The causes of heart failure (HF) include ischemic
cardiomyopathy (ICM) and dilated cardiomyopathy (DCM),
hypertension, valvular heart disease, diabetic cardiomyopathy and
congenital heart disease (CHD) (1).
The pathogenesis of HF is associated with myocardial hypertrophy,
fibrosis or necrosis, cardiomyocyte apoptosis,
renin-angiotensin-aldosterone system imbalance and collagen
changes, as well as several other factors (2–7).
MicroRNAs (miRs) are small (~22 nucleotides in
length), single-strand, non-coding RNA sequences derived from
precursors that control gene expression in a variety of
physiological and developmental processes, which are involved in
post-transcriptional regulation of gene expression (8). miR disorders are associated with a
number of human diseases, including diabetes, myocardial infarction
and cardiovascular disease, obesity and cancer. Several studies
have demonstrated that miRs may affect different aspects of the
occurrence and development of HF (9–14). The
association between miRs and HF is discussed in detail below.
Circulating miRs are increasingly recognized as
promising biomarkers, given their stability and resistance to
endogenous RNase (15); these miRs,
to some degree, may also be used as diagnostic biomarkers for
angiocardiopathy. In addition, miRNAs and various types of HF have
complex relationships, as described below.
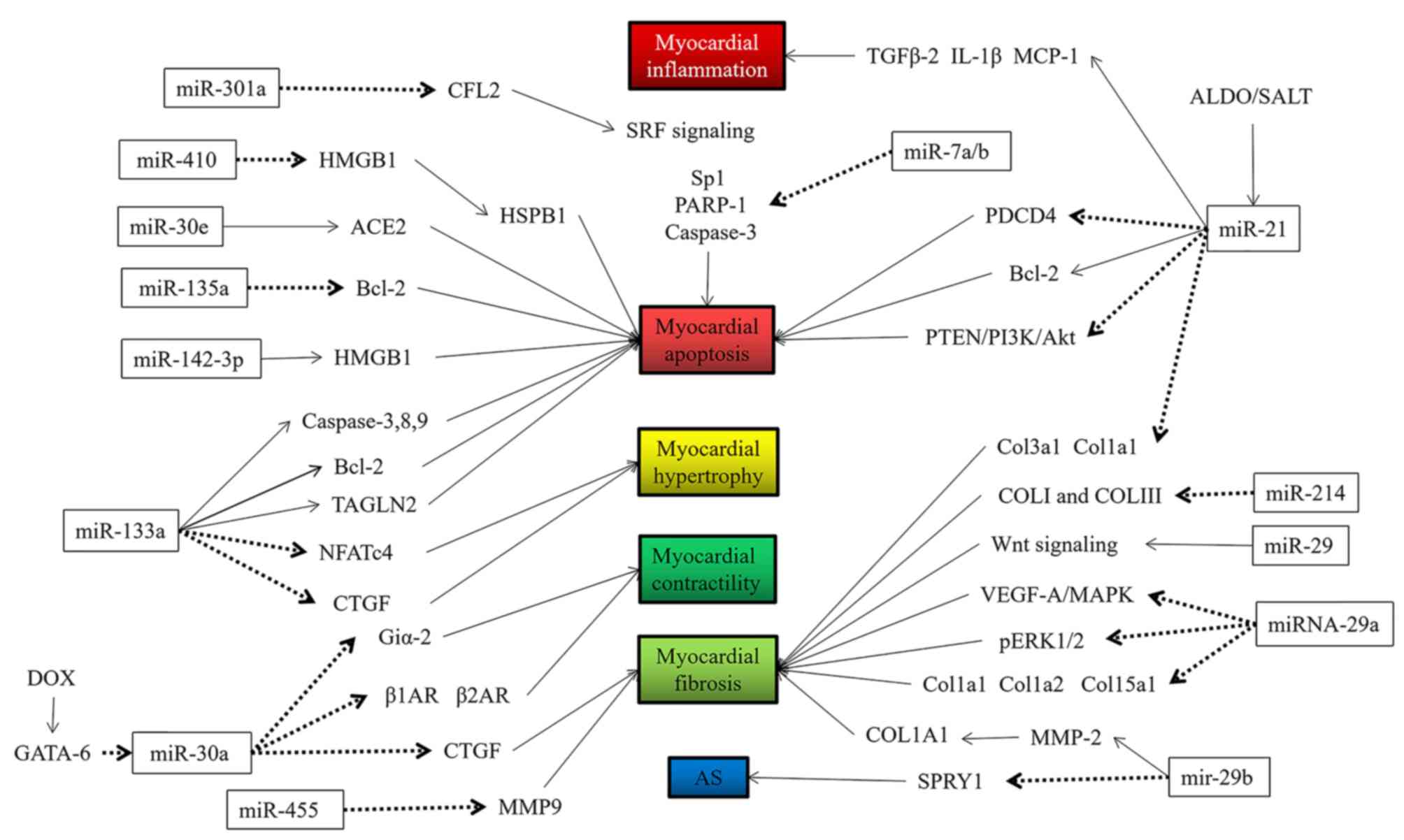
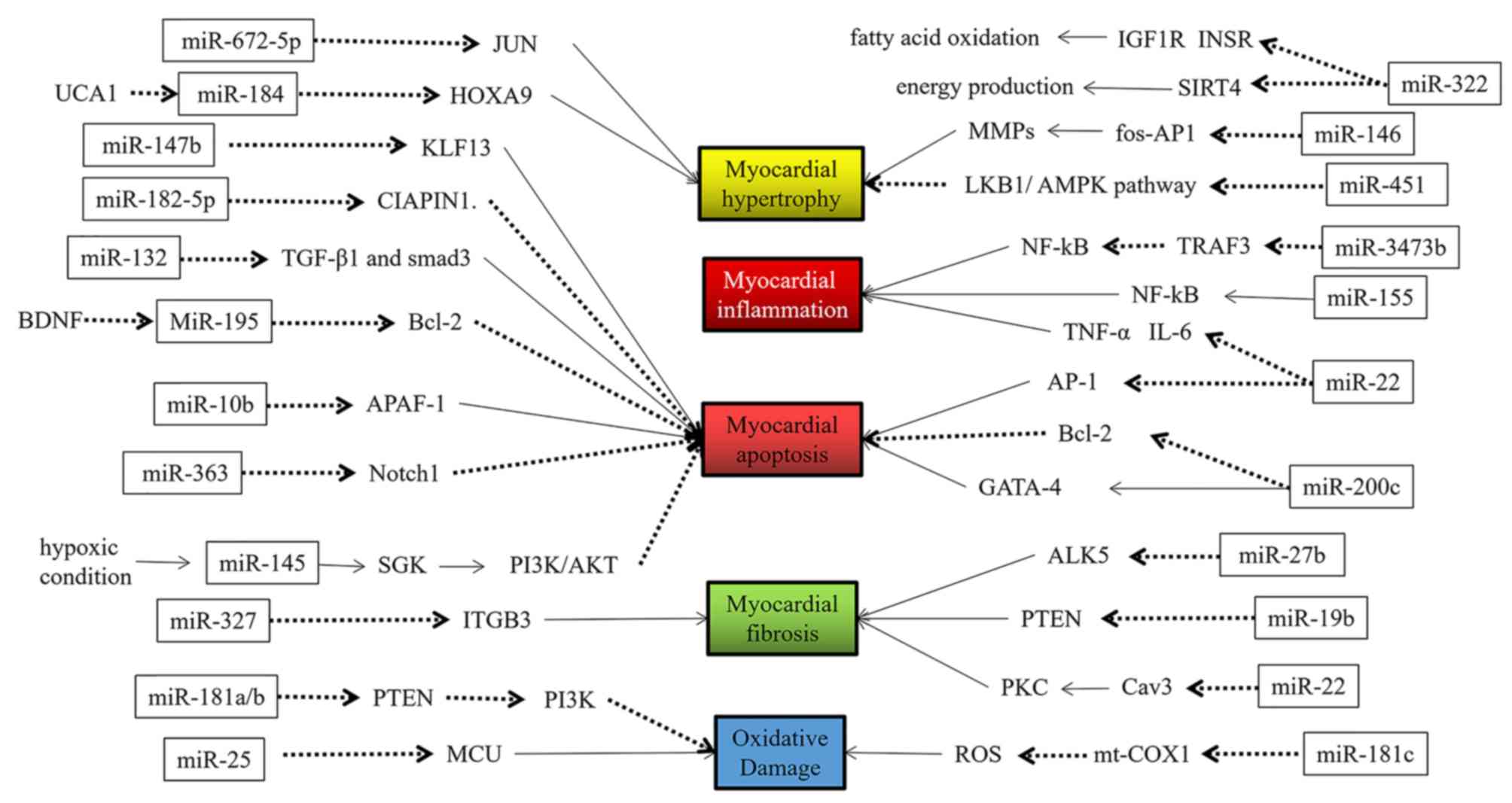
miRs may be involved in several aspects of the
occurrence and development of HF, such as cardiomyocyte apoptosis,
hypertrophy, fibrosis, inflammation, oxidative damage and hypoxic
damage (9–14), among others. The specific regulatory
functions of miRs are indicated in Figs.
1 and 2 and are summarized in
Table I (16–66).
HF is primarily caused by cardiomyopathy,
hypertension, diabetes and CHD, among other causes (15). The different etiology is associated
with several miRs.
Hypertension is an independent risk factor for
cardiac and cerebrovascular disease (84). It has been reported that at least 50%
of patients with long-term hypertension will likely undergo cardiac
remodeling, particularly left ventricular remodeling (85). Myocardial cell hypertrophy is among
the primary causes underlying the occurrence of HF (86). Notably, it has been demonstrated that
miR-208 can induce cardiac hypertrophy and results in the
overexpression of β-myosin heavy chain in myocardial fibrosis
(87). Several miRs were indicated
to be differentially expressed in hypertension, including
miR-296-5p, let-7e and human cytomegalovirus (HCMV)-miR-UL112, as
encoded by HCMV in previous studies of the hypertension-associated
miR spectrum (88–90). Interferon regulatory factor 1, which
is involved in the regulation of blood pressure by acting on nitric
oxide synthase and vascular angiotensin (Ang) receptor, was
demonstrated to be a direct target of HCMV-miR-UL112 (91). However, in hypertension, HCMV titers
are considered to reflect the expression level of HCMV-miR-UL 112
(91), which is an independent risk
factor for hypertension. HCMV has been reported to inhibit
vasodilation by impairing nitric oxide synthase function (92) and causing endothelial cell
dysfunction (93). However, further
research is warranted due to the elusive association between HCMV
infection and endothelial dysfunction.
CHD is a multi-gene genetic disease resulting from
structural or functional cardiovascular abnormalities present at
birth that are caused by congenital abnormalities (119). Disrupted miR expression may result
in CHD via specific protein regulation. miR-133 and miR-1 are
present in the same transcription unit (120); miR-1 is the most abundant miR and
is highly conserved in human myocardial cells (121). Mature miR-1-1 and miR-1-2 have the
same gene sequence; the miR-13 family includes miR-133a-1,
miR-133a-2 and miR-133b (122,123).
During heart development, the deletion or mutation of the essential
gene Hand2 of muscle precursor cells in early embryonic development
may lead to cardiac hypoplasia and even cardiac arrest (124). Mukai et al (125) revealed that miR-486-3p, miR-155-5p
and miR-486-5p were increased in patients with cyanotic heart
disease compared with those without heart disease. Furthermore,
let-7e-5p and miR-1260a were decreased in patients with early-stage
acyanotic heart disease compared with those without heart disease,
suggesting that these miRs may be used for early diagnosis.
The abovementioned data summarize the differences in
expression of miRs in patients with HF (including cardiomyopathy,
hypertension, D-HF and CHD). Their clinical significance as HF
biomarkers were analyzed (Table
II).
Establishing an accurate, reliable circulating miR
system for HF diagnosis, prognosis and prediction of response to
treatment is challenging, from sample collection and processing to
data analysis. First, overlapping between various failure
mechanisms leads to difficulties in assessing which mechanisms
underlie the expression changes in circulating miRs. Second, serum
or plasma are the first choices for sample selection and handling,
but the level of circulating biomarker miRs was low, which to some
degree impedes the detection of miRs (130). Serum hemolysis may result in waste
of samples (131). Furthermore, the
serum level of miRs was higher than for circulating plasma,
indicating that serum samples can prevent potential interference
caused by platelets and leukocytes during sample preparation
(132). Therefore, use of the same
type of material and synchronous sampling is important for the
patient and control groups, as well as a standard scheme to avoid
sample hemolysis, minimizing differences between patient selection
and classification. Third, some studies have reported fluctuation
of miR levels in patients with HF following treatment (133,134).
Blood samples were collected at three stages, namely prior to,
during and following treatment. A fourth factor was the choice of
measurement platform for miR. As indicated in Fig. 2, all research techniques have
advantages and disadvantages, but the most commonly used method is
RT-qPCR. This method is more sensitive and more cost-effective
compared with other methods, but its primary limitation is the
inability to detect new miRs. In addition, the standardization of
miR expression level may be difficult, as the expression levels of
miRs fluctuate with changes in physiological and pathological
conditions. Therefore, standard methods are commonly used for the
experiments, including the use of equal amounts of starting
material (such as serum or plasma), which is more reliable for
endogenous miRs for data normalization.
As observed in the present study, the clinical
manifestations of HF caused by expression changes of different miRs
are similar, and the changes in miR expression caused by different
types of HF may also be similar, reflecting the complexity of miR
biology.
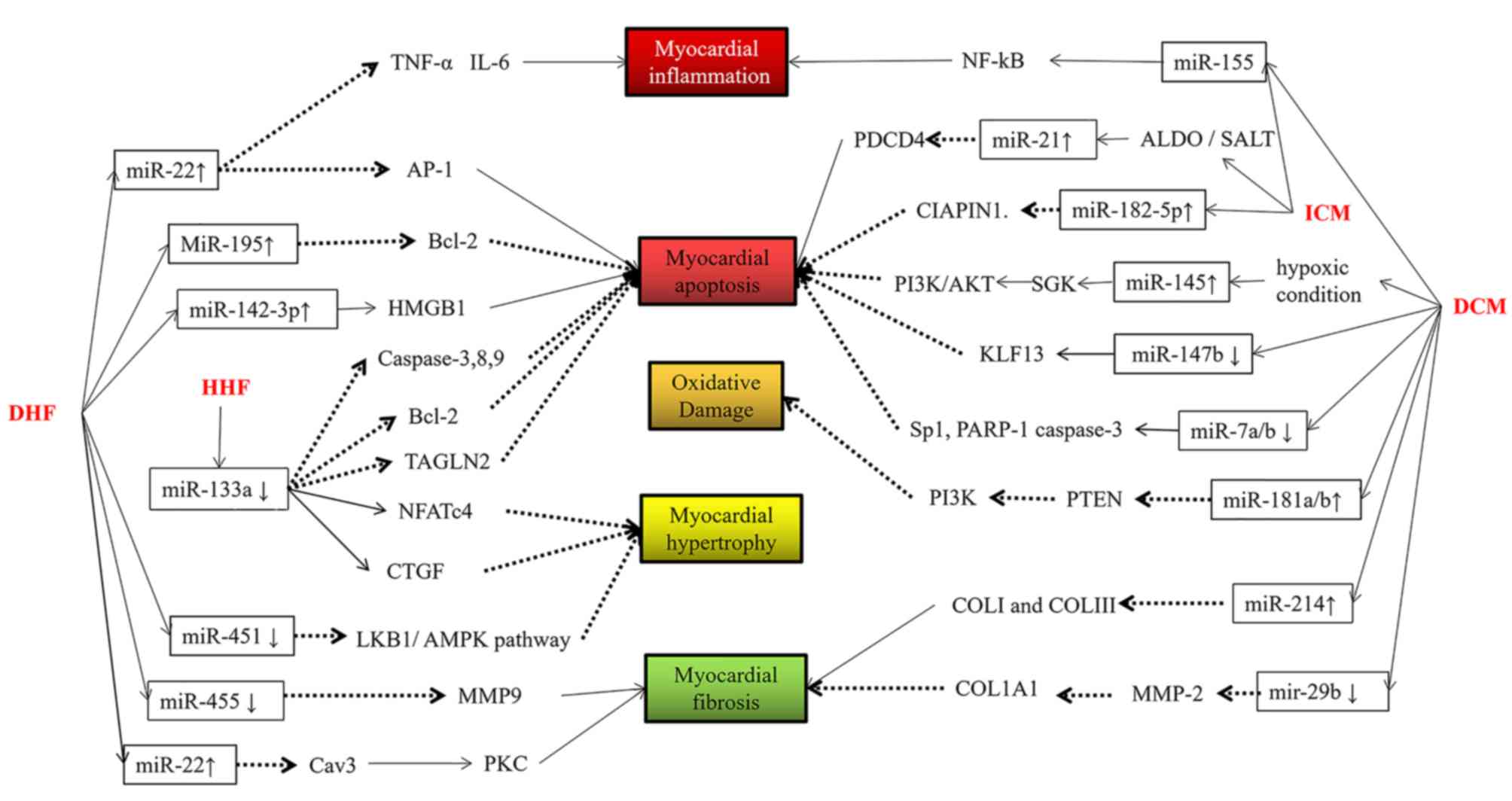
In ischemic HF, upregulation of miR-155 intensified
cardiomyocyte inflammation, and upregulation of miR-182 promoted
apoptosis, which may be an early indicator of this condition.
Upregulation of miR-21 alleviated apoptosis via negative feedback
regulation. Thus, miR-21 may be a late-age indicator in ischemic
HF.
In hypertensive HF, downregulation of miR-133
inhibited cardiomyocyte hypertrophy and promoted cardiomyocyte
apoptosis, which may be a late-stage decompensation.
In D-HF, upregulation of miR-22 reduced
cardiomyocyte fibrosis, apoptosis and inflammation, and
downregulation of miR-455 restrained cell fibrosis, which may be a
late indicator of diabetic heart failure, whereas the upregulation
of miR-195 and miR-142 aggravated apoptosis and miR-451
downregulation exacerbated cardiomyocyte hypertrophy, which may be
an early indicator.
In conclusion, miR-155, miR-22 and miR-133 appear to
be promising markers of the development, diagnosis and prognosis of
HF. However, further research is required to determine whether
there is an efficient miR template for application in clinical
oncology practice.
Not applicable.
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372150 to BHL and
grant nos. nos. 91739301 and 91849102 to MH).
Not applicable.
MH and BHL designed and conceived the study. YMH,
WWL and JW provided advice and assistance. YMH wrote the
manuscript. All the authors have contributed to and approved the
final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Cordes KR and Srivastava D: MicroRNA
regulation of cardiovascular development. Circ Res. 104:724–732.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson ME, Brown JH and Bers DM: CaMKII
in myocardial hypertrophy and heart failure. J Mol Cell Cardiol.
51:468–473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J, Savvatis K, Kang JS, Fan P, Zhong
H, Schwartz K, Barry V, Mikels-Vigdal A, Karpinski S, Kornyeyev D,
et al: Targeting LOXL2 for cardiac interstitial fibrosis and heart
failure treatment. Nat Commun. 7:137102016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kawakami H, Kubota Y, Takeno S, Miyazaki
Y, Wada T, Hamada R and Nanashima A: Gastrointestinal: Severe
congestive heart failure and acute gastric mucosal necrosis. J
Gastroenterol Hepatol. 32:9492017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petrovic D: Cytopathological basis of
heart failure-cardiomyocyte apoptosis, interstitial fibrosis and
inflammatory cell response. Folia Biol (Praha). 50:58–62.
2004.PubMed/NCBI
|
|
6
|
Orsborne C, Chaggar PS, Shaw SM and
Williams SG: The renin-angiotensin-aldosterone system in heart
failure for the non-specialist: The past, the present and the
future. Postgrad Med J. 93:29–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polyakova V, Loeffler I, Hein S, Miyagawa
S, Piotrowska I, Dammer S, Risteli J, Schaper J and Kostin S:
Fibrosis in endstage human heart failure: Severe changes in
collagen metabolism and MMP/TIMP profiles. Int J Cardiol.
151:18–33. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Romaine SP, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Tong Z, Chen K, Hu X, Jin H and Hou
M: The role of miRNA-132 against apoptosis and oxidative stress in
heart failure. Biomed Res Int. 2018:34527482018.PubMed/NCBI
|
|
10
|
Gómez AM, Valdivia HH, Cheng H, Lederer
MR, Santanaet LF, Cannel MB, McCune SA, Altschuld RA and Lederer
WJ: Defective excitation-contraction coupling in experimental
cardiac hypertrophy and heart failure. Science. 276:800–806. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumar R, Woo MA, Birrer BV, Macey PM,
Fonarow GC, Hamilton MA and Harper RM: Mammillary bodies and fornix
fibers are injured in heart failure. Neurobiol Dis. 33:236–242.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neupane B, Zhou Q, Gawaz M and Gramlich M:
Personalized medicine in inflammatory cardiomyopathy. Per Med.
15:127–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dludla PV, Dias SC, Obonye N, Johnson R,
Louw J and Nkambule BB: A systematic review on the protective
effect of N-acetyl cysteine against diabetes-associated
cardiovascular complications. Am J Cardiovasc Drugs. 18:283–298.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Güven Bağla A, Içkin Gülen M, Ercan F,
Aşgün F, Ercan E and Bakar C: Changes in kidney tissue and effects
of erythropoietin after acute heart failure. Biotech Histochem.
93:340–353. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lindner K, Haier J, Wang Z, Watson DI,
Hussey DJ and Hummel R: Circulating microRNAs: Emerging biomarkers
for diagnosis and prognosis in patients with gastrointestinal
cancers. Clin Sci (Lond). 128:1–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li R, Geng HH, Xiao J, Qin XT, Wang F,
Xing JH, Xia YF, Mao Y, Liang JW and Jia XP: miR-7a/b attenuates
post-myocardial infarction remodeling and protects H9c2
cardiomyoblast against hypoxia-induced apoptosis involving Sp1 and
PARP-1. Sci Rep. 6:290822016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ball JP, Syed M, Marañon RO, Hall ME, Kc
R, Reckelhoff JF, Yanes Cardozo LL and Romero DG: Role and
regulation of MicroRNAs in aldosterone-mediated cardiac injury and
dysfunction in male rats. Endocrinology. 158:1859–1874. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng W, Wang Y, Long X, Zhao R, Wang Z,
Liu Z, Cao S and Shi B: miR-21 reduces hydrogen peroxide-induced
apoptosis in c-kit+ cardiac stem cells in vitro through
PTEN/PI3K/Akt signaling. Oxid Med Cell Longev. 2016:53891812016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng M, Wu G, Song Y, Wang L, Tu L, Zhang
L and Zhang C: Celastrol-induced suppression of the MiR-21/ERK
signalling pathway attenuates cardiac fibrosis and dysfunction.
Cell Physiol Biochem. 38:1928–1938. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao J, Pan Y, Li XH, Yang XY, Feng YL,
Tan HH, Jiang L, Feng J and Yu XY: Cardiac progenitor cell-derived
exosomes prevent cardiomyocytes apoptosis through exosomal miR-21
by targeting PDCD4. Cell Death Dis. 7:e22772016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tao H, Chen ZW, Yang JJ and Shi KH:
MicroRNA-29a suppresses cardiac fibroblasts proliferation via
targeting VEGF-A/MAPK signal pathway. Int J Biol Macromol.
88:414–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu CZ, Zhong Q and Huang YQ: Elevated
plasma miR-29a levels are associated with increased carotid
intima-media thickness in atherosclerosis patients. Tohoku J Exp
Med. 241:183–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu Z, Wang F, Yu P, Wang X, Wang Y, Tang
ST and Zhu HQ: Inhibition of miR-29b suppresses MAPK signaling
pathway through targeting SPRY1 in atherosclerosis. Vascul
Pharmacol. 102:29–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sassi Y, Avramopoulos P, Ramanujam D,
Grüter L, Werfel S, Giosele S, Brunner A, Esfandyari D,
Papadopoulou AS, De Strooper B, et al: Cardiac myocyte miR-29
promotes pathological remodeling of the heart by activating Wnt
signaling. Nat Commun. 8:16142017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Panizo S, Carrillo-López N, Naves-Díaz M,
Solache-Berrocal G, Martínez-Arias L, Rodrigues-Díez RR,
Fernández-Vázquez A, Martínez-Salgado C, Ruiz-Ortega M, Dusso A, et
al: Regulation of miR-29b and miR-30c by vitamin D receptor
activators contributes to attenuate uraemia-induced cardiac
fibrosis. Nephrol Dial Transplant. 32:1831–1840. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heid J, Cencioni C, Ripa R, Baumgart M,
Atlante S, Milano G, Scopece A, Kuenne C, Guenther S, Azzimato V,
et al: Age-dependent increase of oxidative stress regulates
microRNA-29 family preserving cardiac health. Sci Rep. 7:168392017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Ji Q, Zhu H, Ren Y, Fan Z and Tian
N: miR-30a attenuates cardiac fibrosis in rats with myocardial
infarction by inhibiting CTGF. Exp Ther Med. 15:4318–4324.
2018.PubMed/NCBI
|
|
28
|
Roca-Alonso L, Castellano L, Mills A,
Dabrowska AF, Sikkel MB, Pellegrino L, Jacob J, Frampton AE, Krell
J, Coombes RC, et al: Myocardial MiR-30 downregulation triggered by
doxorubicin drives alterations in β-adrenergic signaling and
enhances apoptosis. Cell Death Dis. 6:e17542015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lai L, Chen J, Wang N, Zhu G, Duan X and
Ling F: MiRNA-30e mediated cardioprotection of ACE2 in rats with
Doxorubicin-induced heart failure through inhibiting cardiomyocytes
autophagy. Life Sci. 169:69–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Middendorp LB, Kuiper M, Munts C,
Wouters P, Maessen JG, van Nieuwenhoven FA and Prinzen FW: Local
microRNA-133a downregulation is associated with hypertrophy in the
dyssynchronous heart. ESC Heart Fail. 4:241–251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Q, Lin X, Yang X and Chang J: NFATc4 is
negatively regulated in miR-133a-mediated cardiomyocyte
hypertrophic repression. Am J Physiol Heart Circ Physiol.
298:H1340–H1347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li AY, Yang Q and Yang K: miR-133a
mediates the hypoxia-induced apoptosis by inhibiting TAGLN2
expression in cardiac myocytes. Mol Cell Biochem. 400:173–181.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rangrez AY, Hoppe P, Kuhn C, Zille E,
Frank J, Frey N and Frank D: MicroRNA miR-301a is a novel cardiac
regulator of Cofilin-2. PLoS One. 12:e01839012017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dong H, Dong S, Zhang L, Gao X, Lv G, Chen
W and Shao S: MicroRNA-214 exerts a Cardio-protective effect by
inhibition of fibrosis. Anat Rec (Hoboken). 299:1348–1357. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chaturvedi P, Kalani A, Medina I,
Familtseva A and Tyagi SC: Cardiosome mediated regulation of MMP9
in diabetic heart: Role of mir29b and mir455 in exercise. J Cell
Mol Med. 19:2153–2161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu N, Shi YF, Diao HY, Li YX, Cui Y, Song
XJ, Tian X, Li TY and Liu B: MicroRNA-135a regulates apoptosis
induced by hydrogen peroxide in rat cardiomyoblast cells. Int J
Biol Sci. 13:13–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Ouyang M, Wang Q and Jian Z:
MicroRNA-142-3p inhibits hypoxia/reoxygenation-induced apoptosis
and fibrosis of cardiomyocytes by targeting high mobility group box
1. Int J Mol Med. 38:1377–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang F, Li T, Dong Z and Mi R:
MicroRNA-410 is involved in mitophagy after cardiac
ischemia/reperfusion injury by targeting high-mobility group box 1
protein. J Cell Biochem. 119:2427–2439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang S, Zhang R, Wu F and LI X:
MicroRNA-208a regulates H9c2 cells simulated ischemia-reperfusion
myocardial injury via targeting CHD9 through Notch/NF-kappa B
signal pathways. Int Heart J. 59:580–588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan ZG, Qu XL, Chu P, Gao YL, Gao XF, Chen
SL and Tian NL: MicroRNA-210 promotes angiogenesis in acute
myocardial infarction. Mol Med Rep. 17:5658–5665. 2018.PubMed/NCBI
|
|
41
|
Zhang Y, Fang J and Ma H: Inhibition of
miR-182-5p protects cardiomyocytes from hypoxia-induced apoptosis
by targeting CIAPIN1. Biochem Cell Biol. 96:646–654. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu X, Tong Z, Chen K, Hu X, Jin H and Hou
M: The role of miRNA-132 against apoptosis and oxidative stress in
heart failure. Biomed Res Int. 2018:34527482018.PubMed/NCBI
|
|
43
|
Zhou G, Li C, Feng J, Zhang J and Fang Y:
lncRNA UCA1 is a novel regulator in cardiomyocyte hypertrophy
through targeting the miR-184/HOXA9 axis. Cardiorenal Med.
8:130–139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rubiś P, Totoń-Żurańska J,
Wiśniowska-Śmiałek S, Holcman K, Kołton-Wróż M, Wołkow P, Wypasek
E, Natorska J, Rudnicka-Sosin L, Pawlak A, et al: Relations between
circulating microRNAs (miR-21, miR-26, miR-29, miR-30 and
miR-133a), extracellular matrix fibrosis and serum markers of
fibrosis in dilated cardiomyopathy. Int J Cardiol. 231:201–206.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ji Y, Qiu M, Shen Y, Gao L, Wang Y, Sun W,
Li X, Lu Y and Kong X: MicroRNA-327 regulates cardiac hypertrophy
and fibrosis induced by pressure overload. Int J Mol Med.
41:1909–1916. 2018.PubMed/NCBI
|
|
46
|
Lu Y and Wu F: A new miRNA regulator,
miR-672, reduces cardiac hypertrophy by inhibiting JUN expression.
Gene. 648:21–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y, Cai H, Li H, Gao Z and Song K:
Atrial overexpression of microRNA-27b attenuates angiotensin
II-induced atrial fibrosis and fibrillation by targeting ALK5. Hum
Cell. 31:251–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang J, Chen L, Ding J, Zhang J, Fan Z,
Yang C, Yu Q and Yang J: Cardioprotective effect of miRNA-22 on
hypoxia/reoxygenation induced cardiomyocyte injury in neonatal
rats. Gene. 579:17–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang L, Yin H, Jiao L, Liu T, Gao Y, Shao
Y, Zhang Y, Shan H, Zhang Y and Yang B: Abnormal downregulation of
caveolin-3 mediates the pro-fibrotic action of MicroRNA-22 in a
model of myocardial infarction. Cell Physiol Biochem. 45:1641–1653.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng L, Lin S and Lv C: MiR-26a-5p
regulates cardiac fibroblasts collagen expression by targeting
ULK1. Sci Rep. 8:21042018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gu M, Wang J, Wang Y, Xu Y, Zhang Y, Wu W
and Liao S: MiR-147b inhibits cell viability and promotes apoptosis
of rat H9c2 cardiomyocytes via down-regulating KLF13 expression.
Acta Biochim Biophys Sin (Shanghai). 50:288–297. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun N, Meng F, Xue N, Pang G, Wang Q and
Ma H: Inducible miR-145 expression by HIF-1a protects
cardiomyocytes against apoptosis via regulating SGK1 in simulated
myocardial infarction hypoxic microenvironment. Cardiol J.
25:268–278. 2018.PubMed/NCBI
|
|
53
|
Chen Z, Zhang S, Guo C, Li J and Sang W:
Downregulation of miR-200c protects cardiomyocytes from
hypoxia-induced apoptosis by targeting GATA-4. Int J Mol Med.
39:1589–1596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Meng X, Ji Y, Wan Z, Zhao B, Feng C, Zhao
J, Li H and Song Y: Inhibition of miR-363 protects cardiomyocytes
against hypoxia-induced apoptosis through regulation of Notch
signaling. Biomed Pharmacother. 90:509–516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li T, Yang GM, Zhu Y, Wu Y, Chen XY, Lan
D, Tian K and Liu LM: Diabetes and hyperlipidemia induce
dysfunction of VSMCs: Contribution of the metabolic
inflammation/miRNA pathway. Am J Physiol Endocrinol Metab.
308:E257–E269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gallego I, Beaumont J, López B, Ravassa S,
Gómez-Doblas JJ, Moreno MU, Valencia F, de Teresa E, Díez J and
González A: Potential role of microRNA-10b down-regulation in
cardiomyocyte apoptosis in aortic stenosis patients. Clin Sci
(Lond). 130:2139–2149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hang P, Sun C, Guo J, Zhao J and Du Z:
BDNF-mediates down-regulation of MicroRNA-195 inhibits ischemic
cardiac apoptosis in rats. Int J Biol Sci. 12:979–989. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Blumensatt M, Fahlbusch P, Hilgers R,
Bekaert M, Herzfeld de Wiza D, Akhyari P, Ruige JB and Ouwens DM:
Secretory products from epicardial adipose tissue from patients
with type 2 diabetes impair mitochondrial β-oxidation in
cardiomyocytes via activation of the cardiac renin-angiotensin
system and induction of miR-208a. Basic Res Cardiol. 112:22017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Marchand A, Atassi F, Mougenot N, Clergue
M, Codoni V, Berthuin J, Proust C, Trégouët DA, Hulot JS and Lompré
AM: miR-322 regulates insulin signaling pathway and protects
against metabolic syndrome-induced cardiac dysfunction in mice.
Biochim Biophys Acta. 1862:611–621. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhong C, Wang K, Liu Y, Lv D, Zheng B,
Zhou Q, Sun Q, Chen P, Ding S, Xu Y and Huang H: miR-19b controls
cardiac fibroblast proliferation and migration. J Cell Mol Med.
20:1191–1197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pan L, Huang BJ, Ma XE, Wang SY, Feng J,
Lv F, Liu Y, Liu Y, Li CM, Liang DD, et al: MiR-25 protects
cardiomyocytes against oxidative damage by targeting the
mitochondrial calcium uniporter. Int J Mol Sci. 16:5420–5433. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Das S, Kohr M, Dunkerly-Eyring B, Lee DI,
Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA and
Steenbergen C: Divergent effects of miR-181 family members on
myocardial function through protective cytosolic and detrimental
mitochondrial microRNA targets. J Am Heart Assoc. 6(pii):
e0046942017.PubMed/NCBI
|
|
63
|
Palomer X, Capdevila-Busquets E, Botteri
G, Davidson MM, Rodríguez C, Martínez-González J, Vidal F, Barroso
E, Chan TO, Feldman AM, et al: miR-146a targets Fos expression in
human cardiac cells. Dis Model Mech. 8:1081–1091. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Khamaneh AM, Alipour MR, Sheikhzadeh
Hesari F and Ghadiri Soufi F: A signature of microRNA-155 in the
pathogenesis of diabetic complications. J Physiol Biochem.
71:301–309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Fang Y, Chen H, Hu Y, Li Q, Hu Z, Ma T and
Mao X: Burkholderia pseudomallei-derived miR-3473 enhances NF-κB
via targeting TRAF3 and is associated with different inflammatory
responses compared to Burkholderia thailandensis in murine
macrophages. BMC Microbiol. 16:2832016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kuwabara Y, Horie T, Baba O, Watanabe S,
Nishiga M, Usami S, Izuhara M, Nakao T, Nishino T, Otsu K, et al:
MicroRNA-451 exacerbates lipotoxicity in cardiac myocytes and
high-fat diet-induced cardiac hypertrophy in mice through
suppression of the LKB1/AMPK Pathway. Circ Res. 116:279–288. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cohen-Solal A, Beauvais F and Logeart D:
Heart failure and diabetes mellitus: Epidemiology and management of
an alarming association. J Card Fail. 14:615–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Nargesi AA, Esteghamati S, Heidari B,
Hafezi-Nejad N, Sheikhbahaei S, Pajouhi A, Nakhjavani M and
Esteghamati A: Nonlinear relation between pulse pressure and
coronary heart disease in patients with type 2 diabetes or
hypertension. J Hypertens. 34:974–980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Puntmann VO, Carr-White G, Jabbour A, Yu
CY, Gebker R, Kelle S, Hinojar R, Doltra A, Varma N, Child N, et
al: T1-mapping and outcome in nonischemic cardiomyopathy: All-cause
mortality and heart failure. JACC Cardiovasc Imaging. 9:40–50.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cahill TJ, Ashrafian H and Watkins H:
Genetic cardiomyopathies causing heart failure. Circ Res.
113:660–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ortega A, Roselló-Lletí E, Tarazón E,
Molina-Navarro MM, Martínez-Dolz L, González-Juanatey JR, Lago F,
Montoro-Mateos JD, Salvador A, Rivera M and Portolés M: Endoplasmic
reticulum stress induces different molecular structural alterations
in human dilated and ischemic cardiomyopathy. PLoS One.
9:e1076352014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yeung F, Chung E, Guess MG, Bell ML and
Leinwand LA: Myh7b/miR-499 gene expression is transcriptionally
regulated by MRFs and Eos. Nucleic Acids Res. 40:7303–7318. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Abraityte A, Lunde IG, Askevold ET,
Michelsen AE, Christensen G, Aukrust P, Yndestad A, Fiane A,
Andreassen A, Aakhus S, et al: Wnt5a is associated with
rightventricular dysfunction and adverse outcome in dilated
cardiomyopathy. Sci Rep. 7:34902017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yamamoto S, Yang G, Zablocki D, Liu J,
Hong C, Kim SJ, Soler S, Odashima M, Thaisz J, Yehia G, et al:
Activation of Mst1 causes dilated cardiomyopathy by stimulating
apoptosis without compensatory ventricular myocyte hypertrophy. J
Clin Invest. 111:1463–1474. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang Y, Kanter EM and Yamada KA:
Remodeling of cardiac fibroblasts following myocardial infarction
results in increased gap junction intercellular communication.
Cardiovasc Pathol. 19:e233–e240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Naga Prasad SV, Gupta MK, Duan ZH,
Surampudi VS, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM,
Sen S, et al: A unique microRNA profile in end-stage heart failure
indicates alterations in specific cardiovascular signaling
networks. PLoS One. 12:e01704562017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Enes C, oşkun M, Kervancıoğlu M, Öztuzcu
S, Yılmaz Coşkun F, Ergün S, Başpınar O, Kılınç M, Temel L and
Coşkun MY: Plasma microRNA profiling of children with idiopathic
dilated cardiomyopathy. Biomarkers. 21:56–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Miyamoto SD, Karimpour-Fard A, Peterson V,
Auerbach SR, Stenmark KR, Stauffer BL and Sucharov CC: Circulating
microRNA as a biomarker for recovery in pediatric dilated
cardiomyopathy. J Heart Lung Transplant. 34:724–733. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Leger KJ, Singh S, Canseco D, VonGrote EC,
Karim-Ud-Din S, Collins SC, Thibodeau JT, Mishkin JD, Patel PC,
Markham DW, et al: Abstract 13120: Identification of novel
circulating microRNAs in ischemic cardiomyopathy utilizing whole
blood microRNA profiling. Circulation. 128 Suppl 22:A131202013.
|
|
80
|
Zeng X, Li X and Wen H: Expression of
circulating microRNA-182, CITED2 and HIF-1 in ischemic
cardiomyopathy and their correlation. J Clin Cardiol. 33:119–122.
2017.(In Chinese).
|
|
81
|
Olson E and Rooij EV: Dual targeting of
miR-208 and miR-499 in the treatment of cardiac disorders. US
Patent 14104886. Filed December 12, 2013; issued. June 26–2014.
|
|
82
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li X, Liu CY, Li YS, Xu J, Li DG, Li X and
Han D: Deep RNA sequencing elucidates microRNA-regulated molecular
pathways in ischemic cardiomyopathy and nonischemic cardiomyopathy.
Genet Mol Res. 15:gmr74652016.
|
|
84
|
Phelan D, Watson C, Martos R, Collier P,
Patle A, Donnelly S, Ledwidge M, Baugh J and McDonald K: Modest
elevation in BNP in asymptomatic hypertensive patients reflects
sub-clinical cardiac remodeling, inflammation and extracellular
matrix changes. PLoS One. 7:e492592012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mohammed SF, Hussain S, Mirzoyev SA,
Edwards WD, Maleszewski JJ and Redfield MM: Coronary microvascular
rarefaction and myocardial fibrosis in heart failure with preserved
ejection fraction. Circulation. 131:550–559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Shyu KG, Wang BW, Cheng WP and Lo HM:
MicroRNA-208a increases myocardial endoglin expression and
myocardial fibrosis in acute myocardial infarction. Can J Cardiol.
31:679–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cengiz M, Karatas OF, Koparir E, Yavuzer
S, Ali C, Yavuzer H, Kirat E, Karter Y and Ozen M: Differential
expression of hypertension-associated microRNAs in the plasma of
patients with white coat hypertension. Medicine (Baltimore).
94:e6932015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Fu M, Gao Y, Zhou Q, Zhang Q, Peng Y, Tian
K, Wang J and Zheng X: Human cytomegalovirus latent infection
alters the expression of cellular and viral microRNA. Gene.
536:272–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Stern-Ginossar N, Saleh N, Goldberg MD,
Prichard M, Wolf DG and Mandelboim O: Analysis of human
cytomegalovirus-encoded microRNA activity during infection. J
Virol. 83:10684–10693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li S, Zhu J, Zhang W, Chen Y, Zhang K,
Popescu LM, Ma X, Lau WB, Rong R, Yu X, et al: Signature microRNA
expression profile of essential hypertension and its novel link to
human cytomegalovirus infection. Circulation. 124:175–184. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ding M, Wang X, Wang C, Liu X, Zen K, Wang
W, Zhang CY and Zhang C: Distinct expression profile of HCMV
encoded miRNAs in plasma from oral lichen planus patients. J Transl
Med. 15:1332017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kellawan JM, Johansson RE, Harrell JW,
Sebranek JJ, Walker BJ, Eldridge MW and Schrage WG: Exercise
vasodilation is greater in women: Contributions of nitric oxide
synthase and cyclooxygenase. Eur J Appl Physiol. 115:1735–1746.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dolcino M, Puccetti A, Barbieri A, Bason
C, Tinazzi E, Ottria A, Patuzzo G, Martinelli N and Lunardi C:
Infections and autoimmunity: Role of human cytomegalovirus in
autoimmune endothelial cell damage. Lupus. 24:419–432. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kontaraki JE, Marketou ME, Zacharis EA,
Parthenakis FI and Vardas PE: MiR-1, miR-9 and miR-126 levels in
peripheral blood mononuclear cells of patients with essential
hypertension associate with prognostic indices of ambulatory blood
pressure monitoring. Eur Heart J. 34 Suppl 1:S51582013. View Article : Google Scholar
|
|
95
|
Kontaraki JE, Marketou ME, Zacharis EA,
Parthenakis FI and Vardas PE: Mir-143/mir-145 levels in peripheral
blood mononuclear cells associate with ambulatory blood pressure
monitoring parameters in patients with essential hypertension. Eur
Heart J. 34 Suppl 1:S56562013. View Article : Google Scholar
|
|
96
|
Dickinson BA, Semus HM, Montgomery RL,
Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG and
van Rooij E: Plasma microRNAs serve as biomarkers of therapeutic
efficacy and disease progression in hypertension-induced heart
failure. Eur J Heart Fail. 15:650–659. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hou YL, LI SL and Liu LL: Effects of
MicroRNA-137 and AngII on cardiac remodeling in spontaneously
hypertensive rats. Chin J Comp Med. 7–2016.(In Chinese).
|
|
98
|
Li JZ, Tang XN, Li TT, Liu LJ, Yu SY, Zhou
GY, Shao QR, Sun HP, Wu C and Yang Y: Paeoniflorin inhibits
doxorubicin-induced cardiomyocyte apoptosis by downregulating
microRNA-1 expression. Exp Ther Med. 11:2407–2412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yang Q, Jia C, Wang P, Xiong M, Cui J, Li
L, Wang W, Wu Q, Chen Y and Zhang T: MicroRNA-505 identified from
patients with essential hypertension impairs endothelial cell
migration and tube formation. Int J Cardiol. 177:925–934. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li Y, Wu H, Zhu M, Shelat H, Qu J, Zheng
M, Yuan J, Yuan G, Xu J, Wang H and Geng YJ: Insulin-like growth
factor prevents diabetes induced cardiomyopathy mediated by
MICRORNA-1. J Am College Cardiol. 55:A21.E1962010. View Article : Google Scholar
|
|
101
|
Finn NA, Eapen D, Manocha P, Al Kassem H,
Lassegue B, Ghasemzadeh N, Quyyumi A and Searles CD: Coronary heart
disease alters intercellular communication by modifying
microparticle-mediated microRNA transport. FEBS Lett.
587:3456–3463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Dickstein K: Is substantial renal
dysfunction in patients with heart failure no longer a
contraindication for RAS inhibition? The power of a large,
high-quality registry to illuminate major clinical issues. Eur
Heart J. 36:2279–2280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shang F, Wang SC, Hsu CY, Miao Y, Martin
M, Yin Y, Wu CC, Wang YT, Wu G, Chien S, et al: MicroRNA-92a
mediates endothelial dysfunction in CKD. J Am Soc Nephrol.
28:3251–3261. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang C, Fan F, Cao Q, Shen C, Zhu H, Wang
P, Zhao X, Sun X, Dong Z, Ma X, et al: Mitochondrial aldehyde
dehydrogenase 2 deficiency aggravates energy metabolism disturbance
and diastolic dysfunction in diabetic mice. J Mol Med (Berl).
94:1229–1240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wong AK, AlZadjali MA, Choy AM and Lang
CC: Insulin resistance: A potential new target for therapy in
patients with heart failure. Cardiovasc Ther. 26:203–213. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX,
Lin SG and Li Y: Glucose induces apoptosis of cardiomyocytes via
microRNA-1 and IGF-1. Biochem Biophys Res Commun. 376:548–552.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Horie T, Ono K, Nishi H, Iwanaga Y, Nagao
K, Kinoshita M, Kuwabara Y, Takanabe R, Hasegawa K, Kita T and
Kimura T: MicroRNA-133 regulates the expression of GLUT4 by
targeting KLF15 and is involved in metabolic control in cardiac
myocytes. Biochem Biophys Res Commun. 389:315–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Latronico MV, Catalucci D and Condorelli
G: Emerging role of microRNAs in cardiovascular biology. Circ Res.
101:1225–1236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Nandi SS, Duryee MJ, Shahshahan HR, Thiele
GM, Anderson DR and Mishra PK: Induction of autophagy markers is
associated with attenuation of miR-133a in diabetic heart failure
patients undergoing mechanical unloading. Am J Transl Res.
7:683–696. 2015.PubMed/NCBI
|
|
112
|
Deng X, Liu Y, Luo M and Wu J, Ma R, Wan Q
and Wu J: Circulating miRNA-24 and its target YKL-40 as potential
biomarkers in patients with coronary heart disease and type 2
diabetes mellitus. Oncotarget. 8:63038–63046. 2017.PubMed/NCBI
|
|
113
|
Chavali V, Tyagi SC and Mishra PK:
Differential expression of dicer, miRNAs, and inflammatory markers
in diabetic Ins2+/− Akita hearts. Cell Biochem Biophys. 68:25–35.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Izarra A, Moscoso I, Cañón S, Carreiro C,
Fondevila D, Martín-Caballero J, Blanca V, Valiente I, Díez-Juan A
and Bernad A: miRNA-1 and miRNA-133a are involved in early
commitment of pluripotent stem cells and demonstrate antagonistic
roles in the regulation of cardiac differentiation. J Tissue Eng
Regen Med. 11:787–799. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Liu H, Yang L, Chen KH, Sun HY, Jin MW,
Xiao GS, Wang Y and Li GR: SKF-96365 blocks human
ether-à-go-go-related gene potassium channels stably expressed in
HEK 293 cells. Pharmacol Res. 104:61–69. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
van Solingen C, Bijkerk R, de Boer HC,
Rabelink TJ and van Zonneveld AJ: The Role of microRNA-126 in
vascular homeostasis. Curr Vasc Pharmacol. 13:341–351. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Škrha P, Hajer J, Anděl M, Hořínek A and
Korabečná M: miRNA as a new marker of diabetes mellitus and
pancreatic carcinoma progression. Cas Lek Cesk. 154:122–126.
2015.(In Czech). PubMed/NCBI
|
|
119
|
Talmud PJ: How to identify
gene-environment interactions in a multifactorial disease: CHD as
an example. Proc Nutr Soc. 63:5–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Carè A, Catalucci D, Felicetti F, Bonci D,
Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al:
MicroRNA-133 controls cardiac hypertrophy. Nat Med. 13:613–618.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wang L, Tian D, Hu J, Xing H, Sun M, Wang
J, Jian Q and Yang H: MiRNA-145 regulates the development of
congenital heart disease through targeting FXN. Pediatr Cardiol.
37:629–636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Feng Y, Niu LL, Wei W, Zhang WY, Li XY,
Cao JH and Zhao SH: A feedback circuit between miR-133 and the
ERK1/2 pathway involving an exquisite mechanism for regulating
myoblast proliferation and differentiation. Cell Death Dis.
4:e9342013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Liu N, Bezprozvannaya S, Williams AH, Qi
X, Richardson JA, Bassel-Duby R and Olson EN: microRNA-133a
regulates cardiomyocyte proliferation and suppresses smooth muscle
gene expression in the heart. Genes Dev. 22:3242–3254. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Shan ZX, Lin QX, Deng CY, Zhou ZL, Zhang
XC, Fu YH and Yu XY: Plasmid-mediated miRNA-1-2 specifically
inhibits Hand2 protein expression in H9C2 cells. Nan Fang Yi Ke Da
Xue Xue Bao. 28:1559–1561. 2008.(In Chinese). PubMed/NCBI
|
|
125
|
Mukai N, Nakayama Y, Murakami S, Tanahashi
T, Sessler DI, Ishii S, Ogawa S, Tokuhira N, Mizobe T, Sawa T and
Nakajima Y: Potential contribution of erythrocyte microRNA to
secondary erythrocytosis and thrombocytopenia in congenital heart
disease. Pediatr Res. 83:866–873. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhao Y, Samal E and Srivastava D: Serum
response factor regulates a muscle-specific microRNA that targets
Hand2 during cardiogenesis. Nature. 436:214–220. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lu CX, Gong HR, Liu XY, Wang J, Zhao CM,
Huang RT, Xue S and Yang YQ: A novel HAND2 loss-of-function
mutation responsible for tetralogy of Fallot. Int J Mol Med.
37:445–451. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ferreira LR, Frade AF, Santos RH, Teixeira
PC, Baron MA, Navarro IC, Benvenuti LA, Fiorelli AI, Bocchi EA,
Stolf NA, et al: MicroRNAs miR-1, miR-133a, miR-133b, miR-208a and
miR-208b are dysregulated in chronic chagas disease cardiomyopathy.
Int J Cardiol. 175:409–417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chen W and Li S: Circulating microRNA as a
novel biomarker for pulmonary arterial hypertension due to
congenital heart disease. Pediatr Cardiol. 38:86–94. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Patrick DM, Montgomery RL, Qi X, Obad S,
Kauppinen S, Hill JA, van Rooij E and Olson EN: Stress-dependent
cardiac remodeling occurs in the absence of microRNA-21 in mice. J
Clin Invest. 120:3912–3916. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang Y, Gu J, Roth JA, Hildebrandt MA,
Lippman SM, Ye Y, Minna JD and Wu X: Pathway-based serum microRNA
profiling and survival in patients with advanced-stage non-small
cell lung cancer. Cancer Res. 73:4801–4809. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Hamam R, Hamam D, Alsaleh KA, Kassem M,
Zaher W, Alfayez M, Aldahmash A and Alajez NM: Circulating
microRNAs in breast cancer: Novel diagnostic and prognostic
biomarkers. Cell Death Dis. 8:e30452017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Duttagupta R, Jiang R, Gollub J, Getts RC
and Jones KW: Impact of cellular miRNAs on circulating miRNA
biomarker signatures. PLos One. 6:e207692011. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Sassi Y, Avramopoulos P, Ramanujam D,
Grüter L, Werfel S, Giosele S, Brunner AD, Esfandyari D,
Papadopoulou AS, De Strooper B, et al: Cardiac myocyte miR-29
promotes pathological remodeling of the heart by activating Wnt
signaling. Nat Commun. 8:16142017. View Article : Google Scholar : PubMed/NCBI
|