Introduction
Oral lichen planus (OLP) is a common mucosal
inflammatory disease. It has the potential for malignant
transformation (1,2); it has been reported that 1–2% of
patients with OLP develop oral squamous cell carcinoma (3). Although the pathogenesis of OLP remains
to be fully elucidated, immune factors are thought to have
important roles. A wide range of studies have indicated the
importance of the immune response in OLP and have suggested that
OLP is a T cell-mediated chronic inflammatory disease (4,5). Various
studies have suggested that microRNAs (miRNAs/miRs) also have an
important role in the pathogenesis of OLP (6). miRNAs are short, single-stranded RNAs
that do not encode proteins; they have been linked to inflammatory
diseases and immune diseases via the dysregulation of target mRNA
expression (7,8). In an miRNA microarray analysis of
mucosal tissues in patients with OLP and healthy controls, ~70
miRNAs were identified to be significantly dysregulated (6,9–11). Gassling et al (10) identified 16 miRNAs that were
differentially expressed in mucosal tissues between OLP patients
and healthy individuals. A total of 6 miRNAs, including miR-31,
−146a, −155 and −21, exhibited a >2-fold increase, while miR-923
and −30a were downregulated in OLP patients (10). Our group has previously reviewed
miRNAs associated with OLP, and it was identified that miR-155 is
closely linked to cytokines associated with OLP, rendering it a
strong candidate miRNA regarding the involvement in OLP (9). Arão et al (6) reported that miR-155 expression is
upregulated in patients with OLP. However, the precise role of
miR-155 in this disease has remained largely elusive. miR-155 is a
multi-functional miRNA and is closely associated with inflammation,
tumors and immune regulation (12).
Considering the importance of the immune response in the
pathogenesis of OLP, the present study comprehensively reviewed the
role of miR-155 in immune system regulation and its potential
association with OLP. The present review aims to provide a source
of ideas for future research.
OLP and miR-155
OLP is a common oral mucosal disease. Its clinical
manifestations are reticular, ulcerative and plaque-like lesions
(1). OLP predominately affects
females with a prevalence of 0.1–4%, and the World Health
Organization (WHO) has defined it as a potentially malignant
condition (13,14). A recent meta-analysis of data from
20,095 patients demonstrated that 1.1% of cases of OLP develop oral
squamous cell carcinoma (15). The
pathological features of OLP are degeneration of basal cells,
basement membrane disruption and a dense infiltration of
lymphocytes in the sub-epithelial layer of connective tissue
(16). Although the cause of OLP is
remains uncertain, immune dysregulation is important in the
development of this disease. CD4+ and CD8+
lymphocyte-mediated local immune responses are involved in the
pathogenesis of OLP (4,17,18).
Despite extensive research regarding the pathogenesis of OLP, the
current knowledge remains limited and further studies are required.
Detailed study of the pathogenesis of OLP is indispensable.
miRNAs target the 3′-untranslated region (3′-UTR) of
specific mRNAs and thereby regulate gene expression. Approximately
60% of all human protein-coding genes are predicted to contain
miRNA-binding sites in their 3′-UTR. Since their discovery <2
decades ago, >800 miRNAs have been identified in mammals, and a
number of them are conserved across species. Their functions are
only beginning to be elucidated. The roles of miRNAs are being
intensively studied in a wide range of physiological and
pathological processes, including proliferation, apoptosis,
differentiation and oncogenesis (19,20). In
addition, miRNAs have been reported to regulate the immune system
as well as inflammatory networks by affecting associated signaling
pathways. The abnormal expression of miRNAs leads to inflammation
and immune diseases (21,22). Various studies have indicated that
miRNA expression profiles are altered in OLP (9,10). Our
group has previously reviewed the miRNAs associated with OLP,
revealing that miR-155 is closely linked to the cytokines
associated with OLP and is therefore a good candidate for further
research (9).
miR-155 is derived from an exon of the B-cell
integration cluster gene. It has numerous functions and is closely
linked to inflammation, tumor development and immune regulation
(12). The expression of miR-155 is
usually positively correlated with cytokine release. It may also
target genes that encode proteins associated with inflammation
(23). Furthermore, miR-155 is the
first miRNA that was identified as an oncogene. In various cancer
types, including breast, colon, cervical and lung cancer, miR-155
expression is increased. This miRNA is important for tumor
development, functioning mainly as a tumor-promoting factor
(24). Of particular importance,
miR-155 has pivotal roles in immune responses. miR-155 is
upregulated in activated immune cells and has a significant impact
on these cells. Therefore, the abnormal expression of miR-155
results in impaired immune responses and is associated with a
variety of diseases (25). Numerous
studies have suggested that miR-155 is upregulated in patients with
OLP. Liu et al (11) reported
that miR-155 expression is upregulated in peripheral blood
mononuclear cells and lesions of patients with OLP compared with
that in the controls. In addition, its expression was identified to
be closely associated with the severity of the disease. Hu et
al (26) examined an erosive
type of OLP, revealing that miR-155 negatively regulates suppressor
of cytokine signaling 1 (SOCS1) in CD4+ T lymphocytes. A
positive interaction was identified between miR-155 and interferon
(IFN)-γ, which may result in a T helper type 1 cell (Th1)-mediated
immune response in OLP. However, to date, in-depth research on the
mechanisms by which miR-155 is involved in OLP is limited. As
mentioned above, immune factors are of great significance in the
pathogenesis of OLP. miR-155 is closely linked to immune system
regulation. Therefore, its roles in immune system regulation and
its relevance in OLP were herein systematically reviewed.
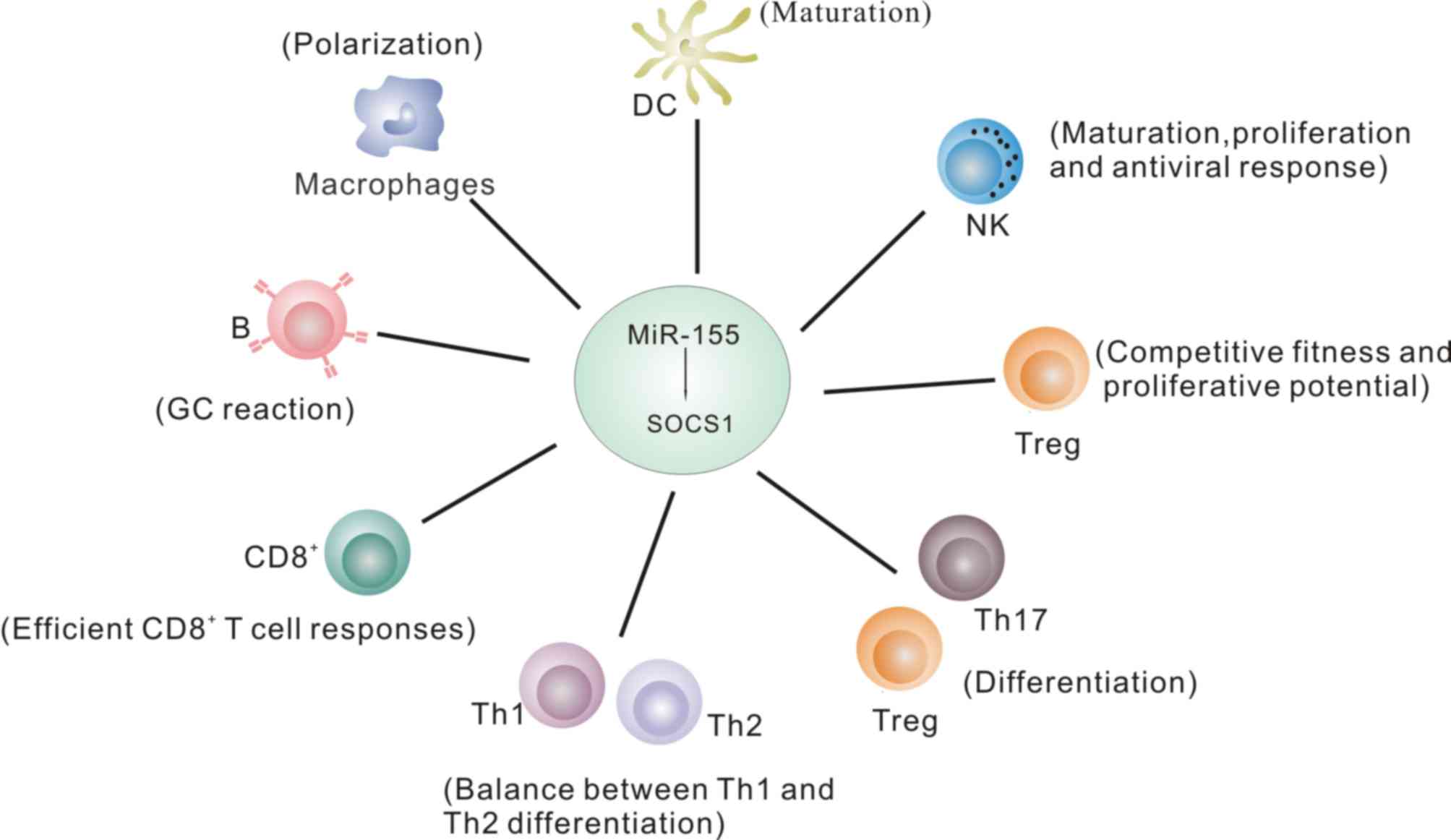
Regulation of the immune system by
miR-155
miR-155 is an important immune system regulator and
loss of miR-155 leads to impaired immune system responses (12). miR-155 targets specific genes to
influence signaling pathways and cellular phenotypes. An increasing
number of studies have focused on signaling pathways and cytokines
regulated by miR-155 in the immune system, which are summarized in
Table I.
 | Table I.Targets of microRNA-155 in the immune
system. |
Table I.
Targets of microRNA-155 in the immune
system.
| Immune system/cell
type | Function | Targets | (Refs.) |
|---|
| Innate immune
system |
|
Macrophages | Regulation of M1
and M2 macrophage polarization | SOCS1, IL13Rα1 | (32,33) |
|
| Decrease of
transforming growth factor-β-dependent gene expression | SMAD2 | (34) |
|
Dendritic cells | Inhibition of
inflammatory response in human DCs | TAB2 | (41) |
|
| Decrease of the
pathogen-binding ability of DCs | PU.1 | (42) |
|
| Promotion of DC
function | SHIP1 | (46,47) |
|
| Activation of the
ability of DCs to break self-tolerance | SOCS1 | (48) |
|
| Essential for DC
maturation and function | c-FOS | (50) |
| NK
cells | Increase of IFN-γ
production | SHIP1 | (54) |
|
| Promotion of the
maturation, the homeostasis proliferation and antiviral response of
NK cells | PMAIP1, SOCS1 | (55) |
|
| Promotion of tumor
cell proliferation and inhibition of apoptosis in NK cell
lymphoma | FOXO3a | (57) |
| Adaptive immune
system |
| B
lymphocytes | Reduction of
apoptosis of GC B cells; promotion of GC response and affinity
maturation. | JARID2 | (65) |
|
| Promotion of
IgG1+B-cell generation and control of GC reaction in B
cells | PU.1 | (63) |
|
| Decrease of extent
of DNA DSB and p53 activation associated with the GC reaction | AICDA, SOCS1 | (67) |
|
| Promotion of the
development of mature B cells | SMAD5 | (68,69) |
| T
lymphocytes | Increase of T cells
proliferation | Arginase-2 | (43) |
|
| Suppression of
differentiation of naive cells into Th2 cells | c-Maf | (69) |
|
| Facilitation of
Treg and Th17 differentiation | SOCS1 | (75) |
|
| Promotion of the
competitive fitness and proliferative potential of Tregs |
| (76) |
|
| Induction of naive
cells to differentiate into Th1 cells |
| (77) |
|
| Promotion of
efficient CD8+T-cell responses |
| (78) |
Innate immune system
Innate immunity has a key role in the defence
against pathogens. As the first line of defense, it provides the
basis and pre-condition for the adaptive immune system (27). Various studies have indicated that
miR-155 has a substantial influence on innate immunity (28).
Macrophages
When a host becomes infected, macrophages are the
first to fight the pathogens. Based on their function, they may be
classified into M1 and M2 macrophages (29). M1 macrophages promote inflammation
and cause tissue lesions. M2 macrophages induce the restoration of
tissues (30). miR-155 regulates the
dynamic balance of M1 and M2 macrophages (31). SOCS1, interleukin (IL) 13 receptor α1
(IL13Rα1) and SMAD2 are targets of miR-155 in macrophages (32–34). The
polarization of M1 and M2 is controlled by SOCS1 (35). Previous studies have demonstrated
that miR-155 directly regulates SOCS1 to induce macrophage
activation and has an important role in innate immunity (32). IL-13, a representative Th2-type
cytokine, regulates the balance between the M1 and M2 states. The
activation of IL-13 and IL-13Rα1 leads to the differentiation of
Th2 types (36). miR-155 directly
targets IL13Rα1, which makes macrophages prone to differentiation
into M1 cells (33). Transforming
growth factor-β (TGF-β) acts as an immunosuppressive and
anti-inflammatory cytokine in the inflammatory process (37). SMAD2 is a key signaling factor in the
TGF-β pathway, and TGF-β regulates gene expression via SMAD
(38). Evidence has indicated that
SMAD2 is a direct target of miR-155 in macrophages. miR-155
decreases TGF-β-dependent gene expression by targeting SMAD2,
thereby promoting the inflammatory response (34).
Dendritic cells (DCs)
DCs are specialized antigen-presenting cells that
recognize, capture and respond to antigens. They are generally
classified into the following types: CD11b+-like and
CD8a+-like, skin-resident Langerhans cells (LCs),
plasmacytoid DCs (pDCs) and monocyte-derived inflammatory DCs
(39). As a consequence of the
uptake and presentation of antigens, DCs initiate the adaptive
immune response. They are the bridge that links innate and adaptive
immunity (40). Numerous studies
have demonstrated that DCs are controlled by miR-155; this miRNA
has a key role in the maturation and function of DCs (41–43).
Martinez-Nunez et al (42)
reported that the maximum miR-155 expression levels reached in DCs
during maturation is 136-fold higher in than that in immature DCs.
Ceppi et al (41) monitored
the expression of immunological miRNAs in activated myeloid (m)DCs
and confirmed that miR-155 expression exhibited the most obvious
change. Surface molecules, including major histocompatibility
complex (MHC)-II, MHC-I, CD86 and CD83, are typically defining
features of DC maturation. In lipopolysaccharide (LPS) or specific
antigen-treated RAW264.7 cells, the expression of these molecules
was induced by miR-155 (44). In
addition, miR-155 knock-out mice exhibited abnormal functioning of
antigen-presenting cells. The induction of T-cell proliferation and
differentiation is weakened when the expression of miR-155 in DCs
is decreased. IL-12, which has an important role in the maturation
and function of DCs, may be induced by miR-155 (44). Collectively, these results indicate
that miR-155 is highly associated with the maturation and function
of DCs. Multiple targets of miR-155 associated with DCs have been
identified. In the Toll-like receptor (TLR)/IL-1 transduction
pathway, TGF-β-activated kinase 1-binding protein 2 (TAB2) is a
crucial signaling molecule. Upon TLR4 or IL-1 receptor activation,
it can induce the activation of the inflammatory response. During
monocyte-derived DC maturation, the levels of TAB2 were increased.
A previous study has demonstrated that TAB2 is directly regulated
by miR-155. Accordingly, miR-155 may have an essential role in
reducing the inflammatory response in human DCs by directly
targeting TAB2 (41). PU.1 is a
master transcription factor; it has an important role in DCs
(42). DC-specific intercellular
adhesion molecule-3-grabbing non-integrin (DC-SIGN), a C-type
lectin, is able to bind to a large array of pathogens. It also has
a substantial capacity to trigger intracellular signaling molecules
that modulate DC maturation (45).
PU.1 binds to motifs of the DC-SIGN promoter region to regulate its
expression. Previous studies have proven that PU.1 is a target of
miR-155. Martinez-Nunez (42)
indicated that miR-155 indirectly inhibits DC-SIGN expression and
impairs DC pathogen-binding ability by directly targeting PU.1. Src
homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) is an
inositol polyphosphatase that regulates DC function and efficiently
triggers Th2-type responses (46).
It has been evidenced that SHIP1 is a target of miR-155 (47). DCs lacking miR-155 exhibit an
impaired capacity to induce a functional T-cell response, and the
downregulation of SHIP1 by miR-155 may activate DC function in
vivo (46). SOCS1 is able to
impair the antigen recognition ability of DCs and cytokine
reactions. When SOCS1 expression is inhibited, DC functions are
augmented. SOCS1 is able to control IL-12 secretion and associated
signaling pathways in DCs to regulate tolerance against self
tumor-associated antigens (otherwise known as self-tolerance) of
these cells. The inhibition of SOCS1 regulated by miR-155 is
crucial for the ability of DCs to break self-tolerance (48). Cyclic AMP is able to curb the
secretion of inflammatory cytokines, and c-fos is closely linked to
this inhibitory process. It inhibits tumor necrosis factor (TNF-α)
expression and has an inhibitory role in DCs (49). It has been indicated that c-Fos mRNA
is indeed targeted by miR-155. The suppression of c-Fos by miR-155
is beneficial for DC maturation and function (50).
Natural killer (NK) cells
NK cells have important roles in the defence against
infection in the absence of specific immunization. They have
anti-infection roles via cytolytic killing, cytokine secretion, and
interactions with antigen-presenting cells and activated T
lymphocytes. Several studies have reported that miR-155 is involved
in the control of NK cell function (51). Upon stimulation, human NK cells
exhibit upregulation of miR-155. Trotta et al (52) indicated that miR-155 positively
regulates NK cell proliferation, development and effector
functions. miR-155 also promotes IFN-γ production in NK cells.
These results demonstrate that miR-155 is important in NK cells.
Several targets of miR-155 in NK cells have been identified. In
human NK cells, SHIP1 negatively regulates the secretion of IFN-γ
(53). Previous studies have
validated that SHIP1 is a direct target of miR-155. Another study
by Trotta et al (54)
demonstrated that miR-155 promotes the production of IFN-γ in NK
cells by decreasing the expression of SHIP1.
Phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1-) is a
member of the BH3-only subfamily and is able to induce cell
apoptosis. PMAIP1 and SOCS1 have been identified as direct targets
of miR-155. NK cells require miR-155 to suppress PMAIP1 and SOCS1
levels in order to increase cell survival and homeostasis.
Therefore, the suppression of PMAIP1 and SOCS1 by miR-155
contributes to the promotion of NK-cell function (55). Forkhead box (FOX)O3a is able to
inhibit malignant transformation and tumorigenesis (56). Studies have identified FOXO3a as a
target of miR-155. In NK-cell lymphoma, overexpression of miR-155
suppresses FOXO3a expression, which increases cancer cell
proliferation. The miR155/FOXO3a interaction may and serve as a
useful marker and provide a theoretical basis for gene therapy for
NK-cell lymphoma (57).
Adaptive immune system
A proper adaptive immune response relies on the
function of B and T lymphocytes. Rodriguez et al (58) examined B and T lymphocyte responses
in miR-155-deficient mice. They identified that immunoglobulin M
(IgM) was decreased in miR-155-deficient mice. The levels of IL-2
and IFN-γ production were markedly lower in these mice than in
healthy controls, indicating that miR-155-deficient mice were
immune-deficient (58). These
results clearly demonstrate that miR-155 has a crucial influence on
the adaptive immune system.
B lymphocytes
Upon antigen stimulation, B cells may differentiate
into plasma cells, which are able to synthesize and secrete Ig
antibodies and are mainly involved in humoral immunity. The
function of B cells depends on the normal expression of miR-155
(59). Once B cells are activated,
the expression of miR-155 is upregulated. In miR-155 knockout mice,
the number of germinal center (GC) B cells is decreased, which is
indicative of defective mature B cells (60). Compared with those of wild-type
equivalents, miR-155−/− mice presented with a decreased
GC response and reduced production of IgM, IgG, and IgA antibodies,
indicating that B-cell responses were impaired (61). The germinal center is a primary
lymphoid follicle region. In this region, mature antibodies and
memory B cells are generated (62).
Several studies have indicated that miR-155 is indispensable for GC
responses and the production of Ig class-switched plasma cells
(63). Compared with wild-type mice,
miR-155-deficient mice were demonstrated to have fewer and smaller
GCs (62). In addition, the
secretion of antigen-specific antibodies was significantly
decreased. The production of TNF-γ and lymphotoxin (LT)-α was
significantly reduced in miR-155−/− B cells, and TNF-γ
and LT-α were reported to be important for GC formation (58). These results demonstrate that
appropriate B-cell function depends on the normal expression of
miR-155. Several potential targets of miR-155 have been identified
in B lymphocytes.
GC can be distinguished to two distinct areas,
called light and dark zones. Light zone B cells capture and present
the processed antigen on MHC complexes to T cells. The recruitment
of polycomb repressive complex 2 to its specific targets may be
disturbed by Jarid2, and Jarid2 has the potential to control the
proliferation and differentiation of cells (64). Studies have demonstrated that it is a
target of miR-155. Nakagawa et al (65) revealed that miR-155 reduces apoptosis
in GC B cells and promotes the survival of c-MYC+ light
zone B cells by directly inhibiting Jarid2 expression; thus,
miR-155 activates the GC response and affinity maturation. Various
studies have suggested that PU.1 is required in the early stage of
B-cell differentiation. It is located downstream of the B-cell
receptor signaling pathway (66). In
wild-type B cells, the expression of Pu.1 was low and difficult to
detect. By contrast, its expression was readily detected in
miR-155-deficient B cells. Overexpression of PU.1 may impair
class-switch recombination and lead to defective generation of
IgG1+ cells in stimulated wild-type B cells (59,63).
PU.1 is presumably a negative regulator of plasma-cell
differentiation and its repression by miR-155 is necessary for the
GC reaction in B cells and for IgG1+ B-cell generation
(63). Somatic hypermutation and
class-switch recombination are two critical steps in the GC
reaction to generate memory B cells and plasma cells. These two
processes involve DNA mutagenesis and double-strand DNA breaks,
which must be fine-tuned for a successful GC response (59). Bouamar et al (67) discovered an important role of miR-155
in decreasing double-strand DNA breaks and the p53-induced DNA
damage response in the GC reaction. Further studies have revealed
that miR-155 exerts these effects by directly targeting
activation-induced cytidine deaminase (AICDA) and SOCS1 (67). In miR-155 knockdown mice, SMAD5
contributes to the impaired GC response and a decrease in the
number of GC B cells. It has been demonstrated that miR-155
directly targets SMAD5, thus contributing to mature B-cell
development (68,69).
T lymphocytes
T cells are functional immune cells; they induce B
cells and adjust the role of immune cells. CD4+ T cells
mainly consist of the following types: Th1, Th2, T-regulatory cells
(Tregs) and Th17 (16). Effector
CD8+ T cells are cytotoxic and secrete pro-inflammatory
factors (70). The relative
frequency of T-cell subgroups and relatively stable conditions are
crucial for the immune system balance. Several analyses have
demonstrated that miR-155 is involved in the regulation of
T-lymphocyte function (71). One
study has indicated that CD4+ T cells are most likely to
differentiate into Th2 cells when miR-155 is knocked down (58). Next, several mechanisms and targets
of miR-155 associated with the regulation of T lymphocytes will be
discussed. miR-155 may affect T-cell proliferation via direct
targeting of arginase (Arg)2, a semi-essential amino acid involved
in multiple metabolic and cellular processes. Dunand-Sauthier et
al (43) identified Arg2 mRNA as
a direct target of miR-155. In activated DCs, miR-155 represses the
expression of Arg2 and thus prevents excessive arginine depletion
in the extracellular microenvironment, which is critical for mature
DCs to initiate T-cell responses. They confirmed that miR-155
actively regulates the function of T cells by directly targeting
Arg2 (28,43). c-Maf induces the differentiation of T
cells into Th2 cells. Furthermore, it also induces Th2 cells to
produce IL-4, IL-5 and IL-10 (72).
Rodriguez et al (58)
demonstrated that miR-155 directly targets c-Maf. CD4+ T
cells with sufficient miR-155 were able to suppress the
differentiation of naive cells into Th2 cells by downregulating
c-Maf. SOCS1 is an important target of miR-155 in T lymphocytes.
CD4+CD25+ Tregs are a repressive Th cell
subgroup and secrete anti-inflammatory cytokines. They are able to
maintain self-tolerance and prevent inflammatory and immune
disorders. Th17 mainly promotes the inflammatory response and
contributes to immune defence (73,74). Yao
et al (75) confirmed that
miR-155 positively drives Treg/Th17-cell differentiation by
directly targeting SOCS1. Furthermore, Lu et al (76) have demonstrated that miR-155
contributes to the competitive fitness and proliferation of Tregs
by targeting SOCS1. In T cells, the ratio of Th1 and Th2 cell
differentiation is regulated by the levels of SOCS1. Higher amounts
of SOCS1 curb naive cell differentiation into Th1 cells. miR-155
can induce Th1 cell differentiation by down-regulating SOCS1
(77). CD8+ T cells are the major
effector cells in the immune system. Dudda et al (78) concluded that miR-155 is a critical
regulator of SOCS1 in CD8+ T cells. The impaired
antiviral response in miR-155−/−CD8+ T cells
may be due to increased expression of SOCS1. The downregulation of
SOCS1 mediated by miR-155 is indispensable for efficient
CD8+ T-cell responses (78).
Pathways associated with the miR-155/SOCS1
axis in the immune system
The studies mentioned above indicate that the
miR-155/SOCS1 axis has vital roles in immune system regulation by
mediating a variety of pathways, which are illustrated in Fig. 1 and further discussed below.
The Janus kinase (JAK)/signal
transducer and activator of transcription (STAT) pathway
The JAK/STAT pathway mediates cell development,
differentiation and apoptosis. This pathway leads to the expression
of IFN-inducible genes and drives M1 polarization (35). In DCs, it regulates IL-12 production
and DC maturation (48,79). Furthermore, the JAK/STAT pathway
contributes to Treg and Th17 differentiation, and regulates the
competitive ability and proliferation of Tregs (75,76).
SOCS1 negatively controls this signaling pathway. When STATs are
activated, they induce the transcription of SOCS1 and,
subsequently, SOCS1 proteins bind to phosphorylated JAKs to turn
off the pathway (80). miR-155
activates the JAK/STAT pathway by directly targeting SOCS1
(Fig. 2).
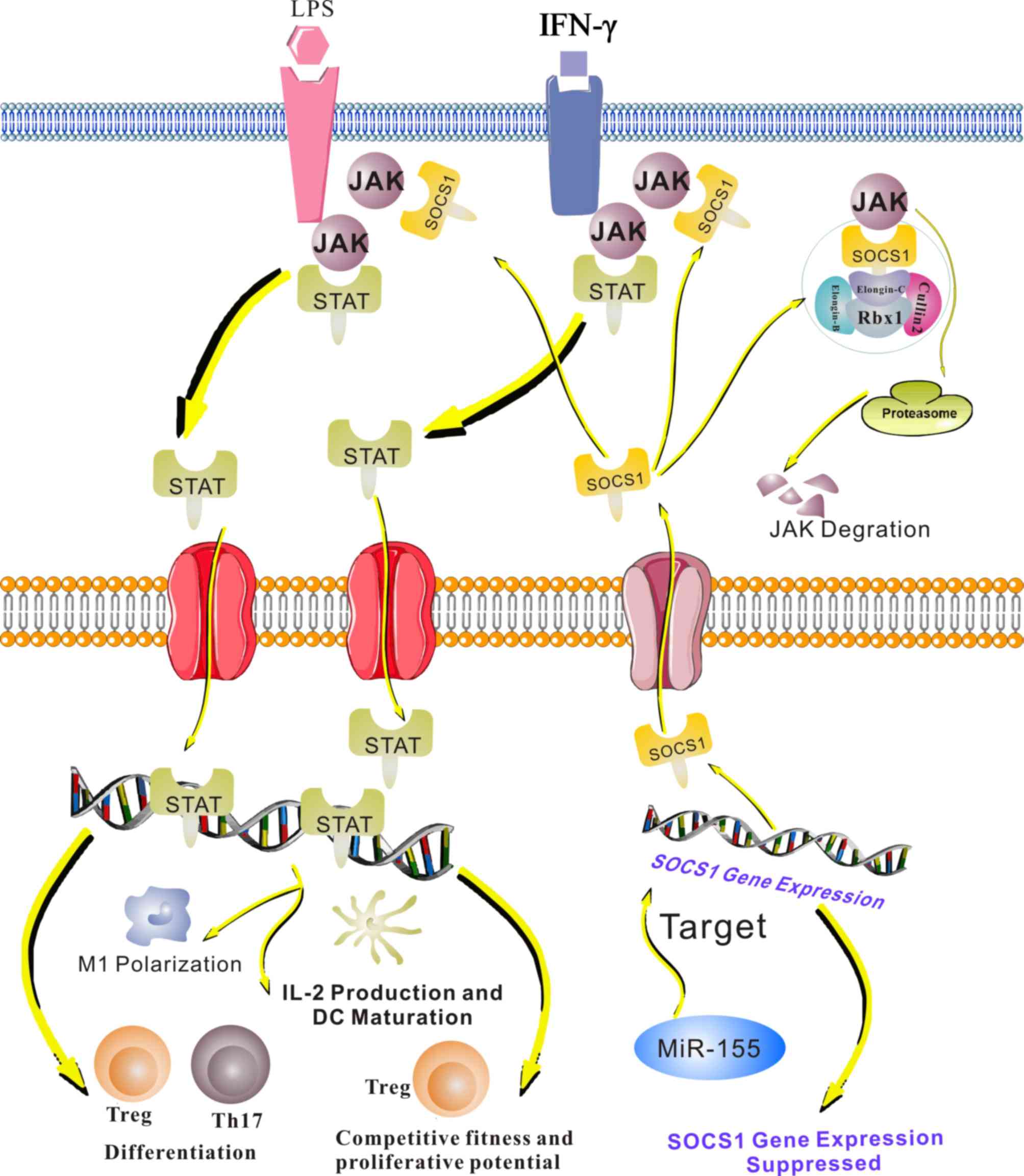 | Figure 2.The JAK/STAT pathway is involved in
the miR155/SOCS1 axis in the immune system. When immune cells are
stimulated by cytokines, including IFN-γ and LPS, the JAK/STAT
pathway is activated. Phosphorylated STATs form a complex and are
translocated to the nucleus, where they activate or repress the
transcription of target genes. In macrophages, this pathway induces
M1 polarization and promotes the production of tumor necrosis
factor-α, IL-6 and IL-1β. JAK/STAT signaling also regulates IL-12
production and DC maturation. Furthermore, the JAK-STAT pathway
contributes to Treg and Th17 differentiation and regulates the
competitive ability and proliferative potential of Tregs. SOCS1
negatively regulates the JAK/STAT pathway by binding to
phosphorylated JAKs. miR-155 targets SOCS1 and inhibits it
expression, leading to decreased binding of SOCS1 to phosphorylated
JAKs. Therefore, miR-155 positively regulates the JAK/STAT pathway.
JAK, Janus kinase; STAT, signal transducer and activator of
transcription; IFN, interferon; LPS, lipopolysaccharide; miR,
microRNA; SOCs, suppressor of cytokine signaling; Th, T-helper
cell; Treg, T-regulatory cell; IL, interleukin; rbx, ring box; DC,
dendritic cell. |
The nuclear factor (NF)-κB
pathway
The NF-κB pathway is essential for the development
of lymphoid organ structures and immune responses. LPS, TNF and
IL-1 are able to activate the NF-κB pathway (81). In the innate immune system, TLRs
identify viral and bacterial products, resulting in NF-κB pathway
activation (82). NF-κB molecules
translocate into the nucleus, where they regulate target gene
expression. In macrophages, the NF-κB pathway drives M1
polarization (83). In T cells, the
activation of this pathway induces the expression of
NF-κB-dependent genes, which are required for T-cell activation and
proliferation (84). P65 is a
subunit of NF-κB. TIR domain-containing adaptor protein (TIRAP) and
IL-1R-associated kinase 1 (IRAK) are adaptor molecules of TLR.
SOCS1 is able to bind to P65, TIRAP and IRAK to suppress the NF-κB
pathway. miR-155 promotes this pathway by directly targeting SOCS1
(Fig. 3) (85,86).
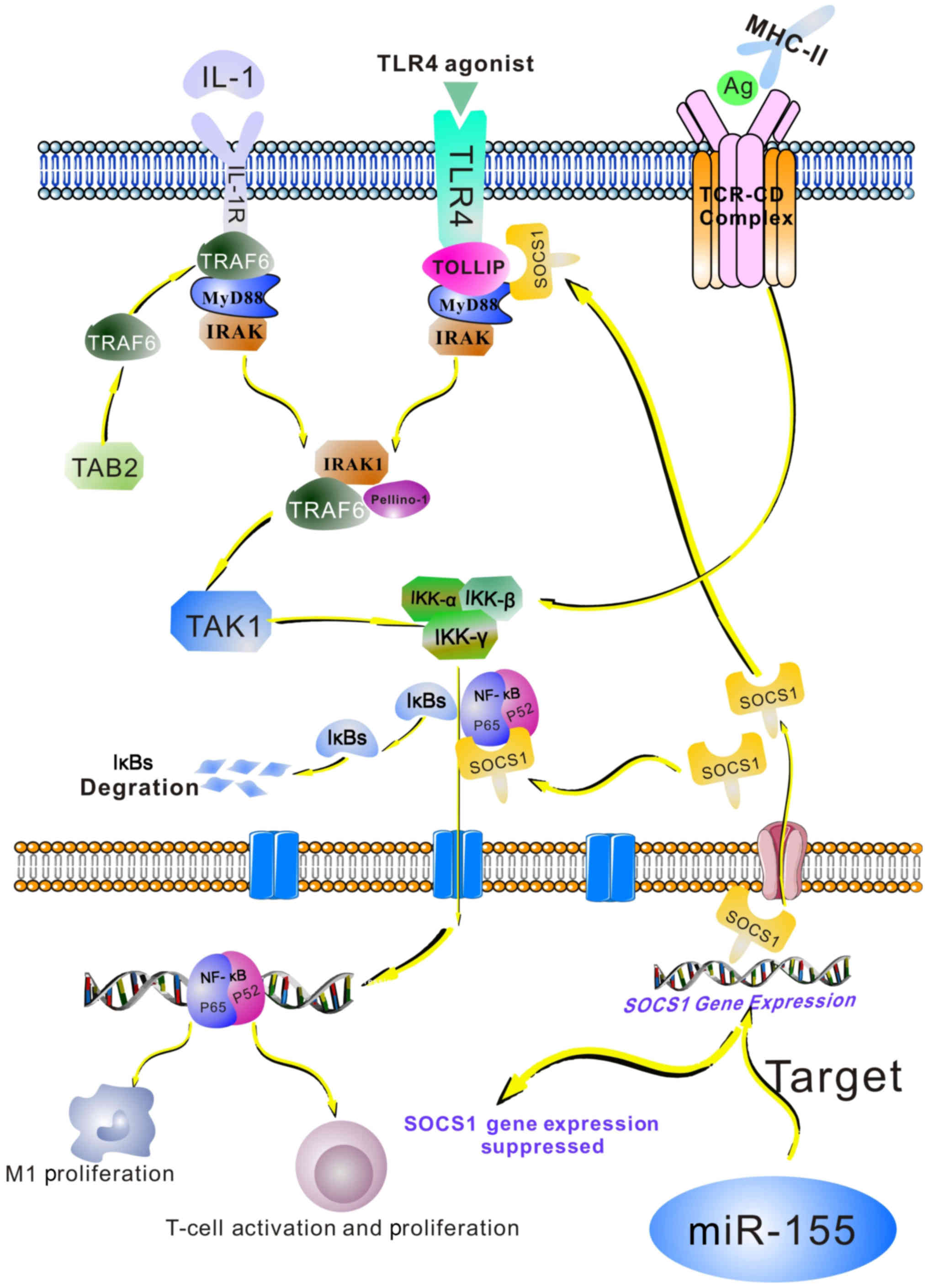 | Figure 3.Role of the NF-κB pathway in the
miR-155/SOCS1 axis in the immune system. Exposure of cells to
lipopolysaccharide or inflammatory cytokines, including TNF or
IL-1, as well as the recognition of bacterial and viral products by
Toll-like receptors on immune cells, results in NF-κB induction.
The interactions between T-cell receptors and CD28 receptors with
their ligands, MHC class II, and the co-stimulatory molecules CD80
and CD86 on the surface of antigen-presenting cells, trigger the
NF-κB pathway in T-cells. NF-κB becomes activated and translocates
to the nucleus, where it induces the expression of its target
genes. This pathway may induce M1 macrophage polarization. In
T-cells, it may lead to the expression of NF-κB-dependent genes,
including IL-2, IL-2R and interferon, which are required for T-cell
activation and proliferation. SOCS1 may regulate NF-κB signaling by
binding to the p65 subunit of NF-κB, the TLR adaptor molecule
TIRAP, as well as IRAK. miR-155 targets SOCS1 and inhibits its
expression, therefore indirectly promoting the NF-κB pathway. Ag,
antigen; TCR-CD, T cell receptor CD; NF, nuclear factor; IκB,
inhibitor of NF-κB; TLR, Toll-like receptor; miR, microRNA; SOCs,
suppressor of cytokine signaling; MHC, major histocompatibility
complex; IL, interleukin; IL-2R, IL-2 receptor; TIRAP, TIR
domain-containing adaptor protein; IRAK, IL-1R-associated kinase 1;
TOLLIP, Toll-interacting protein; MyD88, innate immune signal
transduction adaptor; TRAG6, TNF receptor-associated factor 6; TNF,
tumor necrosis factor; TAB2, TAK1-binding protein 2; TAK1,
transforming growth factor-β-activated kinase 1; IKK, IκB
kinase. |
The P53 pathway
P53 inhibits the occurrence and development of
tumors. It is involved in the regulation of cell growth and
differentiation. Furthermore, it promotes DNA repair. In cells
under stress associated with DNA damage, hypoxia, cytokines or
oncogenes, p53 accumulates in the nucleus to exert its functions
(87). The maturation of B cells
involves DNA mutations, double-strand DNA breaks and the activation
of p53 (88). SOCS1 may combine with
p53 and improve its phosphorylation, transcription and activity,
providing a safeguard role in limiting the extent of DNA damage and
p53 activity. MiR-155 directly targets SOCS1 and regulates p53
function to appropriate levels in B cells (67).
Research on immune factors associated with
OLP
Immune factors have important roles in the
pathogenesis of OLP. Numerous studies have demonstrated that immune
cells are closely linked to OLP.
Innate immune system
Macrophages are normally present in the inflammatory
infiltrate of lesions in chronic OLP (89). M1 macrophages induce pro-inflammatory
agents, including TNF-α, which may trigger the apoptosis of basal
keratinocytes and induce basement membrane damage. In addition, M1
macrophages may increase inflammatory cells in lesions by inducing
keratinocytes to secrete more chemokines (90). Therefore, macrophages actively
participate in the pathogenesis of OLP. In addition, OLP-associated
macrophages may induce matrix metalloproteinases (MMPs) and TNF to
influence oral epithelial and sub-epithelial stromal cells. In
vitro, MMPs have been demonstrated to be associated with
extracellular matrix damage and activation. Furthermore, they also
promote tumorigenesis. TNF is involved in tumor growth and
invasion, and in cutaneous tumors, it contributes to tumor
development in the early phase. Taken together, OLP-associated
macrophages may participate in processes of the progression of OLP
to cancer (91).
Previous studies have revealed an increase in DCs in
OLP lesions, suggesting that these cells are involved in the
occurrence of OLP (92–94). Santoro et al (92) observed that skin-resident LCs, pDCs
and DC-SIGN+ DCs were markedly upregulated in OLP
lesions compared with those in a healthy control group. LCs are
mainly situated in the epithelium. As a guardian of the oral
mucosa, they may influence the responses of the immune system to
external and endogenous antigens. Gueiros et al (93) evaluated the distribution and
concentration of LCs in OLP and observed more LCs in OLP lesions
than in adjacent non-inflammatory tissues and normal mucosae. Once
LCs identify antigens, they may be activated to stimulate T cells.
pDCs may be detected in most patients with OLP and they are
principally located in single epithelial cells and the lymphoid
infiltrate zone (92). One of their
functions is to secrete IFN-α. An imbalance between pDCs and IFN-α
is linked to the occurrence of OLP (95). Yamauchi et al (94) reported that thymic stromal
lymphopoietin (TSLP) receptor+ mDCs are responsible for
abnormal Th2 immune responses and the occurrence of OLP by
regulating TSLP production. These studies suggest that DCs may be
involved in the pathogenesis of OLP.
A limited number of studies have assessed the impact
of NK cells in OLP. In the inflamed skin of the lichen planus,
substantial NK cells may be detected. Furthermore, in lesions of
OLP, they co-exist with pDCs in a chemerin-dependent manner.
NK-cell dysfunction is closely linked to the occurrence of OLP
(96).
Adaptive immune system
Local immune factors, including IgG and IgA
responses, have defense functions in OLP. There are two types of
Ig, i.e., membrane-bound receptors located on the cytomembrane of B
cells and dissoluble molecules produced by plasma cells. The
occurrence of OLP is thought to be associated with Ig levels. IgG
and IgA are the predominant Igs in normal serum (97). The levels of IgG and IgA in OLP
remain to be clarified, and differences exist among previous
studies. Certain studies have identified elevated serum IgG and IgA
levels in OLP (98). However, Divya
and Sathasivasubramanian (97)
reported that the average serum levels of IgG were slightly
increased and the average levels of IgA were somewhat reduced in
OLP compared with those in the controls. They also reported that
the amounts of salivary IgG and IgA were low in patients with OLP
and healthy individuals (97).
However, other studies have indicated that the levels of salivary
IgG and IgA are higher in patients with OLP than those in healthy
individuals (99). Ghaleyani et
al (100) also observed
significantly higher salivary IgG and IgA levels in an OLP group
compared with those in a control group. Differences in the stage of
OLP at which samples were obtained may explain for the differences
among studies. Taken together, the observed alterations in the
levels of IgG and IgA implicate Igs in the occurrence of OLP.
OLP is a chronic mucosal inflammatory disease
mediated by T cells. The quantity of CD4+ T cells in OLP
was obviously greater than that in control subjects, suggesting
that CD4+ T cells are involved in the pathogenesis of
OLP. CD4+ T cells are mainly distributed in the lamina
propria and subepithelial layer of OLP lesions (16). Once activated, CD4+ T
cells bind to receptors located in the cytomembrane of
CD8+ T lymphocytes, and they secrete pro-inflammatory
cytokines. Finally, cytotoxic T lymphocytes are induced and
contribute to chronic inflammation in local lesions (90). In addition, Tregs maintain peripheral
immunological tolerance and control the proliferation of
conventional T lymphocytes. A recent study indicated that, despite
increased Tregs in OLP, the functions of these cells were
defective. This may explain, at least in part, why the increased
Tregs do not reduce the occurrence of OLP (101). In the intra-epithelial area and the
basement membrane of OLP lesions, CD8+ T cells are
present and are associated with apoptotic keratinocytes in OLP
(102). These results suggested
that CD8+ T cells contribute to the occurrence of OLP,
and that CD8+ T cells induce apoptosis in keratinocytes
in this disease. CD8+ T lymphocytes identify and
potentially eliminate antigens presented by class I MHC molecules.
Once CD8+ T cells are activated, they may induce
keratinocyte apoptosis via secretion of TNF-α and granzyme B, as
well as Fas ligand receptor expression (90). In brief, these results suggest that T
lymphocytes are involved in the pathogenesis of OLP.
Conclusions
OLP is a chronic oral mucosal inflammatory disease
and has been identified as a potentially malignant condition by the
WHO. Extensive evidence suggests that immunologic factors have
vital roles in the pathogenesis of OLP. An improved understanding
of the mechanisms underlying OLP may provide novel approaches for
potential therapeutic strategies for this disease. Several studies
have indicated that miR-155 expression is significantly upregulated
in OLP (6). However, the roles of
miR-155, a multi-functional miRNA associated with immune system
regulation, in OLP have remained to be fully elucidated. miR-155
controls macrophage differentiation and function by directly
targeting SOCS1, IL13Rα1 and SMAD2 (32–34). In
DCs, miR-155 targets TAB2, PU.1, SHIP1, SOCS1 and c-Fos to regulate
cell maturation and function (41,42,46–48,50). In
addition, miR-155 promotes the production of IFN-γ in NK cells by
decreasing the expression of SHIP1 (54). It also suppresses PMAIP1 and SOCS1
levels to increase NK cell survival and homeostasis (55). miR-155 has a crucial role in the
adaptive immune system. It directly targets Jarid2, PU.1, AICDA,
SOCS1 and SMAD5 to maintain an adequate B-cell function (63,65,67–69).
Furthermore, c-Maf, SOCS1 and Arg2 have been identified as targets
of miR-155 in T lymphocytes (43,59,75–78).
Previous studies have demonstrated that these immune cells actively
participate in the pathogenesis of OLP. The miR-155/SOCS1 axis has
important roles in immune system regulation by mediating a variety
of signaling pathways, including the JAK/STAT, NF-κB and P53
pathways. NF-κB and associated cytokines, including IL-1α, IL-6,
IL-8 and TNF-α, have beneficial effects in the inflammatory
microenvironment of OLP (103). The
expression of p53 is upregulated in OLP, but the reason for this
remains to be determined. The increased p53 expression may be due
to DNA damage occurring. In addition, the increased cell
proliferation in OLP may lead to the upregulation of p53
expression. An elevated expression of p53 is thought to be
associated with the pathogenesis of OLP (104).
miR-155 is upregulated in OLP, but the underlying
mechanisms remain to be fully elucidated. It is well known that
miR-155 is important for immune system regulation. Immune
regulation has an important role in the pathogenesis of OLP.
Therefore, it is possible that miR-155 is involved in the
pathogenesis of OLP by regulating the immune system and associated
targets, including SOCS1. Ostensibly, this interaction requires
further experimental validation. With this regard, the present
review provides a basis for future research and may facilitate
further investigation of the pathogenesis of OLP, particularly with
regard to the roles of miR-155.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the
Non-profit Industry Research Specific Fund of the National Health
and Family Planning Commission of China (grant no. 201502018), the
National Natural Science Foundation of China (grant nos. 81470747,
81520108009, 81621062, 81200791, 81472533, 81102060 and 81270040)
and the 111 Project of the Ministry of Education, China (grant no.
B14038).
Availability of data and materials
Not applicable.
Authors' contributions
YT collected and analyzed all literature, and wrote
the manuscript. RA and YH also collected the literature. LJ, HD, NJ
and XZ analyzed the data. YZ and QC designed and conceived the
current study. All authors have read and approved the final version
of the manuscript.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Alrashdan MS, Cirillo N and McCullough M:
Oral lichen planus: A literature review and update. Arch Dermatol
Rese. 308:539–551. 2016. View Article : Google Scholar
|
|
2
|
Sanketh DS, Patil S and Swetha B: Oral
lichen planus and epithelial dysplasia with lichenoid features: A
review and discussion with special reference to diagnosis. J
Investig Clin Dent. 8:e122332017. View Article : Google Scholar
|
|
3
|
Gonzalez-Moles MA, Scully C and
Gil-Montoya JA: Oral lichen planus: Controversies surrounding
malignant transformation. Oral Dis. 14:229–243. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kurago ZB: Etiology and pathogenesis of
oral lichen planus: An overview. Oral Surg Oral Med Oral Pathol
Oral Radiol. 122:72–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roopashree MR, Gondhalekar RV, Shashikanth
MC, George J, Thippeswamy SH and Shukla A: Pathogenesis of oral
lichen planus-a review. J Oral Pathol Med. 39:729–734. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arão TC, Guimarães AL, de Paula AM, Gomes
CC and Gomez RS: Increased miRNA-146a and miRNA-155 expressions in
oral lichen planus. Arch Dermatol Res. 304:371–375. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma H, Wu Y, Yang H, Liu J, Dan H, Zeng X,
Zhou Y, Jiang L and Chen Q: MicroRNAs in oral lichen planus and
potential miRNA-mRNA pathogenesis with essential cytokines: A
review. Oral Surg Oral Med Oral Pathol Oral Radiol. 122:164–173.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gassling V, Hampe J, Açil Y, Braesen JH,
Wiltfang J and Häsler R: Disease-associated miRNA-mRNA networks in
oral lichen planus. PLoS One. 8:e630152013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu F, Wu J and Ye F: Expression of
miRNA-155 and miRNA-146a in peripheral blood mononuclear cells and
plasma of oral lichen planus patients. Zhonghua Kou Qiang Yi Xue Za
Zhi. 50:23–27. 2015.(In Chinese). PubMed/NCBI
|
|
12
|
Moffett HF and Novina CD: A small RNA
makes a Bic difference. Genome Biol. 8:2212007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sagari S, Sanadhya S, Doddamani M and
Rajput R: Molecular markers in oral lichen planus: A systematic
review. J Oral Maxillofac Pathol. 20:115–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eisen D: The clinical features, malignant
potential, and systemic associations of oral lichen planus: A study
of 723 patients. J Am Acad Dermatol. 46:207–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aghbari SMH, Abushouk AI, Attia A,
Elmaraezy A, Menshawy A, Ahmed MS, Elsaadany BA and Ahmed EM:
Malignant transformation of oral lichen planus and oral lichenoid
lesions: A meta-analysis of 20095 patient data. Oral Oncol.
68:92–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eisen D, Carrozzo M, Bagan Sebastian JV
and Thongprasom K: Number V Oral lichen planus: Clinical features
and management. Oral Dis. 11:338–349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Zhang D, Han Q, Zhao X, Zeng X, Xu
Y, Sun Z and Chen Q: Role of distinct CD4(+) T helper subset in
pathogenesis of oral lichen planus. J Oral Pathol Med. 45:385–393.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan YQ, Li Q, Zhang J, Du GF, Lu R and
Zhou G: Increased circulating CXCR5+ CD4+ T
follicular helper-like cells in oral lichen planus. J Oral Pathol
Med. 46:803–809. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Raisch J, Darfeuille-Michaud A and Nguyen
HT: Role of microRNAs in the immune system, inflammation and
cancer. World J Gastroenterol. 19:2985–2996. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Contreras J and Rao DS: MicroRNAs in
inflammation and immune responses. Leukemia. 26:404–413. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cremer TJ, Ravneberg DH, Clay CD,
Piper-Hunter MG, Marsh CB, Elton TS, Gunn JS, Amer A, Kanneganti
TD, Schlesinger LS, et al: MiR-155 induction by F. novicida but not
the virulent F. tularensis results in SHIP down-regulation and
enhanced pro-inflammatory cytokine response. PLoS One. 4:e85082009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tili E, Michaille JJ and Croce CM:
MicroRNAs play a central role in molecular dysfunctions linking
inflammation with cancer. Immunol Rev. 253:167–184. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Faraoni I, Antonetti FR, Cardone J and
Bonmassar E: miR-155 gene: A typical multifunctional microRNA.
Biochim Biophys Acta. 1792:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu JY, Zhang J, Ma JZ, Liang XY, Chen GY,
Lu R, Du GF and Zhou G: MicroRNA-155-IFN-γ feedback loop in CD4(+)T
cells of erosive type oral lichen planus. Sci Rep. 5:169352015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rasmussen SB, Reinert LS and Paludan SR:
Innate recognition of intracellular pathogens: Detection and
activation of the first line of defense. APMIS. 117:323–337. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vigorito E, Kohlhaas S, Lu D and Leyland
R: miR-155: An ancient regulator of the immune system. Immunol Rev.
253:146–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mills CD, Kincaid K, Alt JM, Heilman MJ
and Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J
Immunol. 164:6166–6173. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mantovani A, Biswas SK, Galdiero MR, Sica
A and Locati M: Macrophage plasticity and polarization in tissue
repair and remodelling. J Pathol. 229:176–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma F, Liu F, Ding L, You M, Yue H, Zhou Y
and Hou Y: Anti-inflammatory effects of curcumin are associated
with down regulating microRNA-155 in LPS-treated macrophages and
mice. Pharm Biol. 55:1263–1273. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martinez-Nunez RT, Louafi F and
Sanchez-Elsner T: The interleukin 13 (IL-13) pathway in human
macrophages is modulated by microRNA-155 via direct targeting of
interleukin 13 receptor alpha1 (IL13Ralpha1). J Biol Chem.
286:1786–1794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Louafi F, Martinez-Nunez RT and
Sanchez-Elsner T: MicroRNA-155 targets SMAD2 and modulates the
response of macrophages to transforming growth factor-{beta}. J
Biol Chem. 285:41328–41336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wilson HM: SOCS proteins in macrophage
polarization and function. Front Immunol. 5:3572014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fuss IJ and Strober W: The role of IL-13
and NK T cells in experimental and human ulcerative colitis.
Mucosal Immunol. 1 Suppl 1:S31–S33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Turner ML, Schnorfeil FM and Brocker T:
MicroRNAs regulate dendritic cell differentiation and function. J
Immunol. 187:3911–3917. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iwasaki A and Medzhitov R: Regulation of
adaptive immunity by the innate immune system. Science.
327:291–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ceppi M, Pereira PM, Dunand-Sauthier I,
Barras E, Reith W, Santos MA and Pierre P: MicroRNA-155 modulates
the interleukin-1 signaling pathway in activated human
monocyte-derived dendritic cells. Proc Natl Acad Sci USA.
106:2735–2740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martinez-Nunez RT, Louafi F, Friedmann PS
and Sanchez-Elsner T: MicroRNA-155 modulates the pathogen binding
ability of dendritic cells (DCs) by down-regulation of DC-specific
intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN).
J Biol Chem. 284:16334–16342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dunand-Sauthier I, Irla M, Carnesecchi S,
Seguín-Estévez Q, Vejnar CE, Zdobnov EM, Santiago-Raber ML and
Reith W: Repression of arginase-2 expression in dendritic cells by
microRNA-155 is critical for promoting T cell proliferation. J
Immunol. 193:1690–1700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ma YL, Ma ZJ, Wang M, Liao MY, Yao R and
Liao YH: MicroRNA-155 induces differentiation of RAW264.7 cells
into dendritic-like cells. Int J Clin Exp Pathol. 8:14050–14062.
2015.PubMed/NCBI
|
|
45
|
Caparrós E, Munoz P, Sierra-Filardi E,
Serrano-Gómez D, Puig-Kröger A, Rodríguez-Fernández JL, Mellado M,
Sancho J, Zubiaur M and Corbí AL: DC-SIGN ligation on dendritic
cells results in ERK and PI3K activation and modulates cytokine
production. Blood. 107:3950–3958. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lind EF, Millar DG, Dissanayake D, Savage
JC, Grimshaw NK, Kerr WG and Ohashi PS: miR-155 upregulation in
dendritic cells is sufficient to break tolerance in vivo by
negatively regulating SHIP1. J Immunol. 195:4632–4640. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Costinean S, Sandhu SK, Pedersen IM, Tili
E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J,
Kauffman LR, et al: Src homology 2 domain-containing
inositol-5-phosphatase and CCAAT enhancer-binding protein beta are
targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice.
Blood. 114:1374–1382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Evel-Kabler K, Song XT, Aldrich M, Huang
XF and Chen SY: SOCS1 restricts dendritic cells' ability to break
self tolerance and induce antitumor immunity by regulating IL-12
production and signaling. J Clin Invest. 116:90–100. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yoshida R, Suzuki M, Sakaguchi R, Hasegawa
E, Kimura A, Shichita T, Sekiya T, Shiraishi H, Shimoda K and
Yoshimura A: Forced expression of stabilized c-Fos in dendritic
cells reduces cytokine production and immune responses in vivo.
Biochem Biophys Res Commun. 423:247–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dunand-Sauthier I, Santiago-Raber ML,
Capponi L, Vejnar CE, Schaad O, Irla M, Seguín-Estévez Q, Descombes
P, Zdobnov EM, Acha-Orbea H and Reith W: Silencing of c-Fos
expression by microRNA-155 is critical for dendritic cell
maturation and function. Blood. 117:4490–4500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sullivan RP, Fogel LA, Leong JW, Schneider
SE, Wong R, Romee R, Thai TH, Sexl V, Matkovich SJ, Dorn GW II, et
al: MicroRNA-155 tunes both the threshold and extent of NK cell
activation via targeting of multiple signaling pathways. J Immunol.
191:5904–5913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Trotta R, Chen L, Costinean S, Josyula S,
Mundy-Bosse BL, Ciarlariello D, Mao C, Briercheck EL, McConnell KK,
Mishra A, et al: Overexpression of miR-155 causes expansion, arrest
in terminal differentiation and functional activation of mouse
natural killer cells. Blood. 121:3126–3134. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Trotta R, Parihar R, Yu J, Becknell B,
Allard J II, Wen J, Ding W, Mao H, Tridandapani S, Carson WE and
Caligiuri MA: Differential expression of SHIP1 in CD56bright and
CD56dim NK cells provides a molecular basis for distinct functional
responses to monokine costimulation. Blood. 105:3011–3018. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Trotta R, Chen L, Ciarlariello D, Josyula
S, Mao C, Costinean S, Yu L, Butchar JP, Tridandapani S, Croce CM
and Caligiuri MA: miR-155 regulates IFN-γ production in natural
killer cells. Blood. 119:3478–3485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zawislak CL, Beaulieu AM, Loeb GB, Karo J,
Canner D, Bezman NA, Lanier LL, Rudensky AY and Sun JC:
Stage-specific regulation of natural killer cell homeostasis and
response against viral infection by microRNA-155. Proc Natl Acad
Sci USA. 110:6967–6972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ikeda J, Tian T, Wang Y, Hori Y, Honma K,
Wada N and Morii E: Expression of FoxO3a in clinical cases of
malignant lymphoma. Pathol Res Pract. 209:716–720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ji WG, Zhang XD, Sun XD, Wang XQ, Chang BP
and Zhang MZ: miRNA-155 modulates the malignant biological
characteristics of NK/T-cell lymphoma cells by targeting FOXO3a
gene. J Huazhong Univ Sci Technolog Med Sci. 34:882–888. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rodriguez A, Vigorito E, Clare S, Warren
MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska
EA, et al: Requirement of bic/microRNA-155 for normal immune
function. Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
de Yebenes VG, Bartolome-Izquierdo N and
Ramiro AR: Regulation of B-cell development and function by
microRNAs. Immunol Rev. 253:25–39. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sandhu SK, Volinia S, Costinean S, Galasso
M, Neinast R, Santhanam R, Parthun MR, Perrotti D, Marcucci G,
Garzon R and Croce CM: miR-155 targets histone deacetylase 4
(HDAC4) and impairs transcriptional activity of B-cell lymphoma 6
(BCL6) in the Emu-miR-155 transgenic mouse model. Proc Natl Acad
Sci USA. 109:20047–20052. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Clare S, John V, Walker AW, Hill JL,
Abreu-Goodger C, Hale C, Goulding D, Lawley TD, Mastroeni P,
Frankel G, et al: Enhanced susceptibility to Citrobacter rodentium
infection in microRNA-155-deficient mice. Infect Immun. 81:723–732.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Thai TH, Calado DP, Casola S, Ansel KM,
Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et
al: Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Vigorito E, Perks KL, Abreu-Goodger C,
Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A,
Bradley A, et al: microRNA-155 regulates the generation of
immunoglobulin class-switched plasma cells. Immunity. 27:847–859.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bolisetty MT, Dy G, Tam W and Beemon KL:
Reticuloendotheliosis virus strain T induces miR-155, which targets
JARID2 and promotes cell survival. J Virol. 83:12009–12017. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Nakagawa R, Leyland R, Meyer-Hermann M, Lu
D, Turner M, Arbore G, Phan TG, Brink R and Vigorito E:
MicroRNA-155 controls affinity-based selection by protecting c-MYC+
B cells from apoptosis. J Clin Invest. 126:377–388. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Busslinger M: Transcriptional control of
early B cell development. Annu Rev Immunol. 22:55–79. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bouamar H, Jiang D, Wang L, Lin AP, Ortega
M and Aguiar RC: MicroRNA 155 control of p53 activity is context
dependent and mediated by Aicda and Socs1. Mol Cell Biol.
35:1329–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jiang D and Aguiar RC: MicroRNA-155
controls RB phosphorylation in normal and malignant B lymphocytes
via the noncanonical TGF-beta1/SMAD5 signaling module. Blood.
123:86–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rai D, Kim SW, McKeller MR, Dahia PL and
Aguiar RC: Targeting of SMAD5 links microRNA-155 to the TGF-beta
pathway and lymphomagenesis. Proc Natl Acad Sci USA. 107:3111–3116.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang N and Bevan MJ: CD8(+) T cells: Foot
soldiers of the immune system. Immunity. 35:161–168. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sanchez-Diaz R, Blanco-Dominguez R,
Lasarte S, Tsilingiri K, Martín-Gayo E, Linillos-Pradillo B, de la
Fuente H, Sánchez-Madrid F, Nakagawa R, Toribio ML and Martín P:
Thymus-derived regulatory T cell development is regulated by C-Type
lectin-mediated BIC/MicroRNA 155 expression. Mol Cell Boil.
37:e003042017.
|
|
72
|
Hwang ES, White IA and Ho IC: An
IL-4-independent and CD25-mediated function of c-maf in promoting
the production of Th2 cytokines. Proc Natl Acad Sci USA.
99:13026–13030. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
O'Garra A and Vieira P: Regulatory T cells
and mechanisms of immune system control. Nat Med. 10:801–805. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bettelli E, Korn T, Oukka M and Kuchroo
VK: Induction and effector functions of T(H)17 cells. Nature.
453:1051–1057. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X
and Liao YH: MicroRNA-155 modulates treg and Th17 cells
differentiation and Th17 cell function by targeting SOCS1. PLoS
One. 7:e460822012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lu LF, Thai TH, Calado DP, Chaudhry A,
Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K and
Rudensky AY: Foxp3-dependent microRNA155 confers competitive
fitness to regulatory T cells by targeting SOCS1 protein. Immunity.
30:80–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Harada M, Nakashima K, Hirota T, Shimizu
M, Doi S, Fujita K, Shirakawa T, Enomoto T, Yoshikawa M, Moriyama
H, et al: Functional polymorphism in the suppressor of cytokine
signaling 1 gene associated with adult asthma. Am J Respir Cell Mol
Biol. 36:491–496. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dudda JC, Salaun B, Ji Y, Palmer DC,
Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM,
Perret R, et al: MicroRNA-155 is required for effector CD8+ T cell
responses to virus infection and cancer. Immunity. 38:742–753.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huffaker TB and O'Connell RM:
miR-155-SOCS1 as a functional axis: Satisfying the burden of proof.
Immunity. 43:3–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Weniger MA, Melzner I, Menz CK, Wegener S,
Bucur AJ, Dorsch K, Mattfeldt T, Barth TF and Möller P: Mutations
of the tumor suppressor gene SOCS-1 in classical hodgkin lymphoma
are frequent and associated with nuclear phospho-STAT5
accumulation. Oncogene. 25:2679–2684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pomerantz JL and Baltimore D: Two pathways
to NF-kappaB. Mol Cell. 10:693–695. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Musikacharoen T, Matsuguchi T, Kikuchi T
and Yoshikai Y: NF-kappa B and STAT5 play important roles in the
regulation of mouse Toll-like receptor 2 gene expression. J
Immunol. 166:4516–4524. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Strebovsky J, Walker P and Dalpke AH:
Suppressor of cytokine signaling proteins as regulators of innate
immune signaling. Front Biosci (Landmark Ed). 17:1627–1639. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li J, Liu Z, Jiang S, Cortesini R,
Lederman S and Suciu-Foca N: T suppressor lymphocytes inhibit
NF-kappa B-mediated transcription of CD86 gene in APC. J Immunol.
163:6386–6392. 1999.PubMed/NCBI
|
|
85
|
Baetz A, Frey M, Heeg K and Dalpke AH:
Suppressor of cytokine signaling (SOCS) proteins indirectly
regulate toll-like receptor signaling in innate immune cells. J
Boil Chem. 279:54708–54715. 2004. View Article : Google Scholar
|
|
86
|
Ryo A, Suizu F, Yoshida Y, Perrem K, Liou
YC, Wulf G, Rottapel R, Yamaoka S and Lu KP: Regulation of
NF-kappaB signaling by Pin1-dependent prolyl isomerization and
ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 12:1413–1426.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ceskova P, Chichger H, Wallace M, Vojtesek
B and Hupp TR: On the mechanism of sequence-specific DNA-dependent
acetylation of p53: The acetylation motif is exposed upon DNA
binding. J Mol Boil. 357:442–456. 2006. View Article : Google Scholar
|
|
88
|
Guikema JE, Linehan EK, Esa N, Tsuchimoto
D, Nakabeppu Y, Woodland RT and Schrader CE: Apurinic/apyrimidinic
endonuclease 2 regulates the expansion of germinal centers by
protecting against activation-induced cytidine
deaminase-independent DNA damage in B cells. J Immunol.
193:931–939. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hirota J, Osaki T and Tatemoto Y:
Immunohistochemical staining of infiltrates in oral lichen planus.
Pathol Res Pract. 186:625–632. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Payeras MR, Cherubini K, Figueiredo MA and
Salum FG: Oral lichen planus: Focus on etiopathogenesis. Arch Oral
Boil. 58:1057–1069. 2013. View Article : Google Scholar
|
|
91
|
Mignogna MD, Fedele S, Lo Russo L, Lo
Muzio L and Bucci E: Immune activation and chronic inflammation as
the cause of malignancy in oral lichen planus: Is there any
evidence ? Oral Oncol. 40:120–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Santoro A, Majorana A, Roversi L, Gentili
F, Marrelli S, Vermi W, Bardellini E, Sapelli P and Facchetti F:
Recruitment of dendritic cells in oral lichen planus. J Pathol.
205:426–434. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gueiros LA, Gondak R, Jorge Junior J,
Coletta RD, Carvalho Ade A, Leão JC, de Almeida OP and Vargas PA:
Increased number of Langerhans cells in oral lichen planus and oral
lichenoid lesions. Oral Surg Oral Med Oral Pathol Oral Radiol.
113:661–666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yamauchi M, Moriyama M, Hayashida JN,
Maehara T, Ishiguro N, Kubota K, Furukawa S, Ohta M, Sakamoto M,
Tanaka A and Nakamura S: Myeloid dendritic cells stimulated by
thymic stromal lymphopoietin promote Th2 immune responses and the
pathogenesis of oral lichen planus. PLoS One. 12:e01730172017.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Trucci VM, Salum FG, Figueiredo MA and
Cherubini K: Interrelationship of dendritic cells, type 1
interferon system, regulatory T cells and toll-like receptors and
their role in lichen planus and lupus erythematosus-a literature
review. Arch Oral Biol. 58:1532–1540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Skrzeczynska-Moncznik J, Stefanska A,
Zabel BA, Kapinska-Mrowiecka M, Butcher EC and Cichy J: Chemerin
and the recruitment of NK cells to diseased skin. Acta Biochim Pol.
56:355–360. 2009.PubMed/NCBI
|
|
97
|
Divya VC and Sathasivasubramanian S:
Estimation of serum and salivary immunoglobulin G and
immunoglobulin A in oral pre-cancer: A study in oral submucous
fibrosis and oral lichen planus. J Nat Sci Biol Med. 5:90–94. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Albanidou-Farmaki E, Kayavis I,
Sideropoulos I, Papanayiotou P and Polymenidis Z: Serum
immunoglobulins IgA, IgG and IgM, and oral lichen planus.
Stomatologia (Athenai). 47:114–120. 1990.(In Greek, Modern).
PubMed/NCBI
|
|
99
|
Sistig S, Vucicevic-Boras V, Lukac J and
Kusic Z: Salivary IgA and IgG subclasses in oral mucosal diseases.
Oral Dis. 8:282–286. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ghaleyani P, Sardari F and Akbari M:
Salivary IgA and IgG in oral lichen planus and oral lichenoid
reactions diseases. Adv Biomed Res. 1:732012.PubMed/NCBI
|
|
101
|
Zhou L, Cao T, Wang Y, Yao H, Du G, Chen
G, Niu X and Tang G: Frequently increased but functionally impaired
CD4+CD25+ regulatory T cells in patients with oral lichen planus.
Inflammation. 39:1205–1215. 2016.PubMed/NCBI
|
|
102
|
Zhou XJ, Sugerman PB, Savage NW, Walsh LJ
and Seymour GJ: Intra-epithelial CD8+ T cells and basement membrane
disruption in oral lichen planus. J Oral Pathol Med. 31:23–27.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Georgakopoulou EA, Achtari MD, Achtaris M,
Foukas PG and Kotsinas A: Oral lichen planus as a preneoplastic
inflammatory model. J Biomed Biotechnol. 2012:7596262012.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ebrahimi M, Boldrup L, Coates PJ, Wahlin
YB, Bourdon JC and Nylander K: Expression of novel p53 isoforms in
oral lichen planus. Oral Oncol. 44:156–161. 2008. View Article : Google Scholar : PubMed/NCBI
|