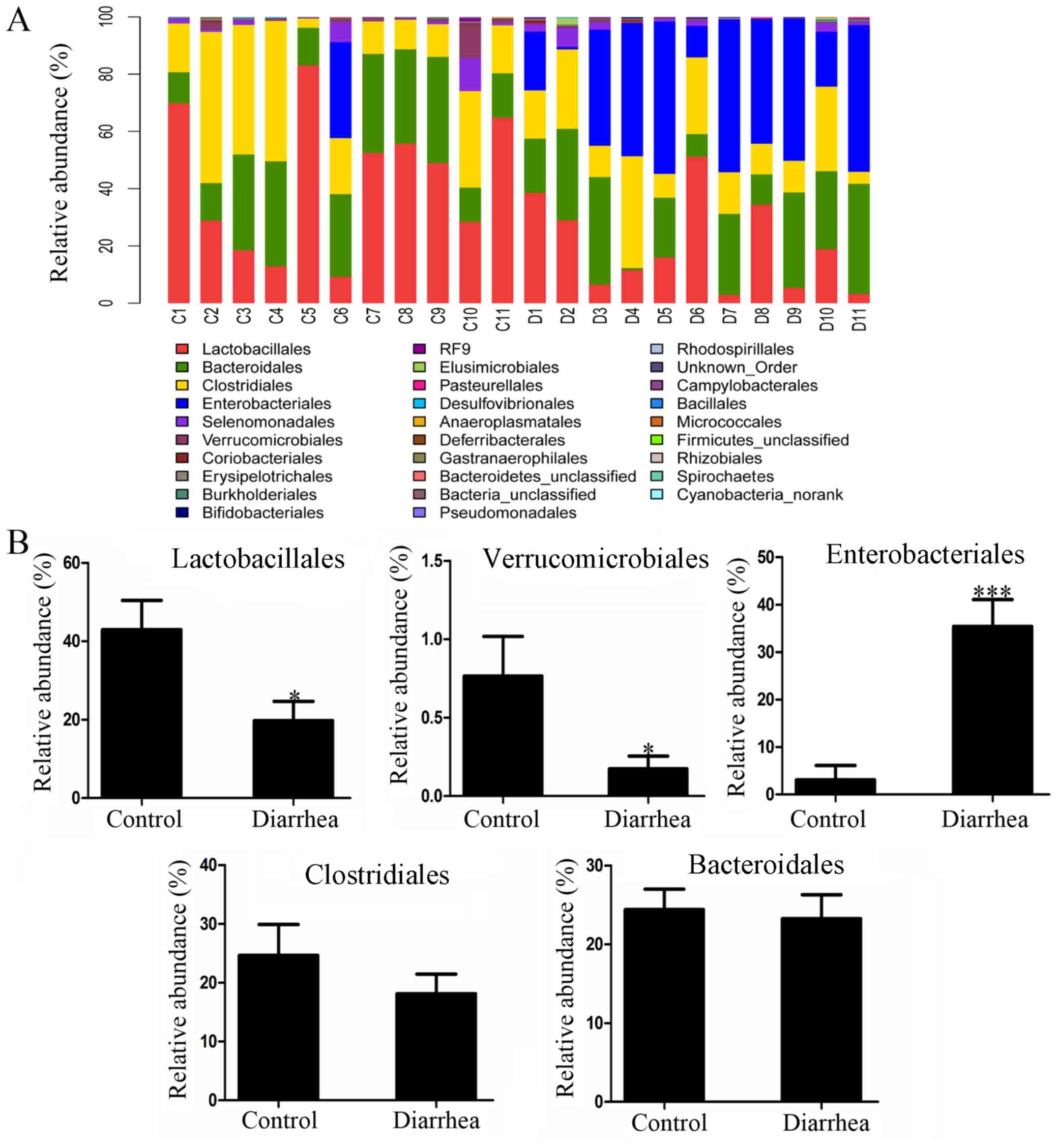
The order-level distribution patterns of the two
groups are shown in Fig. 4A. In the
control group, the orders of the most dominant bacterial
communities included Lactobacillales (42.95±7.47%), Clostridiales
(24.62±5.27%), Bacteroidales (24.39±3.39%), Enterobacteriales
(3.06±3.04%) and Verrucomicrobiales (0.76±0.25%), accounting for a
total of 95.77% of the overall bacteria presented in this group. In
the diarrhea group, the orders of the most dominant bacterial
communities were Enterobacteriales (35.39±5.70%), Bacteroidales
(23.26±3.79%), Lactobacillales (19.75±4.92%), Clostridia
(18.15±3.30%) and Verrucomicrobiales (0.17±0.08%), accounting for a
total of 96.72% of the overall bacteria presented in this group.
Furthermore, statistical analysis demonstrated that the proportion
of Lactobacillales belonging to the phylum Firmicutes and the
proportion of Verrucomicrobiales of the phylum Verrucomicrobia were
significantly decreased in the diarrhea group as compared with that
in the control group (P<0.05). By contrast, the proportion of
Enterobacteriales belonging to the phylum Proteobacteria was
significantly increased in the diarrhea group (P<0.05). However,
the levels of Bacteroidales and Clostridiales did not differ
significantly between the two groups (P>0.05) (Fig. 4B).
Furthermore, the difference of the microbiota
distribution at the genus level was compared between the control
and diarrhea rats. The microbial distribution was significantly
different at the genus level, suggesting that the composition of
microbiota in the intestine of the rats was severely altered due to
E. coli O101 infection. Among the 100 genera that
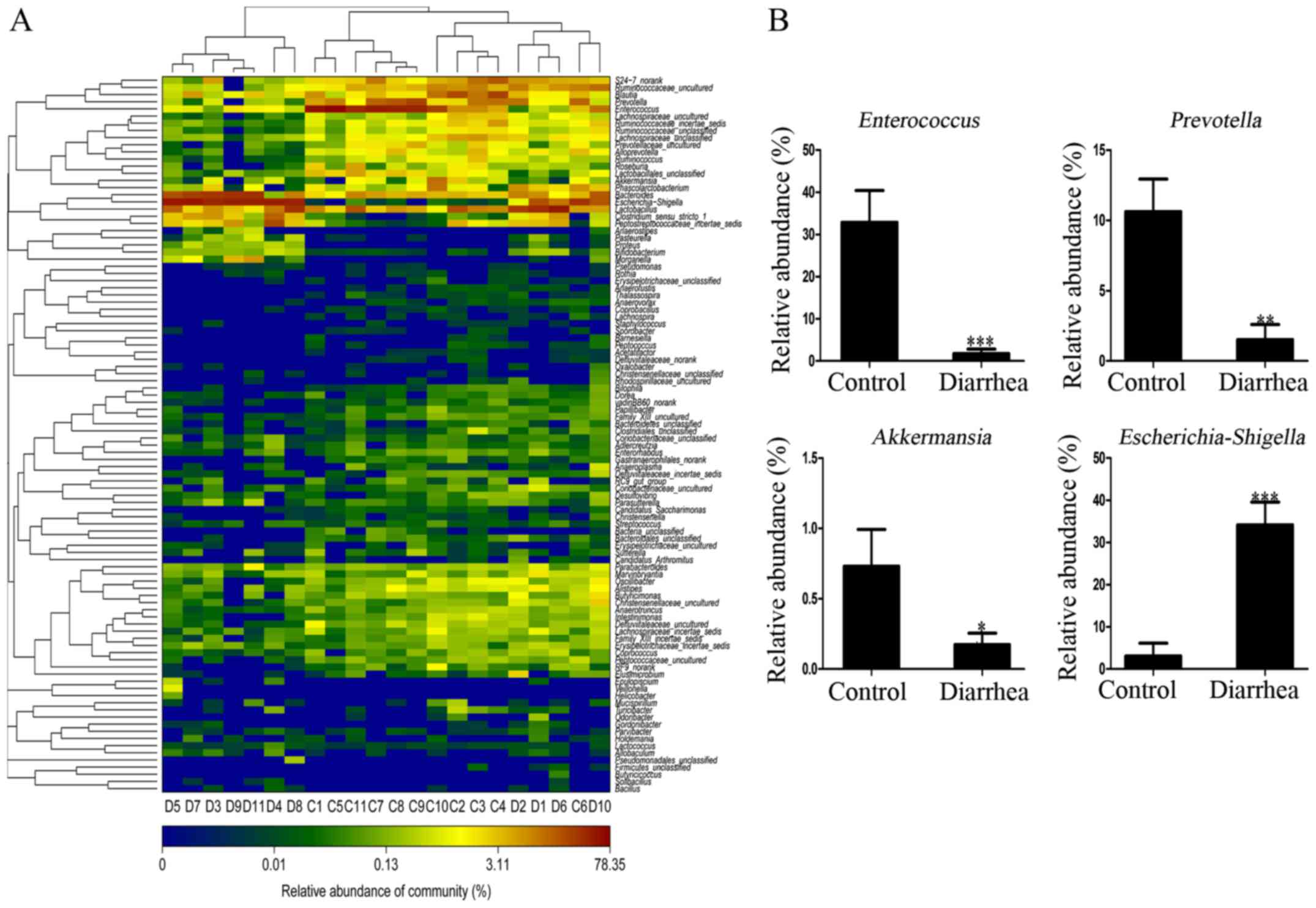
are displayed in the heatmap in Fig.
5A, certain of these exhibited a significant difference between
the control and diarrhea groups, including the genera
Enterococcus, Prevotella, Akkermansia and
Escherichia/Shigella (P<0.05) (Fig. 5B). In addition, the specific
phylotype at the OTUs level was identified in response to E.
coli O101 infection. A total of 18 OTUs of relative
abundance presented in the two groups were selected for comparison.
The proportion of Enterococcus (2 OTUs) belonging to the
order Lactobacillales was significantly lower in the diarrhea group
(mean, 1.74%) as compared with that in the control group (mean,
32.88%; P<0.001). The proportion of Prevotella (14 OTUs)
belonging to the order Bacteroidales was also significantly lower
in the diarrhea group (mean, 1.51%) in comparison with that in the
control group (mean, 10.62%; P<0.01). Furthermore, the
proportion of Akkermansia (1 OTU) belonging to the order
Verrucomicrobia was significantly lower in the diarrhea group
(mean, 0.17%) as compared with the control group (mean, 0.73%;
P<0.001). Finally, the proportion of Escherichia/Shigella
(1 OTU) belonging to the order Enterobacteriales was
significantly higher in the diarrhea group (mean, 34.16%) as
compared with that in the control group (mean, 3.05%; P<0.001)
(Fig. 5B).
Discussion
ETEC strains that produce multiple enterotoxins are
major causes of severe dehydrating diarrhea in humans and animals
(34,35). Several studies have reported that the
diarrhea-associated diseases, such as cholera (36), diarrhea-predominant irritable bowel
syndrome (37) and porcine epidemic
diarrhea (38), can change the
composition of the gut microbiota. This evidence indicated a
potential association between gut microbiota and diarrhea. However,
little is known regarding the status of gut microbiota and the
histopathological changes in intestinal tissues following ETEC
infection in animals. Therefore, elucidating the role of ETEC in
altering the composition of gut microbiota and the intestinal
tissue in a model of ETEC-induced diarrhea is essential.
Notably, the intestinal mucosa is a vital barrier
for protecting the body against infection by pathogenic
microorganisms (39,40). Histological assessment is commonly
used in the diagnosis of gastrointestinal diseases (41,42). In
a previous study, the investigation of histological sections of
intestinal tissue from diarrhea mice revealed damaged surface
epithelium with inflammatory infiltrates in the lamina propria
(43). Similarly, the results of the
present study also revealed that the intestinal mucosa of diarrhea
rats was damaged, and the surface epithelium and villous lamina
propria were disrupted by the inflammatory cellular infiltrates.
According to the histopathological scores, the diarrhea rats
exhibited severe injury in the intestinal tissues.
The gut microbiota in the intestinal mucosa serves a
crucial role in the development and integrity of the mucosal
epithelium (44–46). As fecal microbial communities
represented the highest bacterial diversity in the gut (47), fecal sample were used in the present
study. Barcoded Illumina MiSeq sequencing of the V3-V4
hypervariable region of 16S rRNA was employed, in order to compare
the composition of the fecal microbiota between the normal and
E. coli O101-treated rats. Chao1 and Ace analysis
revealed greater microbial diversity in the control group compared
with that in the diarrhea group, while the Shannon and Simpson
indices also indicated that the bacterial community diversity of
the control group was higher than that of the diarrhea group.
Therefore, it is concluded that E. coli O101
infection reduced the diversity of the gut microbiota in rats.
The phylum-level distribution of the bacterial
communities in the control and diarrhea groups of the present study
revealed that the two prevalent phyla, namely Bacteroidetes and
Firmicutes, are dominant irrespective of E. coli
O101 infection. This observation was in agreement with
the findings of a previous study, which demonstrated that
Bacteroidetes and Firmicutes are the main phyla in rats regardless
of the age (48). In the present
study, Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia
and Actinobacteria were dominant in the groups, which was in
agreement with previous findings (49). However, these dominant bacteria phyla
displayed a different tendency subsequent to E. coli
O101 infection. The proportion of Firmicutes and
Verrucomicrobia was decreased, while that of Proteobacteria and
Actinobacteria was significantly increased. As reported previously,
the phylum Verrucomicrobia was absent in mice treated with
cyclophosphamide, a potent immunosuppressive agent (50,51).
Thus, it can be speculated that E. coli O101
infection decreased the abundance of Verrucomicrobia and that it
may suppress the immune function of the host. Furthermore,
Proteobacteria is the main pathogenic bacterial phylum that is
closely associated with the presence of diarrhea symptoms (52,53). As
a consequence of E. coli O101 infection, the abundance of
Proteobacteria was increased significantly.
The comparison at the order level was in agreement
with that at the phylum level in the current study. For instance,
the orders Lactobacillales and Verrucomicrobiales were
significantly decreased in the diarrhea group, while the order
Enterobacteriales was significantly increased in the diarrhea
group. The phyla of these microbes, namely Firmicutes,
Verrucomicrobia and Proteobacteria, respectively, exhibited the
same tendency of decrease or increase. Thus, the alteration at the
order level contributed to that at the phylum level.
Among the fully classified genera in the present
study, several genera of specific interest were identified. For
instance, Escherichia/Shigella, belonging to the order
Enterobacteriales and the phylum Proteobacteria, exhibited a higher
abundance (11.2-fold) in the diarrhea group as compared with that
in the control group. Thus, it can be inferred that the increase in
the abundance of Escherichia/Shigella caused the increase of
the order Enterobacteriales and the phylum Proteobacteria in the
diarrhea rats. In addition, the genus Prevotella exhibited
the most significant difference in relative abundance between the
diarrhea and control groups. The abundance of Prevotella was
significantly lower (7.0-fold) in the diarrhea group in comparison
with the control group. Similar to the current observations, Pop
et al (54) reported that
diarrhea in young children from low-income countries led to a low
abundance of Prevotella. Kang et al (55) also found a significantly lower
abundance of Prevotella in autistic children. De Filippo
et al (56) further
demonstrated that Prevotella suppresses the pathogenic
Escherichia/Shigella, resulting in a low incidence of
gastrointestinal disorders among African children. These
observations suggested that Prevotella may exert a
beneficial physiological role in human health, particularly in
children. Furthermore, the proportion of Enterococcus, a
large genus of lactic acid bacteria of the order Lactobacillales
and the phylum Firmicutes, was markedly reduced (18.9-fold) in
diarrhea rats as compared with normal rats. The most common species
of Enterococcus spp. include Enterococcus faecium and
Enterococcus faecalis, which can be isolated from the
gastrointestinal tract, the oral cavity and the vagina of animals
as normal commensals (57).
Enterococcus faecium strains are frequently used in pig
production to decrease the incidence of diarrhea and the count of
E. coli in pigs, as well as improve the animals' performance
and feed conversion. Hu et al (58) reported that Enterococcus
faecalis LAB31 effectively reduced the incidence of diarrhea in
weaned piglets by increasing the relative number of
Lactobacilli. The abundance of Akkermansia, belonging
to the order Verrucomicrobiales and phylum Verrucomicrobia, was
also evidently reduced in the diarrhea group. Similarly, Liu et
al (38) revealed that
Akkermansia was highly abundant in the control group than in
the porcine epidemic diarrhea group. Diarrhea patients with
microscopic colitis also had a significantly lower amount of
Akkermansia (59).
Furthermore, several studies suggested that Akkermansia may
be used as a beneficial bacterium to regulate the host immunity, as
well as an indicator for its evaluation. Shin et al
(60) previously reported that
Akkermansia attenuated tissue inflammation by activating the
Foxp3 regulatory T-cells. Akkermansia muciniphila, belonging
to the genus Akkermansia, altered the mucosal gene
expression profiles toward altering immune responses (61). Taken together, the present study
revealed the E. coli O101 infection decreased the
proportion of the beneficial bacteria Prevotella,
Enterococcus and Akkermansia, while increasing the
proportion of the pathogenic bacteria Escherichia/Shigella.
Therefore, the beneficial bacteria Prevotella, Enterococcus
and Akkermansia protected rats against the pathogenic
bacteria Escherichia/Shigella, while E. coli
O101 infection altered the balance.
In an attempt to distinguish
Escherichia/Shigella in feces from rats injected with E.
coli O101, the current study also conducted a
sequence comparison and found a 99% similarity (data not shown),
which suggested that Escherichia/Shigella in feces were
putatively of the same genus of E. coli O101 that
were injected into the peritoneum of the rats. Thus, the present
study hypothesized that injected E. coli O101 may
colonize in the intestinal tract of rats and compete with the
beneficial bacteria, leading to the decreased abundance of the
beneficial bacteria and consequently resulting in an imbalance of
gut microbiota.
The gut microbiome is hypothesized to serve a
critical role in gastrointestinal diseases, such as diarrhea. By
regulating the balance of intestinal flora, increasing the
beneficial bacteria and reducing the harmful bacteria, the symptoms
of diarrhea can be alleviated and diseases can be treated.
Historically, antibiotics were primarily used to treat individuals
with diarrhea. However, the blind use of antibiotics may eliminate
the sensitive beneficial bacteria and aggravate the microbiota
imbalance. Therefore, in order to prevent further aggravation of
the microbiota disorder as identified in the current study,
appropriate use of drugs should be considered for adjuvant therapy.
The efficiency of specific probiotics for the treatment of
infection-associated diarrhea in adults has been supported by
clinical studies (62). Dietary
fiber benefits human health and can also modulate gut microbiota
for treating diarrhea infections (63). Furthermore, the Chinese herbal
formula SLBZS has been demonstrated to have an effect in shifting
the gut microbiome structure during the treatment of rats with
antibiotic-associated diarrhea (64). Taken together, the current study may
provide a theoretical basis for gut microbiota and potential novel
targets for the control of the disease.
In conclusion, given the crucial role of gut
microbiota in maintaining intestinal health, identifying the
changes in systemic gut microbiota and specific microbes is
essential. As a first step to achieve this long-term goal, the
present study established an E. coli O101-induced
diarrheal rat model with increasing diarrhea index and injury in
the intestinal tissues. Next, several key changes in fecal
microbiota subsequent to treatment with E. coli
O101 were identified. It was revealed that the diarrhea
rats tended to have a less diverse gut microbiome, while a shifted
distribution pattern of the bacterial communities was demonstrated
at the phylum and order levels in diarrhea rats. Finally, several
individual genera, primarily the beneficial Prevotella,
Enterococcus and Akkermansia, exhibited significantly
lower abundance, while the pathogenic Escherichia/Shigella
had significantly higher abundance in diarrhea rats as compared
with the control group. Taken together, the data of the present
study provided crucial insights into E. coli
O101-induced dysbiosis in gut microbiota in the fecal
samples. Thus, further genomic studies are necessary to better
characterize the indicative bacteria and assess the potential
development of a microbiological intervention for treating
diarrhea.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 31572558 and 31272144), the
Beijing Nova Program (grant no. Z141105001814041), and the
Profession Scientific Research Special item of Agricultural Public
Welfare, Ministry of Agriculture, China (grant no.
201403051-10).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS and XW performed the DNA extraction and
pyrosequencing, statistical analysis and drafted of manuscript. YH,
YW and BF participated in the design of the study and performed the
statistical analysis. YG participated in drafted of manuscript. YG,
GH and XM revised the manuscript and interpreted the data. HD and
YZ conceived of the study, participated in the study design and
coordination, and helped to draft the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Institutional
Animal Care and Use Committee of the AMMS. All animal care and
experimental procedures were conducted according to the
institutional ethical guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the global
burden of disease study. Lancet. 380:2095–2128. 2010. View Article : Google Scholar
|
|
2
|
Boschi-Pinto C, Velebit L and Shibuya K:
Estimating child mortality due to diarrhoea in developing
countries. Bull World Health Organ. 86:710–717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chowdhury F, Khan IA, Patel S, Siddiq AU,
Saha NC, Khan AI, Saha A, Cravioto A, Clemens J, Qadri F and Ali M:
Diarrheal illness and healthcare seeking behavior among a
population at high risk for diarrhea in Dhaka, Bangladesh. PLoS
One. 10:e1301052015. View Article : Google Scholar
|
|
4
|
Maya L, Puentes R, Reolón E, Acuña P, Riet
F, Rivero R, Cristina J and Colina R: Molecular diversity of bovine
viral diarrhea virus in Uruguay. Arch Virol. 161:529–535. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qadri F, Svennerholm AM, Faruque AS and
Sack RB: Enterotoxigenic Escherichia coli in developing countries:
Epidemiology, microbiology, clinical features, treatment, and
prevention. Clin Microbiol Rev. 18:465–483. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khalil S, Mirdha BR, Sinha S, Panda A,
Singh Y, Joseph A and Deb M: Intestinal parasitosis in relation to
anti-retroviral therapy, CD4(+) T-cell count and diarrhea in HIV
patients. Korean J Parasitol. 53:705–712. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Estrada-Garcia T, Lopez-Saucedo C,
Thompson-Bonilla R, Abonce M, Lopez-Hernandez D, Santos JI, Rosado
JL, DuPont HL and Long KZ: Association of diarrheagenic Escherichia
coli pathotypes with infection and diarrhea among Mexican children
and association of atypical enteropathogenic E. coli with acute
diarrhea. J Clin Microbiol. 47:93–98. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Konishi N, Obata H, Monma C, Nakama A, Kai
A and Tsuji T: Bacteriological and epidemiological characteristics
of enterotoxigenic Escherichia coli isolated in Tokyo, Japan,
between 1966 and 2009. J Clin Microbiol. 49:3348–3351. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duchet-Suchaux M: Protective antigens
against enterotoxigenic Escherichia coli O101:K99,F41 in the infant
mouse diarrhea model. Infect Immun. 56:1364–1370. 1988.PubMed/NCBI
|
|
10
|
Liu B, Wu F, Li D, Beutin L, Chen M, Cao B
and Wang L: Development of a serogroup-specific DNA microarray for
identification of Escherichia coli strains associated with bovine
septicemia and diarrhea. Vet Microbiol. 142:373–378. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kelder T, Stroeve JH, Bijlsma S, Radonjic
M and Roeselers G: Correlation network analysis reveals
relationships between diet-induced changes in human gut microbiota
and metabolic health. Nutr Diabetes. 4:e1222014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tomás-Barberán FA, García-Villalba R,
González-Sarrías A, Selma MV and Espín JC: Ellagic acid metabolism
by human gut microbiota: Consistent observation of three urolithin
phenotypes in intervention trials, independent of food source, age,
and health status. J Agric Food Chem. 62:6535–6538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diamant M, Blaak EE and de Vos WM: Do
nutrient-gut-microbiota interactions play a role in human obesity,
insulin resistance and type 2 diabetes? Obes Rev. 12:272–281. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dollé L, Tran HQ, Etienne-Mesmin L and
Chassaing B: Policing of gut microbiota by the adaptive immune
system. BMC Med. 14:272016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doré J and Blottière H: The influence of
diet on the gut microbiota and its consequences for health. Curr
Opin Biotechnol. 32:195–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Flowers SA and Ellingrod VL: The
microbiome in mental health: Potential contribution of gut
microbiota in disease and pharmacotherapy management.
Pharmacotherapy. 35:910–916. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Zhao Y, Xu J, Xue Z, Zhang M,
Pang X, Zhang X and Zhao L: Modulation of gut microbiota by
berberine and metformin during the treatment of high-fat
diet-induced obesity in rats. Sci Rep. 5:144052015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Collins B, Hoffman J, Martinez K, Grace M,
Lila MA, Cockrell C, Nadimpalli A, Chang E, Chuang CC, Zhong W, et
al: A polyphenol-rich fraction obtained from table grapes decreases
adiposity, insulin resistance and markers of inflammation and
impacts gut microbiota in high-fat-fed mice. J Nutr Biochem.
31:150–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Larsen N, Vogensen FK, van den Berg FW,
Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ,
Hansen LH and Jakobsen M: Gut microbiota in human adults with type
2 diabetes differs from non-diabetic adults. PLoS One. 5:e90852010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ling Z, Liu X, Jia X, Cheng Y, Luo Y, Yuan
L, Wang Y, Zhao C, Guo S, Li L, et al: Impacts of infection with
different toxigenic Clostridium difficile strains on faecal
microbiota in children. Sci Rep. 4:74852014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Monaco MH, Wang M, Comstock SS,
Kuhlenschmidt TB, Fahey GC Jr, Miller MJ, Kuhlenschmidt MS and
Donovan SM: Human milk oligosaccharides shorten rotavirus-induced
diarrhea and modulate piglet mucosal immunity and colonic
microbiota. ISME J. 8:1609–1620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guard BC, Barr JW, Reddivari L,
Klemashevich C, Jayaraman A, Steiner JM, Vanamala J and Suchodolski
JS: Characterization of microbial dysbiosis and metabolomic changes
in dogs with acute diarrhea. PLoS One. 10:e1272592015. View Article : Google Scholar
|
|
23
|
Ansorge WJ: Next-generation DNA sequencing
techniques. N Biotechnol. 25:195–203. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Laboratory Animals' Welfare and Ethics
guidelines (GB/T 35892-2018). http://www.gb688.cn/bzgk/gb/newGbInfo?hcno=9BA619057D5C13103622A10FF4BA5D14
|
|
25
|
De Cupere F, Deprez P, Demeulenaere D and
Muylle E: Evaluation of the effect of 3 probiotics on experimental
Escherichia coli enterotoxaemia in weaned piglets. Zentralbl
Veterinarmed B. 39:277–284. 1992.PubMed/NCBI
|
|
26
|
Zhou G, Hu Z and Wang Y: An inquiry into
preparing diarrhea model of mice and application of diarrhea index.
Chin Tradit Herb Drugs. 4:195–196. 1994.
|
|
27
|
Han D, Hu Y, Li L, Tian H, Chen Z, Wang L,
Ma H, Yang H and Teng K: Highly pathogenic porcine reproductive and
respiratory syndrome virus infection results in acute lung injury
of the infected pigs. Vet Microbol. 169:135–146. 2014. View Article : Google Scholar
|
|
28
|
Cheng J, Chen Y, He T, Liao R, Liu R, Yi
M, Huang L, Yang Z, Fu T and Li X: Soil nitrogen leaching decreases
as biogas slurry DOC/N ratio increases. Appl Soil Ecol.
111:105–113. 2017. View Article : Google Scholar
|
|
29
|
Edgar RC: UPARSE: Highly accurate OTU
sequences from microbial amplicon reads. Nat Methods. 10:996–998.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Edgar RC, Haas BJ, Clemente JC, Quince C
and Knight R: UCHIME improves sensitivity and speed of chimera
detection. Bioinformatics. 27:2194–2200. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Amato KR, Yeoman CJ, Kent A, Righini N,
Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba
M, et al: Habitat degradation impacts black howler monkey (Alouatta
pigra) gastrointestinal microbiomes. ISME J. 7:1344–1353. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schloss PD, Gevers D and Westcott SL:
Reducing the effects of PCR amplification and sequencing artifacts
on 16S rRNA-based studies. PLoS One. 6:e273102011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Sheng HF, He Y, Wu JY, Jiang YX,
Tam NF and Zhou HW: Comparison of the levels of bacterial diversity
in freshwater, intertidal wetland, and marine sediments by using
millions of illumina tags. Appl Environ Microbiol. 78:8264–8271.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Richie E, Punjabi NH, Corwin A, Lesmana M,
Rogayah I, Lebron C, Echeverria P and Simanjuntak CH:
Enterotoxigenic Escherichia coli diarrhea among young children in
Jakarta, Indonesia. Am J Trop Med Hyg. 57:85–90. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park ES, Jo S, Seong JK, Nam TC, Yang IS,
Choi MC and Yoon YS: Effect of acupuncture in the treatment of
young pigs with induced Escherichia coli diarrhea. J Vet Sci.
4:125–128. 2003.PubMed/NCBI
|
|
36
|
Monira S, Nakamura S, Gotoh K, Izutsu K,
Watanabe H, Alam NH, Nakaya T, Horii T, Ali SI, Iida T and Alam M:
Metagenomic profile of gut microbiota in children during cholera
and recovery. Gut Pathog. 5:12013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rigsbee L, Agans R, Shankar V, Kenche H,
Khamis HJ, Michail S and Paliy O: Quantitative profiling of gut
microbiota of children with diarrhea-predominant irritable bowel
syndrome. Am J Gastroenterol. 107:1740–1751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu S, Zhao L, Zhai Z, Zhao W, Ding J, Dai
R, Sun T and Meng H: Porcine epidemic diarrhea virus infection
induced the unbalance of gut microbiota in piglets. Curr Microbiol.
71:643–649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tanabe S: Short peptide modules for
enhancing intestinal barrier function. Curr Pharm Des. 18:776–781.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martínez-Augustin O, Rivero-Gutiérrez B,
Mascaraque C and de Medina Sánchez F: Food derived bioactive
peptides and intestinal barrier function. Int J Mol Sci.
15:22857–22873. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hartfield D, Turner J, Huynh H, Lidman P,
Chaba T and Lacson A: The role of histopathology in diagnosing
protracted diarrhea of infancy. Fetal Pediatr Pathol. 29:144–157.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Simmerson SM, Armstrong PJ, Wünschmann A,
Jessen CR, Crews LJ and Washabau RJ: Clinical features, intestinal
histopathology, and outcome in protein-losing enteropathy in
yorkshire terrier dogs. J Vet Intern Med. 28:331–337. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Saha DR, Guin S, Krishnan R, Nag D, Koley
H, Shinoda S and Ramamurthy T: Inflammatory diarrhea due to
enteroaggregative Escherichia coli: Evidence from clinical and mice
model studies. Gut Pathog. 5:362013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hooper LV: Bacterial contributions to
mammalian gut development. Trends Microbol. 12:129–134. 2004.
View Article : Google Scholar
|
|
45
|
Zhang K, Hornef MW and Dupont A: The
intestinal epithelium as guardian of gut barrier integrity. Cell
Microbiol. 17:1561–1569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Roselli M, Finamore A, Britti MS, Bosi P,
Oswald I and Mengheri E: Alternatives to in-feed antibiotics in
pigs: Evaluation of probiotics, zinc or organic acids as protective
agents for the intestinal mucosa. A comparison of in vitro and in
vivo results. Anim Res. 54:203–218. 2005. View Article : Google Scholar
|
|
47
|
Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan
LP, Nie Y and Wu XL: Bacterial community mapping of the mouse
gastrointestinal tract. PLoS One. 8:e749572013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Y, Liao Q, Lin M, Zhong D, Wei L, Han
Bo B, Miao H, Yao Meicun M and Xie Z: An integrated metabonomics
and microbiology analysis of host-microbiota metabolic interactions
in rats with Coptis chinensis-induced diarrhea. RSC Adv.
5:79329–79341. 2015. View Article : Google Scholar
|
|
49
|
Zhang X, Zhao Y, Zhang M, Pang X, Xu J,
Kang C, Li M, Zhang C, Zhang Z, Zhang Y, et al: Structural changes
of gut microbiota during berberine-mediated prevention of obesity
and insulin resistance in high-fat diet-fed rats. PLoS One.
7:e425292012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu X and Zhang X: Effects of
cyclophosphamide on immune system and gut microbiota in mice.
Microbiol Res. 171:97–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jang SE, Joh EH, Ahn YT, Huh CS, Han MJ
and Kim DH: Lactobacillus casei HY7213 ameliorates
cyclophosphamide-induced immunosuppression in mice by activating
NK, cytotoxic T cells and macrophages. Immunopharmacol
Immunotoxicol. 35:396–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reeves AE, Theriot CM, Bergin IL,
Huffnagle GB, Schloss PD and Young VB: The interplay between
microbiome dynamics and pathogen dynamics in a murine model of
Clostridium difficile infection. Gut Microbes. 2:145–158. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lv Z, Peng G, Liu W, Xu H and Su J:
Berberine blocks the relapse of Clostridium difficile infection in
C57BL/6 mice after standard vancomycin treatment. Antimicrob Agents
Chemother. 59:3726–3735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pop M, Walker AW, Paulson J, Lindsay B,
Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I,
et al: Diarrhea in young children from low-income countries leads
to large-scale alterations in intestinal microbiota composition.
Genome Biol. 15:R762014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kang DW, Park JG, Ilhan ZE, Wallstrom G,
Labaer J, Adams JB and Krajmalnik-Brown R: Reduced incidence of
Prevotella and other fermenters in intestinal microflora of
autistic children. PLoS One. 8:e683222013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
De Filippo C, Cavalieri D, Di Paola M,
Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G and
Lionetti P: Impact of diet in shaping gut microbiota revealed by a
comparative study in children from Europe and rural Africa. Proc
Natl Acad Sci USA. 107:14691–14696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kayser FH: Safety aspects of enterococci
from the medical point of view. Int J Food Microbiol. 88:255–262.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hu Y, Dun Y, Li S, Zhang D, Peng N, Zhao S
and Liang Y: Dietary Enterococcus faecalis LAB31 improves growth
performance, reduces diarrhea, and increases fecal Lactobacillus
number of weaned piglets. PLoS One. 10:e1166352015.
|
|
59
|
Fischer H, Holst E, Karlsson F, Benoni C,
Toth E, Olesen M, Lindén M and Sjöberg K: Altered microbiota in
microscopic colitis. Gut. 64:1185–1186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shin NR, Lee JC, Lee HY, Kim MS, Whon TW,
Lee MS and Bae JW: An increase in the Akkermansia spp. population
induced by metformin treatment improves glucose homeostasis in
diet-induced obese mice. Gut. 63:727–735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Derrien M, Van Baarlen P, Hooiveld G,
Norin E, Müller M and de Vos WM: Modulation of mucosal immune
response, tolerance, and proliferation in mice colonized by the
mucin-degrader Akkermansia muciniphila. Front Microbiol. 2:1662011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dylag K, Hubalewska-Mazgaj M, Surmiak M,
Szmyd J and Brzozowski T: Probiotics in the mechanism of protection
against gut inflammation and therapy of gastrointestinal disorders.
Curr Pharm Des. 20:1149–1155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gong J and Yang C: Advances in the methods
for studying gut microbiota and their relevance to the research of
dietary fiber functions. Food Res Int. 48:916–929. 2012. View Article : Google Scholar
|
|
64
|
Lv W, Liu C, Ye C, Sun J, Tan X, Zhang C,
Qu Q, Shi D and Guo S: Structural modulation of gut microbiota
during alleviation of antibiotic-associated diarrhea with herbal
formula. Int J Biol Macromol. 105:1622–1629. 2017. View Article : Google Scholar : PubMed/NCBI
|