Introduction
Glioma is the most common type of primary brain
tumor in adults, accounting for ~46% of intracranial tumors, with
an incidence rate of 3–10/10 million, corresponding to 1–3% of
detected malignancies worldwide (1,2). Grade-I
and -II gliomas are classified as benign and low-grade gliomas,
while grade-III and -IV gliomas are classified as high-grade
gliomas, according to the World Health Organization (WHO) grading
system (3). The current standard
treatment for gliomas mainly includes surgery, followed by
radiotherapy and chemotherapy. However, the therapeutic effect is
not satisfactory. The median survival time of glioblastoma
multiforme (GBM; grade 4 astrocytoma) patients remains poor,
ranging from 12 to 15 months, 2 to 5 years and 6 to 8 years for
grade IV, III and I–II gliomas, respectively according to the WHO
grading system (4) Previous studies
have proven that cytogenetic and molecular analyses have a vital
role in predicting the remission and survival rates of patients.
The mutation status of isocitrate dehydrogenase, NADP+, the
heterozygous deletion at the chromosomal position 1p/19q and the
methylation of the O-6-methylguanine-DNA methyltransferase promoter
(5–7)
have been progressively used as diagnostic and prognostic markers
for the comprehensive assessment of glioma patients. However,
biological differences between individual patients are apparent and
novel biomarkers are required to fully evaluate the prognosis of
patients and predict the effectiveness of glioma treatment.
MicroRNAs (miRNAs/miRs) are a class of short
endogenous non-coding RNA molecules that are 18 to 25 nucleotides
in length and exert a tumor-regulatory function at the
post-transcriptional level by binding to the 3′-untranlated region
(3′-UTR) and frequently to the 5′-UTR of mRNA molecules.
Accumulating studies have identified multiple dysregulated miRNAs,
which may serve as key molecules in cancer progression, and
regulate various biological processes, including proliferation,
differentiation, apoptosis and survival (8–10).
Previous studies have demonstrated a significant difference in the
expression profile of miRNAs between healthy subjects and patients
with glioma, suggesting that the expression levels of certain
miRNAs are associated with the overall survival (OS) of glioma
patients (11–14). Therefore, the identification of
differentially expressed miRNAs is of great importance in order to
evaluate the early prognosis of glioma patients.
In the present study, a microarray-based analysis
was performed to recognize differentially expressed miRNAs in
gliomas by comparing miRNA expression profiles among low-grade and
high-grade gliomas. Furthermore, the differential expression of
miR-374a in human glioma tissues was confirmed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis. The prognostic value of this differentially expressed
miRNA was then investigated. Furthermore, a bioinformatics analysis
based on miRNA target gene databases was used to identify target
genes of miR-374a. The GO and KEGG analysis were performed and the
hub genes were analyzed by construction of a protein-protein
interaction (PPI) network for the target genes. The aim of the
present study was to obtain novel prognostic and predictive
biomarkers for glioma, and to investigate the potential mechanisms
of glioma progression.
Patients and methods
Patients and tissue specimens
Glioma tissues (15 grade II, 13 grade III and 20
grade IV) from a total of 48 patients were collected from the
Department of Neurosurgery of the Second Hospital of the Lanzhou
University (Gansu, China), from January 2013 to December 2016. The
5 normal brain tissues from patients without glioma who underwent
surgery for other reasons, including cerebral trauma. Following
surgical removal, the tissue samples were snap-frozen in liquid
nitrogen and stored at −80°C until used for RNA isolation. Patients
who had received chemotherapy or radiotherapy prior to surgery were
excluded. During the follow-up, OS was observed from the date of
diagnosis to the date of patient death and/or the last census date
in case of the patient being alive.
Microarray
A total of six glioma tissues (3 low-grade and 3
high-grade gliomas) were next analyzed using microarray
methodologies in order to screen for the expression of miRNAs. A
total of 1 µg of total RNA was extracted from each tumor sample
using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), and first-strand complementary (c)DNA
synthesis was performed using the miRNA First-Strand cDNA Synthesis
kit (cat. no. AS-MR-004; Arraystar, Rockville, MD, USA) following
the manufacturer's protocols. The miRStar™ Human Cancer Focus miRNA
PCR Array (cat. no. AS-MR-0033; Arraystar) was applied on the ABI
PRISM7900 system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Each 384-well miRStar™ Human Cancer Focus miRNA PCR Array
contained 184 miRNAs linked to human cancers, nine wells for
different housekeeping miRNAs, a genomic DNA contamination control,
three replicate RT controls and three replicate positive PCR
controls. The values of the quantification cycle (Cq) that were
obtained for quantification were used for the calculation of fold
changes in miRNA abundance according to the 2−ΔΔCq
method (15). The raw data were
processed by the following workflow: Background detection, RMA
global background correlation, quantile normalization, median
polish adjustment was performed and log2-transformation with miRNA
QC tool software (Affymetrix; Thermo Fisher Scientific, Inc.)
(16).
RT-qPCR
Total RNA was extracted from tissues using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. cDNA was randomly synthesized from 2
µg total RNA using the miRcute Plus miRNA First-Strand cDNA
Synthesis Kit (cat. no. KR211; Tiangen Biochemical Technology
Beijing Co. Ltd., Beijing, China) according to the manufacturer's
instructions. RT products were amplified using SYBR Green PCR (cat.
no. FP411; Tiangen Biochemical Technology Beijing Co. Ltd.) on a
Bio-Rad CFX96 real-time PCR system (Bio-Rad Laboratories, Hercules,
CA, USA). The reaction was performed according to the
manufacturer's instructions. The relative mRNA expression data were
acquired and analyzed using the 2−ΔΔCq method, and the
expression values were normalized to U6, which was used as an
internal control. The primers used in the present study were as
follows: miR-374a forward, 5′-CGGCGGTTATAATACAACCTG-3′ and reverse,
5′-AGCTCGAGTGGAAGTCTGTGCA-3′; U6 forward,
5′-CTCGCTTCGGCAGCACATATA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Data analysis
The median value of the expression levels of
miR-374a was used as the cut-off point, and the patients were
divided into a high-level and a low-level group. The expression
levels of miR-374a between glioma and normal brain tissues were
compared with the independent-samples t-test. Pearson's Chi-square
test was used to analyze the association between miR-374a
expression levels and the clinicopathological characteristics of
the patients. The OS rates of the patients in the high-level and
low-level groups were evaluated using the Kaplan-Meier method.
Furthermore, univariate and multivariate Cox proportional hazard
regression models were used to evaluate the prognostic value of
multiple variables, including miR-374a expression, sex, age and
Karnofsky performance status (KPS) score.
Bioinformatics analysis
The target genes of miR-374a were predicted using
TargetScan (http://www.targetscan.org/) (17), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php)
(18), starBase (http://starbase.sysu.edu.cn/browseIntersectTargetSite.php)
(19) and miRDB (http://www. mirdb.org/) (20). To enhance the reliability of the
bioinformatics analysis, the overlapping target genes were
identified using a Venn diagram. To further investigate the
functions of these consensus target genes, the Database for
Annotation, Visualization and Integrated Discovery (DAVID)
bioinformatics tool (https://david.ncifcrf.gov/) and mirPath v.3
(http://snf-515788.vm.okeanos.grnet.gr/) were used to
perform gene ontology (GO) and Kyoto Encyclopedia of genes and
genomes (KEGG) pathway enrichment analyses. These analyses aimed to
predict protein interactions, which included physical and
functional associations. The present study used Search Tool for the
Retrieval of Interacting Genes (STRING; http://string-db.org/) to construct the PPI network
for target genes (minimum required interaction score. >0.4). In
addition, Cytoscape software version 3.6.0 (http://www.cytoscape.org/download-platforms.html)
was used for visualization of the PPI networks.
Statistical analysis
All analyses were performed using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5 software
(GraphPad Software Inc., La Jolla, CA, USA). Data calculations from
each experiment were performed independently at least three times
and values are expressed as the mean ± standard error of the mean.
Differences between groups were assessed by one-way analysis of
variance followed by an LSD post-hoc test and a Student's t-test.
The Kaplan-Meier method was used to estimate the OS curves.
P<0.05 was considered to indicate statistical significance.
Results
miRNA profiling
The miRNA profile was analyzed for low-grade (n=3)
and high-grade gliomas (n=3) using the miRStar™ Human Cancer Focus
miRNA PCR Array, which allowed for the assessment of the expression
levels of 184 miRNAs associated with cancer. The median expression
levels of each miRNA in each of the two groups were calculated, and
the differences between them were determined using the t-test.
P<0.05 was considered to indicate significant differences in
expression and a fold change of >2 was set as the cut-off value.
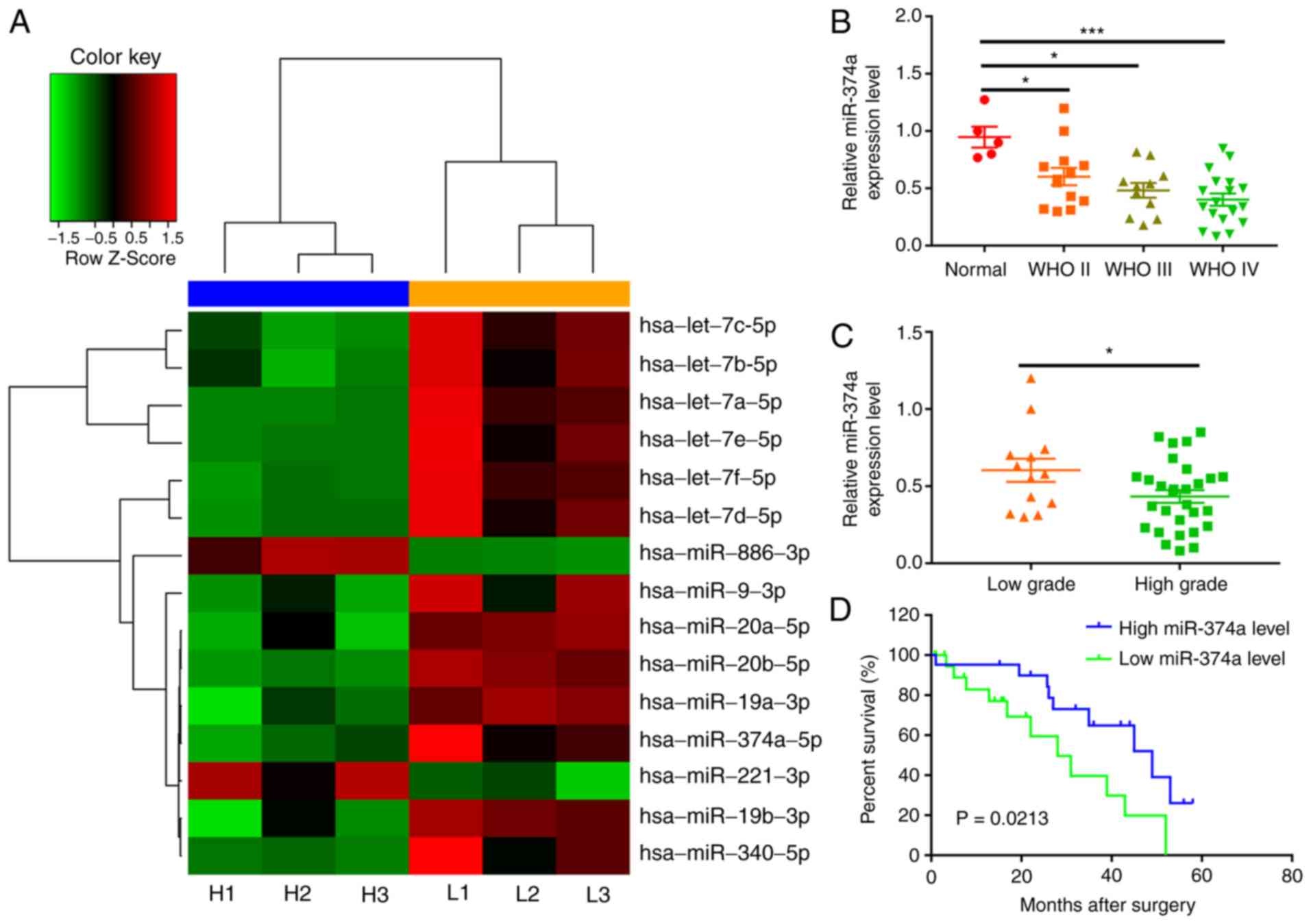
A total of 15 miRNAs, including 13 downregulated and 2 upregulated
miRNAs, were identified between low-grade and high-grade glioma
tissues(Fig. 1A; Table I). As the differences in multiples of
miR-374a were the largest, miR-374a was selected for further study.
It was revealed that the expression levels of miR-374a in the
high-grade glioma tissues were lower than those in the low-grade
glioma tissues (P=0.027).
 | Table I.Differential expression miRNAs
between high-grade and low-grade glioma groups. |
Table I.
Differential expression miRNAs
between high-grade and low-grade glioma groups.
| miRNA ID | log2 fold
change | P-value |
|---|
| hsa-let-7a-5p | −3.10 | 0.010 |
| hsa-let-7b-5p | −2.28 | 0.032 |
| hsa-let-7c-5p | −2.23 | 0.012 |
| hsa-let-7d-5p | −2.82 | 0.018 |
| hsa-let-7e-5p | −3.46 | 0.022 |
| hsa-let-7f-5p | −3.29 | 0.011 |
| hsa-miR-19a-3p | −2.60 | 0.008 |
| hsa-miR-19b-3p | −3.02 | 0.021 |
| hsa-miR-20a-5p | −2.31 | 0.017 |
| hsa-miR-221-3p | 2.58 | 0.026 |
| hsa-miR-340-5p | −4.15 | 0.044 |
|
hsa-miR-374a-5p | −4.43 | 0.027 |
| hsa-miR-886-3p | 3.82 | 0.001 |
| hsa-miR-9-3p | −3.79 | 0.049 |
miR-374a is downregulated in glioma
tissues
To validate the results that were obtained by the
microarray analysis, the expression levels of miRNA-374a we
evaluated in glioma (n=42) and normal brain tissues (n=5) by
RT-qPCR. Significantly lower miR-374a expression levels were
observed in glioma tissues compared with those in the adjacent
normal tissues and the expression of miR-374a decreased with
increasing glioma grade (P<0.05; Fig.
1B and C). These RT-qPCR data confirmed that the results of the
microarray analysis were reliable.
Association of miR-374a expression
levels with clinicopathological characteristics and OS of glioma
patients
The association of the miR-374a expression levels
with the demographic and clinicopathological characteristics of the
glioma patients was then assessed (Table II). The median expression levels of
miR-374a were used as a cut-off value, and all patients were
divided into a high-level group (n=21) and a low-level group
(n=21). The expression levels of miR-374a were associated with the
KPS score (P=0.032) and the WHO grade (P=0.028; Table II). However, the miR-374a expression
levels were not significantly associated with any of the other
parameters assessed, including age and sex (P>0.05).
 | Table II.Association of the expression level
of miR-374a with clinicopathological factors of glioma. |
Table II.
Association of the expression level
of miR-374a with clinicopathological factors of glioma.
|
|
| miR-374a expression
(n) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Patients (n) | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.334 |
|
≥50 | 15 | 6 | 19 |
|
|
<50 | 27 | 15 | 12 |
|
| Sex |
|
|
| 0.533 |
|
Male | 24 | 11 | 13 |
|
|
Female | 18 | 10 | 8 |
|
| WHO grade |
|
|
| 0.028 |
| II | 13 | 5 | 8 |
|
|
III/IV | 29 | 16 | 13 |
|
| KPS score |
|
|
| 0.032 |
|
≥90 | 11 | 2 | 9 |
|
|
<90 | 31 | 9 | 12 |
|
Kaplan-Meier curves and the log-rank test were used
in order to evaluate the prognostic value of miR-374a in glioma.
The results indicated that patients with low expression levels of
miR-374a had a significantly shorter OS than those with high
miR-374a expression levels (P=0.021; Fig. 1D). Furthermore, Cox regression
analysis indicated that the expression levels of miR-374a were
associated with the OS of glioma patients (hazard ratio, 0.472; 95%
confidence interval, 0.125–1.733; P<0.05) and the WHO grade was
also significantly associated with the OS of glioma patients
(hazard ratio, 1.914; 95% confidence interval, 1.362–3.885;
P<0.05; Table III)
Collectively, these data suggest that miR-374a may be a prognostic
factor regarding OS in patients with glioma.
 | Table III.Univariate and multivariate analyses
of prognostic factors in patients with glioma. |
Table III.
Univariate and multivariate analyses
of prognostic factors in patients with glioma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≥50 vs. <50
years) | 1.414 | 0.591–3.384 | 0.436 | 1.031 | 0.421–2.524 | 0.946 |
| Sex (male vs.
female) | 1.19 | 0.497–2.851 | 0.696 | 1.155 | 0.444–3.007 | 0.768 |
| KPS (≥90 vs.
<90) | 0.274 | 0.089–0.844 | 0.024 | 1.025 | 0.390–2.699 | 0.056 |
| WHO grade (II vs.
III/IV) | 1.51 | 1.163–2.98 | 0.027 | 1.914 | 1.362–3.885 | 0.036 |
| miR-374a expression
(low vs. high) | 0.379 | 0.154–0.936 | 0.021 | 0.472 | 0.125–1.733 | 0.016 |
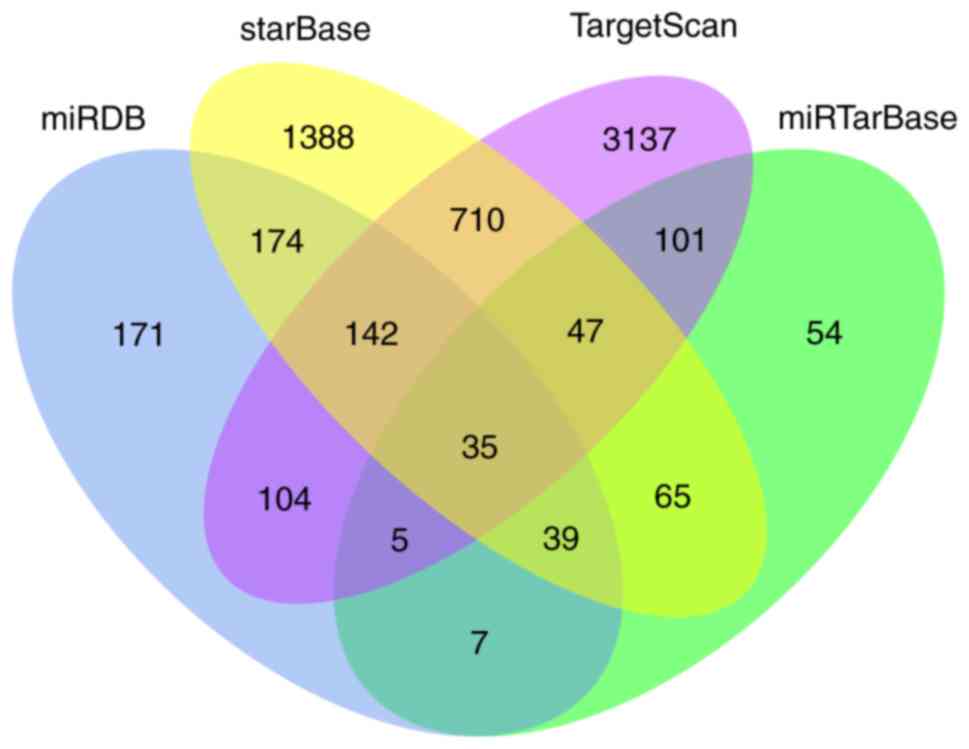
Target prediction and functional
analysis
To elucidate the potential biological function of
miR-374a in glioma, the target genes of miR-374a were predicted
using the TargetScan, miRTarBase, StarBase and miRDB online
analysis tools. A total of 35 overlapping genes among the four
different tools were identified (Fig.
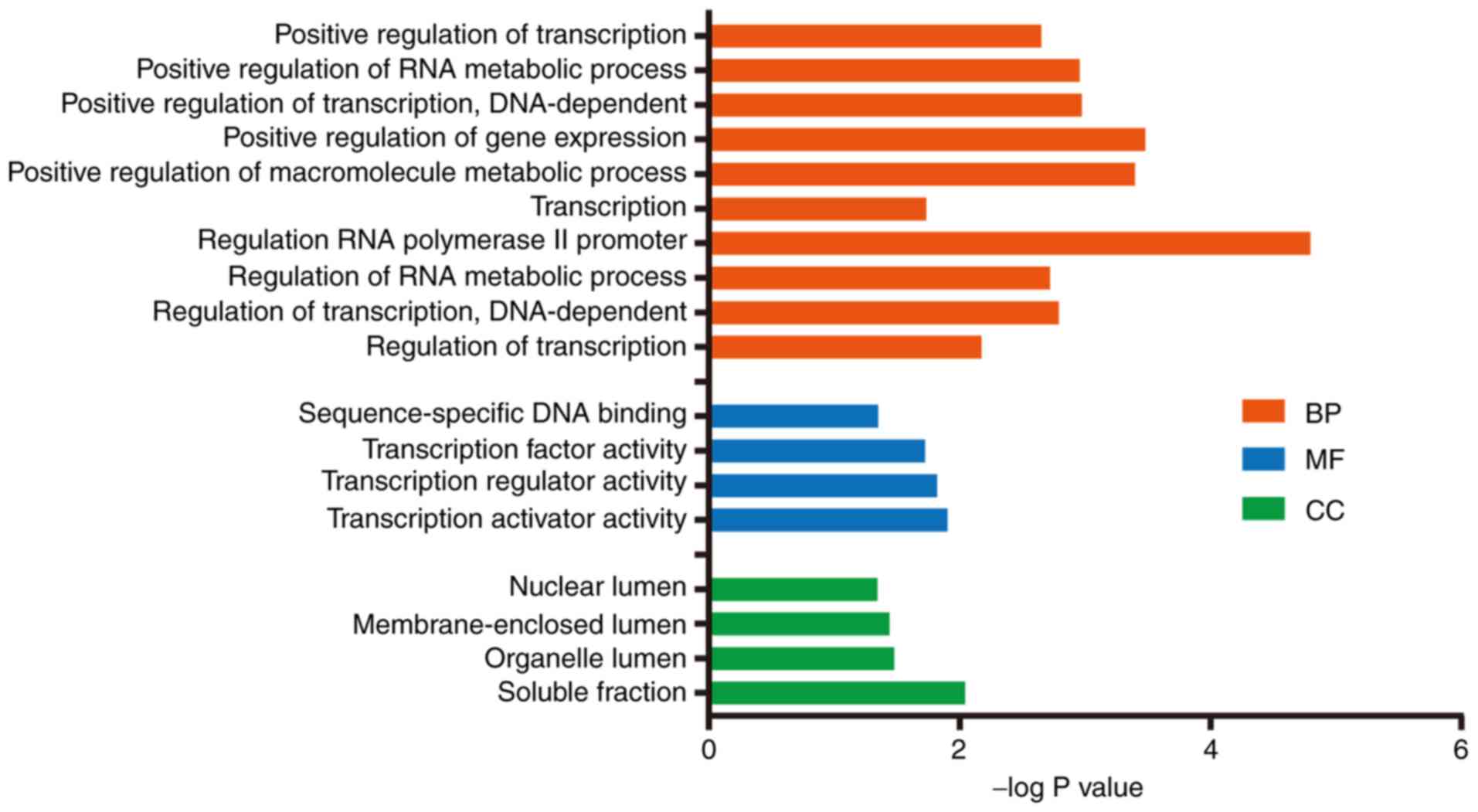
2). Subsequently, a functional enrichment analysis was
performed to investigate the biological functions of these
consensus target genes. The enriched GO terms in the categories
biological process (BP), cellular component and molecular function
were identified. In the category BP, the target genes were mostly
involved in metabolic processes (Fig.
3).
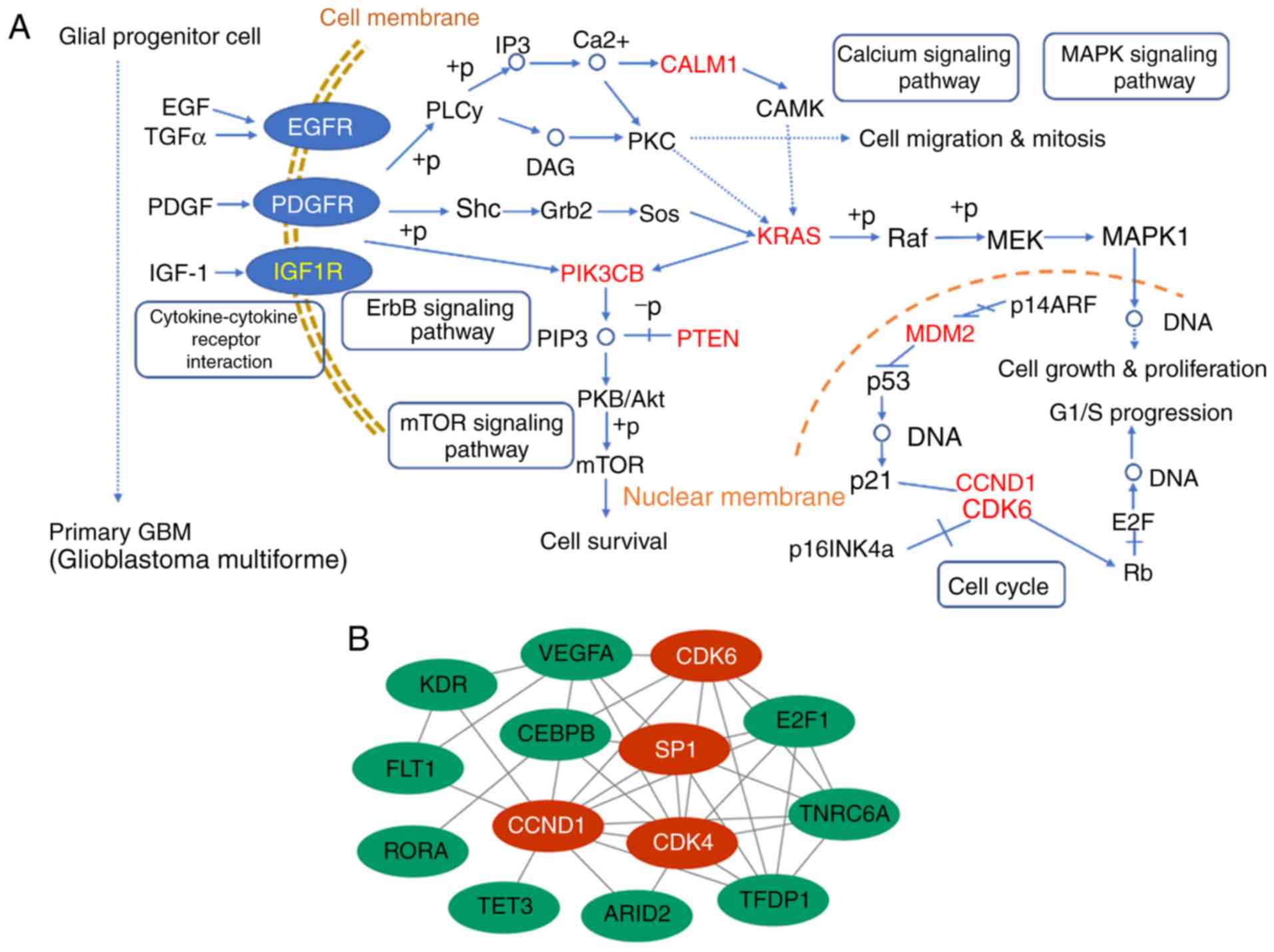
KEGG pathway analysis and PPI
network
The KEGG pathways of the miR-374a target genes were
predicted using DIANA-miRPath in order to identify miRNA-mRNA
regulatory signaling pathways in glioma (21–23). The
most significant biological pathways were associated with tumor
progression from glial progenitor cells to primary GBM, including
calcium metabolism, ErbB, mammalian target of rapamycin (mTOR) and
several cell cycle-associated pathways (Fig. 4A). PPI network analysis of all target
genes using the STRING database revealed 125 interactions involved.
The nodes with a degree of interaction of ≥7 were defined as hub
genes, including cyclin D (CCND)1, specificity protein 1 (Sp1),
cyclin-dependent kinase (CDK)4 and CDK6 (Fig. 4B).
Discussion
miRNAs are regulators of post-transcriptional gene
expression and are involved in multiple complex pathways (24). A large number of studies have
reported that certain miRNA are involved in the regulation of tumor
progression, and may have tumor-promoting or tumor-suppressive
roles with regard (25–27). miRNAs may be used as prognostic
indicators and therapeutic targets in gliomas (28,29). In
the present study, comprehensive miRNA profiling of high-grade vs.
low-grade gliomas was performed by microarray analysis. A total of
15 significantly altered miRNAs in high-grade vs. low-grade gliomas
were identified. In addition, a novel miRNA, miR-374a, was
identified to be differentially expressed between normal and glioma
tissues, which has not been previously reported. Therefore,
miR-374a was selected for subsequent investigation. It was
demonstrated that miR-374a was significantly lower expressed in
high-grade vs. low-grade gliomas. The miRNA expression was verified
by RT-qPCR in order to confirm the results of the microarray
profiling. Furthermore, the present results indicated that miR-374a
may be considered a potential prognostic biomarker in glioma due to
the association of its expression levels with the long-term
survival of glioma patients.
A number of studies have also reported that the
abnormal expression of miR-374a are involved in multiple types of
cancer. Li et al (30)
revealed that deregulation of miR-374a may be involved in the
development and regulation of cisplatin resistance in ovarian
cancer cells. Wu et al (31)
demonstrated that the expression levels of miR-374a were
significantly lower in lung adenocarcinoma compared with those in
the adjacent normal tissues. Furthermore, regulation of
transforming growth factor α gene expression by miR-374a inhibited
the proliferation, migration and invasion of lung adenocarcinoma
cells (31). Slattery et al
(32) demonstrated that the
expression levels of miR-374a were downregulated in colorectal
cancer, whereas low miR-203 expression levels were associated with
worse clinicopathological data and shorter OS time. However, Xu
et al (33) reported that
miR-374a acts as a tumor promoter in gastric cancer, where it and
promotes cell proliferation, migration and invasion via the
regulation of SRC kinase signaling inhibitor 1 expression levels.
In addition, Pan et al (34)
highlighted that the expression levels of miR-374 were decreased in
glioma tissues and were associated with the prognosis of glioma
patients, which is consistent with the results of the present
study.
It has been reported that the aberrant expression of
certain miRNAs is associated with the development of cancer via the
abnormal regulation of multiple BPs and signaling pathways
(35). To further elucidate the
molecular function of miR-374a and its target genes, functional
enrichment analyses of the target genes in GO terms and KEGG
pathways were performed. The GO analysis demonstrated that the
terms in the category BP included the regulation of the RNA
transcription and the nucleic acid metabolic processes.
Furthermore, several of the enriched pathways were associated with
tumorigenesis, including ErbB, mTOR and cell cycle signaling
pathways. The ErbB pathway is associated with tumor progression in
the majority of cancer types, including glioma (36,37).
Furthermore, it was reported that the mTOR pathway is a crucial
signaling pathway involved in the development of glioma (38).
In addition, four hub genes, CCND1, SP1, CDK6 and
CDK4, were identified from the PPI of predicted target genes of
miR-374a, which may be directly or indirectly involved in the
development of glioma. CCND1 is a protein required for the
progression from G1 phase to the S phase of the cell cycle. CCND1
overexpression is associated with early cancer onset (39) and tumor progression and decreased Fas
expression, leading to increased chemotherapeutic resistance and
protection from apoptosis (40).
CDK4 and CDK6 are two members of the CDK family that bind to CCND1.
A dysregulation of CDK4/6 may promote GBM proliferation; however,
CDK4/6 kinase inhibitors were demonstrated to inhibit cell
proliferation in subcutaneous glioma models (41). Sp1 belongs to the Sp/Kruppel-like
factor family of transcription factors and is widely expressed in
gliomas. Sp1 has an important role in the activation of oncogenes
required for tumor survival (42).
Guan et al (43) demonstrated
that SP1 is upregulated in human glioma and may serve as a
prognostic marker.
The primary limitation of the present study is that
no luciferase reporter assays of the hub genes (CCND1, SP1, CDK6
and CDK4) were performed to validate the target genes of miR-374a.
Therefore, further analyses are required to determine the
mechanisms in the processes of malignant progression in gliomas.
Future studies will use PCR to verify the expression levels of
miR-374a in glioma cells and a combination of other molecular
markers to successfully identify patients with OS.
In conclusion, the present study demonstrated that
the downregulation of miR-374a is associated with reduced survival
in glioma patients. miR-374a may be a potential prognostic factor
for patients with glioma. However, the results of the present study
should be verified using a larger sample size and further
experimental research is required to confirm the functions of
miR-374a in the progression of glioma.
Acknowledgements
Not applicable.
Funding
This project was supported by the Natural Science
Foundation of Gansu (grant nos. 18JR3RA365 and 18JR3RA309), the
Research Fund from the Project of the Healthy and Family Planning
Commission of Gansu (grant no. GSWSKY-2014-31/2015-58), the Lanzhou
Science and Technology Bureau Project (grant no. 2018-1-109) and
the doctoral research fund and the Cuiying Science and Technology
fund of Lanzhou University Second Hospital (grant nos.
ynbskyjj2015-1-02/2015-2-11/2015-2-5 and
CY2017-MS12/-MS15/CYXZ-01).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YP and QD initiated the project. QD, ML, GY, MW and
JH performed the experiments. QD, ML, QX and GY analyzed the data.
QX, JH, SL and XM generated the figures. YP, QD, GY and ML wrote
the manuscript. All co-authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the Second Hospital of the Lanzhou University. Written
informed consent was provided by each patient or their guardians
prior his/her participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathologica. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Delgado-López PD and Corrales-García EM:
Survival in glioblastoma: A review on the impact of treatment
modalities. Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kleihues P, Burger PC, Aldape KD, et al;
Louis DN, Ohgaki H, Wiestler OD and Cavenee WK: WHO classification
of tumours of the central nervous system. 2007.
|
|
7
|
Abrunhosa-Branquinho AN, Bar-Deroma R,
Collette S, Clementel E, Liu Y, Hurkmans CW, Feuvret L, Van Beek K,
van den Bent M, Baumert BG and Weber DC: Radiotherapy quality
assurance for the RTOG 0834/EORTC 26053-22054/NCIC CTG CEC.1/CATNON
intergroup trial ‘concurrent and adjuvant temozolomide chemotherapy
in newly diagnosed non-1p/19q deleted anaplastic glioma’:
Individual case review analysis. Radiother Oncol. 127:292–298.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou L, Liu F, Wang X and Ouyang G: The
roles of microRNAs in the regulation of tumor metastasis. Cell
Biosci. 5:322015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aakula A, Kohonen P, Leivonen SK, Mäkelä
R, Hintsanen P, Mpindi JP, Martens-Uzunova E, Aittokallio T,
Jenster G, Perälä M, et al: Systematic identification of MicroRNAs
that impact on proliferation of prostate cancer cells and display
changed expression in tumor tissue. Eur Urol. 69:1120–1128. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu QN, Renaud H and Guo Y:
Bioinformatics-based identification of miR-542-5p as a predictive
biomarker in breast cancer therapy. Hereditas. 155:172018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Man HB, Bi WP and Man HH: Decreased
microRNA-198 expression and its prognostic significance in human
glioma. Genet Mol Res. 15–2016. View Article : Google Scholar
|
|
12
|
Zhang J, Lv J, Zhang F, Che H, Liao Q,
Huang W, Li S and Li Y: MicroRNA-211 expression is down-regulated
and associated with poor prognosis in human glioma. J Neurooncol.
133:553–559. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan GQ, Wei NL, Mu LY, Wang XQ, Zhang YN,
Zhou WN and Pan YW: A 4-miRNAs signature predicts survival in
glioblastoma multiforme patients. Cancer Biomark. 20:443–452. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou MH, Zhou HW, Liu M and Sun JZ: The
role of miR-92b in cholangiocarcinoma patients. Int J Biol Markers.
33:293–300. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sarrion I, Milian L, Juan G, Ramon M,
Furest I, Carda C, Cortijo Gimeno J and Mata Roig M: Role of
circulating miRNAs as biomarkers in idiopathic pulmonary arterial
hypertension: Possible relevance of miR-23a. Oxid Med Cell Longev.
2015:7928462015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ekimler S and Sahin K: Computational
Methods for MicroRNA Target Prediction[J]. Genes. 5(3): 671–683.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:(Database Issue). D163–D169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:(Database Issue). D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40:W498–W504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic eAcids Rse. 43:(Database Issue). D146–D152. 2015.
View Article : Google Scholar
|
|
22
|
Papadopoulos GL, Alexiou P, Maragkakis M,
Reczko M and Hatzigeorgiou AG: DIANA-mirPath: Integrating human and
mouse microRNAs in pathways. Bioinformatics. 25:1991–1993. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fazi B, Felsani A, Grassi L, Moles A, D
Andrea D, Toschi N, Sicari D, De Bonis P, Anile C, Guerrisi MG, et
al: The transcriptome and miRNome profiling of glioblastoma tissues
and peritumoral regions highlights molecular pathways shared by
tumors and surrounding areas and reveals differences between
short-term and long-term survivors. Oncotarget. 6:22526–22552.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dai J, Li Q, Bing Z, Zhang Y, Niu L, Yin
H, Yuan G and Pan Y: Comprehensive analysis of a microRNA
expression profile in pediatric medulloblastoma. Mol Med Rep.
15:4109–4115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Babashah S and Soleimani M: The oncogenic
and tumour suppressive roles of microRNAs in cancer and apoptosis.
Eur J Cancer. 47:1127–1137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang
JH and Wei YQ: Oncogenic and tumor suppressive roles of microRNAs
in apoptosis and autophagy. Apoptosis. 19:1177–1189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xue L, Wang Y, Yue S and Zhang J: The
expression of miRNA-221 and miRNA-222 in gliomas patients and their
prognosis. Neurol Sci. 38:67–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji Y, Wei Y, Wang J, Gong K, Zhang Y and
Zuo H: Correlation of microRNA-10b upregulation and poor prognosis
in human gliomas. Tumour Biol. 36:6249–6254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li N, Yang L, Wang H, Yi T, Jia X, Chen C
and Xu P: MiR-130a and MiR-374a function as novel regulators of
cisplatin resistance in human ovarian cancer A2780 cells. PLoS One.
10:e01288862015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu H, Liu Y, Shu XO and Cai Q: MiR-374a
suppresses lung adenocarcinoma cell proliferation and invasion by
targeting TGFA gene expression. Carcinogenesis. 37:567–575. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Slattery ML, Pellatt AJ, Lee FY, Herrick
JS, Samowitz WS, Stevens JR, Wolff RK and Mullany LE: Infrequently
expressed miRNAs influence survival after diagnosis with colorectal
cancer. Oncotarget. 8:83845–83859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu X, Wang W, Su N, Zhu X, Yao J, Gao W,
Hu Z and Sun Y: miR-374a promotes cell proliferation, migration and
invasion by targeting SRCIN1 in gastric cancer. FEBS Lett.
589:407–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan Z, Shi Z, Wei H, Sun F, Song J, Huang
Y, Liu T and Mao Y: Magnetofection based on superparamagnetic iron
oxide nanoparticles weakens glioma stem cell proliferation and
invasion by mediating high expression of MicroRNA-374a. J Cancer.
7:1487–1496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang C, Zhang C, Ma M and Dai D:
Three-microRNA signature identified by bioinformatics analysis
predicts prognosis of gastric cancer patients. World J
Gastroenterol. 24:1206–1215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Späth F, Andersson U, Dahlin AM, Langseth
H, Hovig E, Johannesen TB, Grankvist K, Björkblom B, Wibom C and
Melin B: Pre-diagnostic serum levels of EGFR and ErbB2 and genetic
glioma risk variants: A nested case-control study. Tumour Biol.
37:11065–11072. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu R, Qu Y, Chen L, Pu J, Ma S, Zhang X,
Yang Q, Shi B, Hou P and Ji M: Genomic copy number gains of ErbB
family members predict poor clinical outcomes in glioma patients.
Oncotarget. 8:92275–92288. 2017.PubMed/NCBI
|
|
38
|
Wang N, Zhang Q, Luo L, Ning B and Fang Y:
β-asarone inhibited cell growth and promoted autophagy via
P53/Bcl-2/Bclin-1 and P53/AMPK/mTOR pathways in Human Glioma U251
cells. J Cell Physiol. 233:24342018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Diehl JA: Cycling to cancer with cyclin
D1. Cancer Biol Ther. 1:226–231. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shintani M, Okazaki A, Masuda T, Kawada M,
Ishizuka M, Doki Y, Weinstein IB and Imoto M: Overexpression of
cyclin DI contributes to malignant properties of esophageal tumor
cells by increasing VEGF production and decreasing Fas expression.
Anticancer Res. 22:639–647. 2002.PubMed/NCBI
|
|
41
|
Yin L, Li H, Liu W, Yao Z, Cheng Z, Zhang
H and Zou H: A highly potent CDK4/6 inhibitor was rationally
designed to overcome blood brain barrier in gliobastoma therapy.
Eur J Med Chem. 144:1–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bajpai R and Nagaraju GP: Specificity
protein 1: Its role in colorectal cancer progression and
metastasis. Crit Rev Oncol Hematol. 113:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J
and Li M: Sp1 is upregulated in human glioma, promotes
MMP-2-mediated cell invasion and predicts poor clinical outcome.
Int J Cancer. 130:593–601. 2012. View Article : Google Scholar : PubMed/NCBI
|