Introduction
Osteosarcoma (OS) is one of the most aggressive and
common types of bone cancer and is the major cause of
tumor-associated mortality among the youth worldwide (1,2). At
present, radical surgery and neo-adjuvant chemotherapy are the main
methods for the treatment of OS, and these approaches markedly
increase the survival of patients with OS (2). However, the clinical outcomes of OS
remain poor due to the high rates of tumor metastasis and
recurrence, making OS a serious threat to human health and life
(3). Therefore, the underlying
mechanism of OS must be elucidated to develop new effective
therapeutic targets.
microRNAs (miRNAs/miRs) are a class of noncoding
transcripts, 18–25 nucleotides in length, that participate in
diverse human diseases, including cancer (4–6). miRNAs
target the 3′-untranslated region (UTR) of specific mRNAs and
regulate gene expression (7).
Through this mechanism, miRNAs regulate numerous biological
processes, including cell proliferation, apoptosis, differentiation
and metastasis (8–10). Scholars reported that miRNAs function
in different types of human cancer. For example, Bai et al
(11) revealed that miRNA-196b
suppressed cell metastasis and proliferation by inhibiting Runx2 in
lung cancer. Wang et al (12)
reported that miR-101 suppressed proliferation and migration of
breast cancer cells by targeting SOX2. Furthermore, it has been
demonstrated that miR-329 inhibited OS development (13), and miR-506 overexpression inhibited
cell proliferation and enhanced apoptosis in OS (14). Nonetheless, the function of numerous
miRNAs in OS remains unclear. Understanding the functions and
regulatory networks of miRNAs may benefit OS intervention.
miR-552-5p acts as an oncogene in certain types of
cancer. For instance, miRNA-552 targets dachshund homolog 1 (DACH1)
to enhance cell proliferation and migration in colorectal cancer
(15). Wang et al (16) revealed that miR-552 promoted
colorectal cancer metastasis. Nevertheless, the role of miR-552-5p
in OS remains to be investigated.
In the current study, miR-552-5p was overexpressed
in OS tissues and cell lines compared with normal tissues and
osteoblast cells. Knockdown of miR-552-5p significantly suppressed
the proliferation, migration and invasion of OS cells. miR-552-5p
inhibitors inhibited the cell cycle and it directly targeted the
3′-UTR of Wnt inhibitory factor 1 (WIF1) mRNA. Furthermore,
miR-552-5p overexpression inhibited the WIF1 expression in OS
cells. WIF1 was downregulated in OS tissues and cell lines. WIF1 is
an inhibitor of the Wnt/β-catenin pathway, which serves a role in
tumor growth and metastasis (17).
By inhibiting WIF1 expression, miR-552-5p promoted the
proliferation, migration and invasion of OS cells.
Materials and methods
Clinical specimens
A total of 51 histologically diagnosed OS tissues
(33 patients <20 years old and 18 patients ≥20 years old; sex:
30 males and 21 females) and 19 adjacent normal tissues were
collected from patients of the Huai'an First People's Hospital,
Nanjing Medical University (Huai'an, China). The samples were
divided into two groups according to the existence of lymph node
metastasis. None of the patients received immunotherapy,
radiotherapy or chemotherapy prior to the surgery. Written informed
consent was obtained from each patient before the clinical
specimens were used. The tissue specimens were conserved in liquid
nitrogen for further investigation. The present study was approved
by the Research Ethics Committee of Nanjing Medical University in
accordance with the Declaration of Helsinki.
Cell culture and transfection
Human osteoblast cell line hFOB1.19
(CRL-11372™) and human OS cell lines, including U2OS,
MG63 and SAOS2 were obtained from the American Type Culture
Collection (Manassas, VA, USA). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine
serum (FBS; HyClone; GE Healthcare Life Sciences) and penicillin
and streptomycin (both 50 U/ml; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a humidified incubator with 5% CO2
at 37°C.
miR-552-5p inhibitors (50 nM;
5′-CCAACAGGCAAAAGGUUAAAC-3′), miR-552-5p mimics (50 nM;
5′-GUUUAACCUUUUGCCUGUUGG-3′) and negative controls (NC; 50 nM;
5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were obtained from GeneCopoeia,
Inc. (Rockville, MD, USA). Specific small interfering (si)RNAs
against WIF1 (100 nM; siWIF1, 5′-GCAAUAUAAUAUAUUGUAAAC-3′) and
scrambled NC (100 nM; siNC, 5′-AAUUCUCCGAACGUGUCACGU-3′) were
synthesized by Wuhan Genesil Biotechnology Co., Ltd (Wuhan, China).
miRNAs and siRNAs were transfected into 5×106 cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol. A
total of 48 h post transfection, the efficiency was validated using
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR).
Cell proliferation assay
Cell proliferation was detected using a Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). The cells were grown in a 96-well plate at a
density of 2×103 cells/well and incubated at 37°C under
5% CO2 until a cell confluence rate of 70% was attained.
Following transfection with the miRNA inhibitors or NC for 48 h,
the cells were cultured for 24, 48 and 72 h. Then, CCK-8 solution,
~10 µl, was added into each well. The absorbance was measured at a
wavelength of 450 nm using a Sunrise™ microplate reader
(Tecan Group, Ltd., Mannedorf, Switzerland).
Colony formation assay
For the colony formation assay, OS cells were
counted at 24 h post-transfection and seeded into 24-well plates at
a density of 1×103 cells/well. The culture medium was
replaced every 3 days. After 14 days, the cells were washed with 1X
PBS, fixed with 4% formaldehyde for 30 min at 25°C, stained with
0.5% crystal violet for 30 min at 25°C, and counted using an
inverted light microscope (IX83; Olympus Corporation, Tokyo, Japan)
at a magnification of ×100.
Migration and invasion assay
The lower chambers (8 µm pore; Corning,
Incorporated, Corning, NY, USA) were filled with 600 µl of DMEM
supplemented with 20% FBS to induce the migration of OS cells. A
total of 2×104 cells were seeded in the upper chamber
filled with 200 µl serum-free DMEM medium. At 24 h after seeding,
OS cells that migrated through the membranes were stained with 0.5%
crystal violet dye for 30 min at 25°C and cell number was counted
under the light microscope at a magnification of ×100. For the
invasion assay, the membrane was covered with 70 µl of Matrigel and
the remaining steps were the same as for the migration assay.
RT-qPCR
RNA was isolated from 1×107 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) in accordance with the manufacturer's protocol. First-strand
cDNA was synthesized using a TIANScript RT kit (Tiangen Biotech
Co., Ltd., Beijing, China). The thermocycling conditions were as
follows: 37°C for 25 min and 85°C for 5 sec. qPCR amplification was
performed using TaqMan Human microRNA assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.) for miRNA analysis and UltraSYBR
Mixture (cat. no. CW0957; Beijing ComWin Biotech, Co., Ltd.,
Beijing, China) for mRNA analysis on a LightCycler® 480
PCR system (Roche Diagnostics, Indianapolis, IN, USA). The
thermocycling conditions were as follows: Denaturation at 95°C for
10 min, followed by 40 cycles of denaturation at 95°C for 15 sec
and elongation at 60°C for 1 min. The gene expression was
calculated using the 2−ΔΔCq method (18). U6 was used as a reference gene to
normalize the miR-552-5p expression. The primers used for
miR-552-5p and U6 were synthesized and purchased from GeneCopoeia,
Inc. (Rockville, MD, USA). GAPDH was used to normalize WIF1
expression. The primer sequences were as follows: WIF1 (forward,
5′-TTGTTTCAGTGCTTTGGGACAG-3′ and reverse,
5′-CCCCCAGACACCATAAATGC-3′), miR-552-5p (forward,
5′-GTTTAACCTTTTGCCTGTTGG-3′ and reverse,
5′-CGAACGCTTCACGAATTTG-3′), U6 (forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-CGAACGCTTCACGAATTTG-3′) and GAPDH (forward,
5′-TCATGGGTGTGAACCATGAGAA-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′).
Luciferase reporter assay
The potential targets of miR-552-5p were analyzed
using the TargetScan7 tool (http://www.targetscan.org/vert_71/). The wild-type
(WT) or mutant (MUT) 3′-UTR of WIF1 was amplified and cloned into a
pmiR-RB-REPORT™ luciferase vector (Promega Corporation,
Madison, WI, USA). The WT or MUT 3′-UTR of WIF1 was co-transfected
with a miR-552-5p mimic or negative control into 5×103
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). A total of 48 h after the
co-transfection, reporter activity was detected using a
Dual-Luciferase® Reporter Assay kit (Promega
Corporation). The firefly luciferase activity was normalized to the
luciferase activity of Renilla.
Cell cycle analysis
A total of 1×106 cells were harvested,
washed twice with ice-cold PBS and then fixed in 70% ethanol for 24
h at 4°C. Cells were subsequently washed three times with ice cold
PBS and incubated with 1 mg/ml RNase A (cat. no. R6148;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at 37°C.
Following this, cells were stained at 25°C for 10 min with 50 µg/ml
propidium iodide (BD Biosciences, Franklin Lakes, NJ, USA) in 0.5%
Tween-20 with PBS and subjected to analysis of cell cycle
distribution using a BD FACScan flow cytometer coupled with Cell
Quest acquisition and analysis programs (version 2; both BD
Biosciences).
Western blotting
Cells were lysed in cold radioimmunoprecipitation
assay buffer (Thermo Fisher Scientific, Inc.) and the protein
concentration was determined using a Bicinchoninic Acid Protein
Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Protein (40
µg/lane) was separated via SDS-PAGE on a 10% gel and then
transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo
Fisher Scientific, Inc.). Following this, the membrane was blocked
using 5% non-fat milk in PBS (Thermo Fisher Scientific, Inc.)
containing 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA) at room
temperature for 3 h. Subsequently, the PVDF membrane was incubated
with anti-Cyclin D1 (cat. no. ab16663), anti- proliferating cell
nuclear antigen (PCNA; cat. no. ab92552), anti-p21 (cat. no.
ab109520) and anti-GAPDH (cat. no. ab9485; all 1:1,000; Abcam,
Cambridge, MA, USA) primary antibodies at room temperature for 2 h.
Following washing with PBS for 10 min, the PVDF membrane was
incubated with horseradish peroxidase-conjugated goat anti-rabbit
secondary antibodies (cat. no. ab7090; 1:5,000; Abcam) at room
temperature for 1 h. Membranes were then washed with PBS for 10 min
and the protein bands were visualized using an Enhanced
Chemiluminescence Western Blotting kit (Pierce; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Protein densitometry was performed using ImageJ software (version
1.41; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Data are presented as the mean ± standard deviation
and were analyzed using GraphPad Prism 5 software (GraphPad
Software, Inc., La Jolla, CA, USA). Pearson's correlation
coefficient analysis was used to determine association between
miR-552-5p and WIF1 expression. Student's t-test and one-way
analysis of variance followed by Tukey's post hoc test were used to
analyze two or multiple groups, respectively. P<0.05 was
considered to indicate a statistically significant difference.
Results
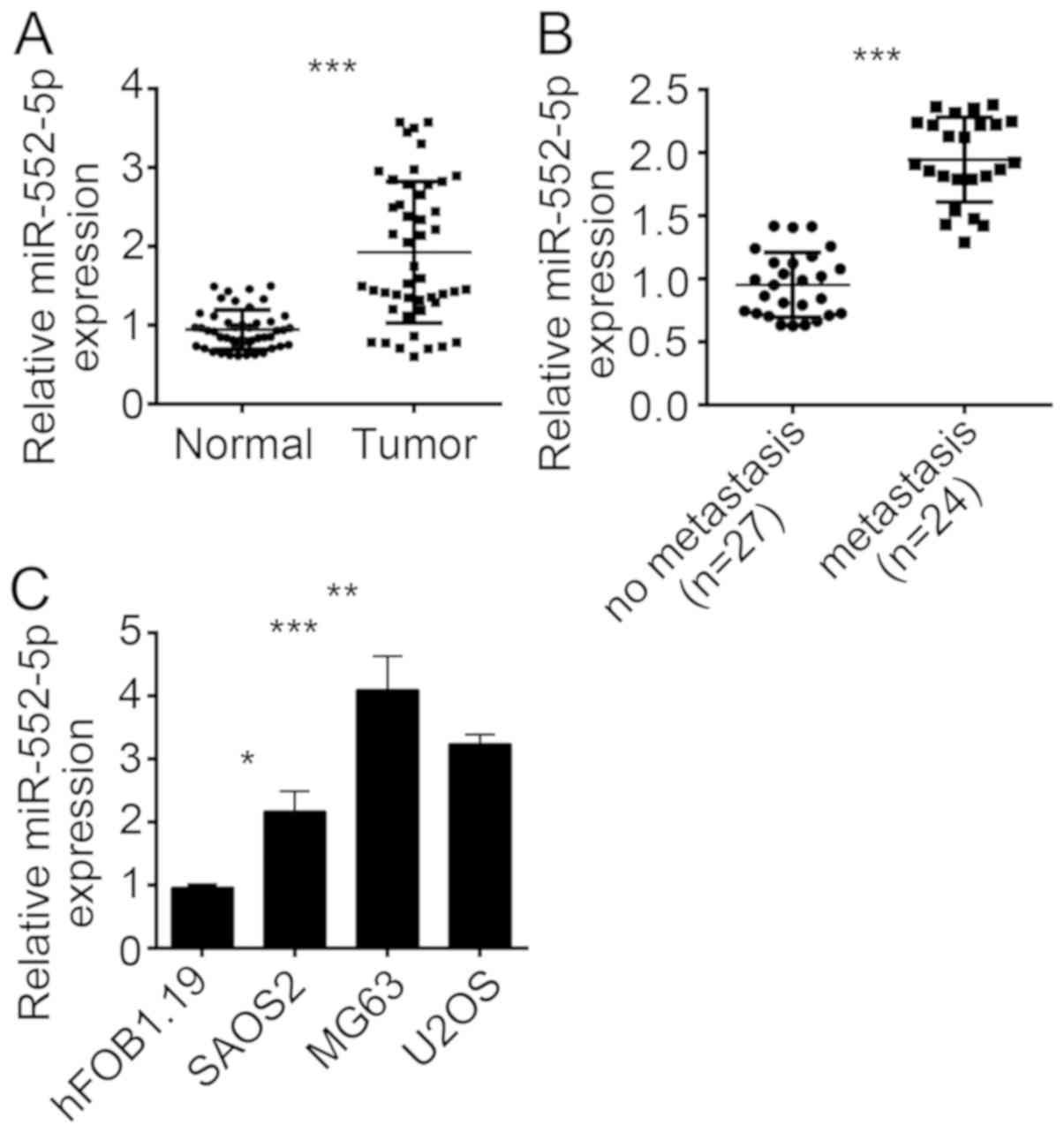
miR-552-5p is overexpressed in
osteosarcoma tissues and cell lines
To investigate the function of miR-552-5p, the
expression of this miRNA was initially determined in OS tissues.
The results of RT-qPCR revealed that the expression of miR-552-5p
was upregulated in OS tissues compared with adjacent normal tissues
(Fig. 1A). OS metastasis is a major
cause of OS-induced mortality and poor outcomes (13). To determine the association between
miR-552-5p expression and OS metastasis, the 51 OS tissues were
divided into the metastasis and non-metastasis groups. The RT-qPCR
result indicated that miR-552-5p exhibited an increased expression
level in metastatic OS tissues compared with non-metastatic tissues
(Fig. 1B). The expression pattern of
miR-552-5p was further determined in OS cell lines. The results
revealed that miR-552-5p was overexpressed in SAOS2, MG63 and U2OS
cells compared with normal osteoblast hFOB1.19 cells (Fig. 1C). The miR-552-5p expression was the
highest in MG63 and U2OS cells. Therefore, MG63 and U2OS cells were
used in the subsequent experiments.
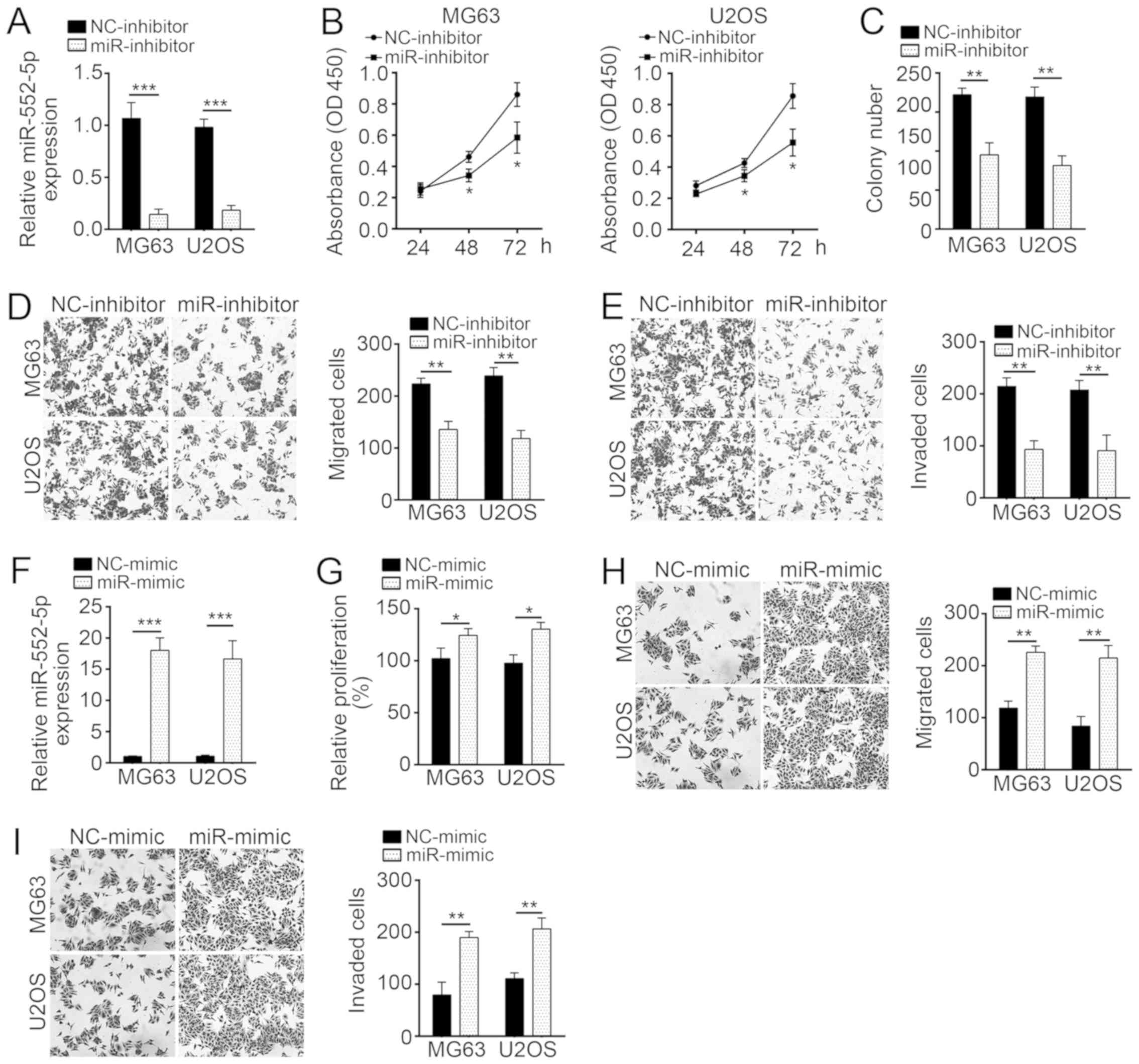
miR-552-5p knockdown suppresses
proliferation, migration and invasion of MG63 and U2OS cells
The expression of miR-552-5p was knocked down by
miR-552-5p inhibitors in MG63 and U2OS cells (Fig. 2A). To evaluate the effect of
miR-552-5p on cell proliferation, CCK-8 and colony formation assays
were performed in MG63 and U2OS cells treated with miR-552-5p
inhibitor or NC. The results indicated that miR-552-5p knockdown
significantly inhibited cell proliferation and colony formation
(Fig. 2B and C). As aforementioned,
metastatic OS tissues exhibited a high miR-552-5p expression
(Fig. 1B). Therefore, the effect of
miR-552-5p on cellular mobility was further assessed in
vitro. Transwell assays revealed that miR-552-5p knockdown
markedly suppressed the migration and invasion of MG63 and U2OS
cells, compared with the corresponding NC-inhibitor groups
(Fig. 2D and E). Furthermore,
ectopic expression of miR-552-5p promoted proliferation, migration
and invasion of OS cells, compared with the NC-mimic treatment
(Fig. 2F-I).
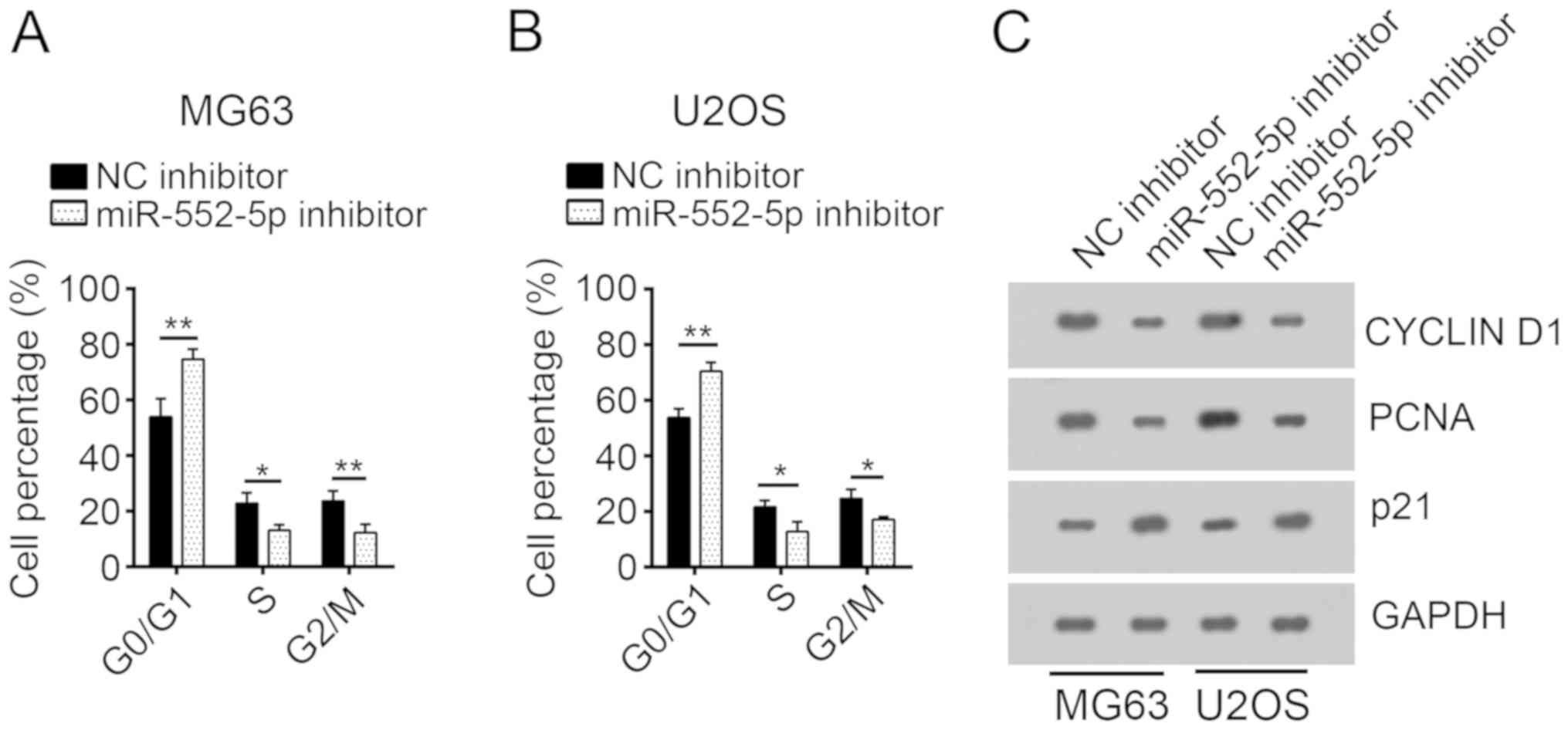
miR-552-5p knockdown inhibits the cell
cycle of osteosarcoma cells
To investigate the mechanism underlying the
decreased cell proliferation following transfection with miR-552-5p
inhibitors, the cell cycle of MG63 and U2OS cells was analyzed.
Following transfection with miR-552-5p inhibitors, a significantly
increased number of MG63 and U2OS cells remained in the G0 phase,
compared with the NC inhibitor group. Furthermore, an increased
number of cells in the NC inhibitor group entered the S and G2/M
phases, compared with cells treated with miR-552-5p inhibitors
(Fig. 3A and B). Protein expression
levels of cell cycle-associated genes, including Cyclin D1, PCNA
and p21 (4), were determined through
western blot analysis. Following miR-552-5p knockdown, the
expression levels of Cyclin D1 and PCNA were decreased, whereas the
expression of p21, a cell-cycle inhibitor (19), was increased, compared with the NC
inhibitor group (Fig. 3C).
WIF1 is a target of miR-552-5p in
osteosarcoma cells
miRNAs regulate gene expression by targeting the
3′-UTR of specific mRNAs (20). To
determine the molecular mechanism of miR-552-5p, downstream target
genes of miR-552-5p were analyzed using bioinformatics methods. The
results of this analysis indicated that WIF1 was a potential target
of miR-552-5p, as this miRNA included a potential binding site in
the 3′-UTR of WIF1 mRNA (Fig. 4A).
Luciferase activity reporter assay indicated that the transfection
with miR-552-5p mimics inhibited luciferase intensity in MG63 and
U2OS cells transduced with the WT 3′-UTR of WIF1 mRNA (Fig. 4B). However, the mutation of the
binding site of miR-552-5p in the 3′-UTR of WIF1 mRNA abrogated the
inhibitory effect of miR-552-5p mimics on the luciferase intensity
(Fig. 4B). These data implied that
miR-552-5p directly targeted the mRNA of WIF1 in MG63 and U2OS
cells. Furthermore, overexpression of miR-552-5p significantly
downregulated the mRNA expression level of WIF1 in MG63 and U2OS
cells (Fig. 4C). In addition, the
expression levels of miR-552-5p and WIF1 were inversely correlated
in OS tissues (Fig. 4D). The WIF1
expression in OS tissues and cell lines was determined by RT-qPCR.
WIF1 was significantly downregulated in OS tissues and cell lines
compared with normal tissues and cells (Fig. 4E and F).
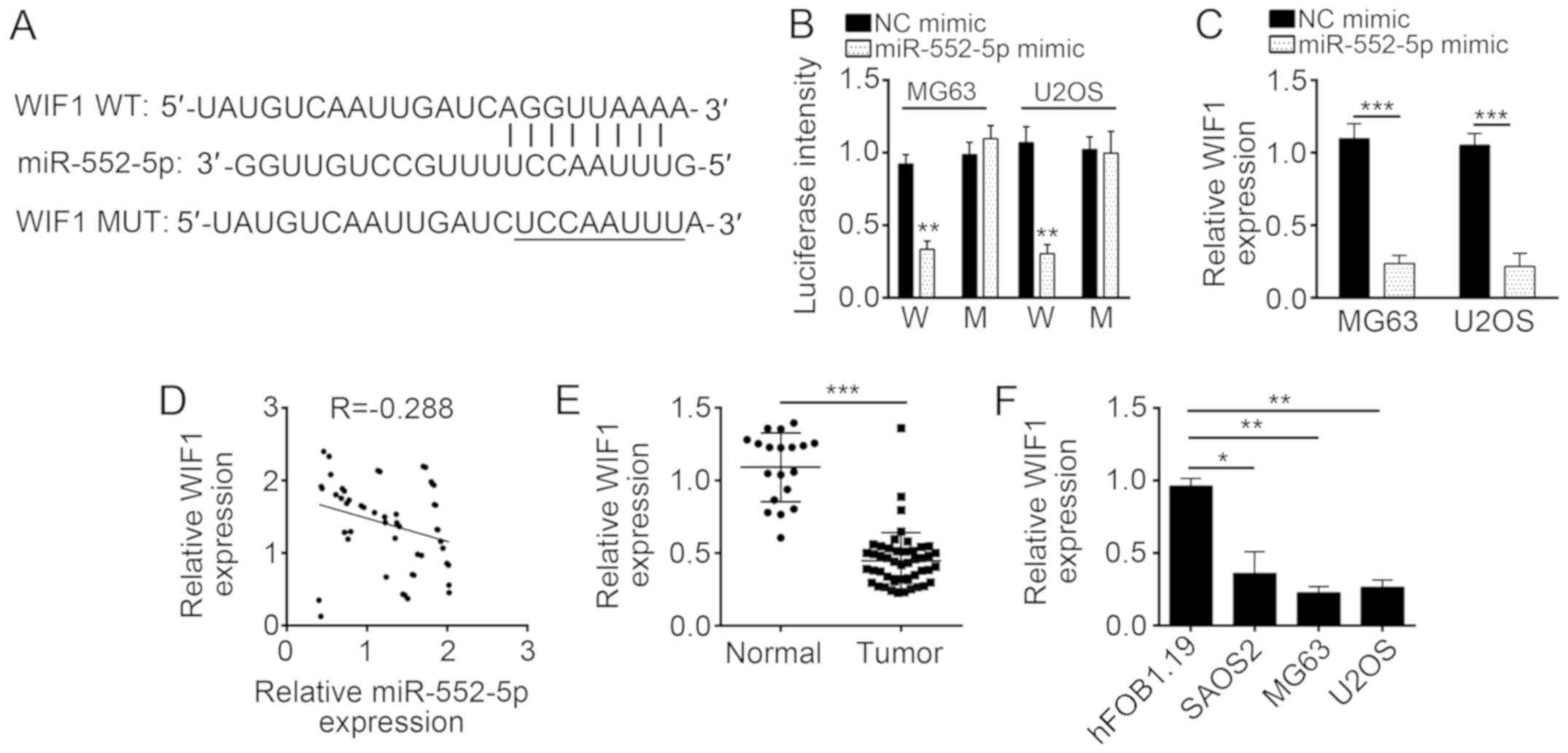 | Figure 4.WIF1 is a target of miR-552-5p in
osteosarcoma cells. (A) miR-552-5p may bind to the 3′-UTR of WIF1
mRNA. The underlined sequence is the mutated site. (B) miR-552-5p
mimics inhibited the luciferase intensity in MG63 and U2OS cells
while mutation of associating element in 3′-UTR of WIF1 mRNA
abolished the effect of miR-552-5p mimic on luciferase intensity.
(C) Overexpression of miR-552-5p decreased the mRNA expression
level of WIF1 in MG63 and U2OS cells. (D) Expression of miR-552-5p
was inversely associated with that of WIF1 in osteosarcoma tissues.
(E) Reverse transcription-quantitative polymerase chain reaction
analysis indicated that the expression of WIF1 was downregulated in
the tumor group (n=51) compared with adjacent normal tissues
(n=19). (F) Expression of WIF1 was downregulated in osteosarcoma
cell lines SAOS2, U2OS and MG63 cells, compared with hFOB1.19
cells. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA;
NC, negative control; WIF1, Wnt inhibitory factor 1; UTR,
untranslated region; WT or W, wild-type 3′-UTR; MUT or M, mutant
3′-UTR; tumor, osteosarcoma tissues. |
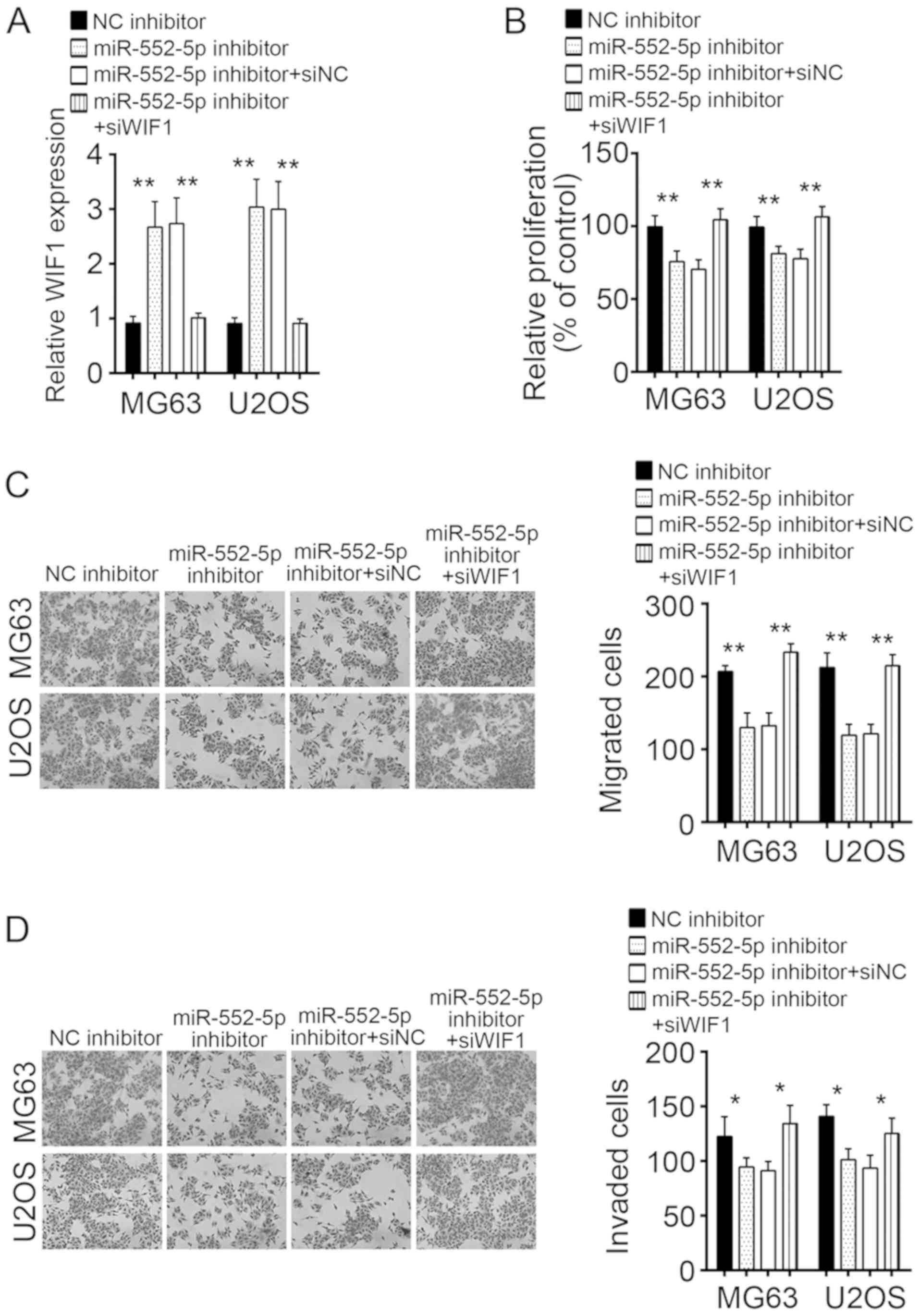
Knockdown of WIF1 in MG63 and U2OS
cells treated with miR-552-5pR inhibitor rescues cell
proliferation, migration, and invasion
To verify whether the regulation of OS cells by
miR-552-5p was dependent on WIF1, MG63 and U2OS cells transfected
with miR-552-5p inhibitors were treated with si-WIF1, which rescued
the expression of WIFI to the control level (Fig. 5A). CCK-8 and Transwell assays were
used to evaluate the effects of miR-552-5p and WIF1 on the
proliferation, migration, and invasion of cells. Knockdown of
miR-552-5p inhibited the proliferation, migration, and invasion of
MG63 and U2OS cells (Fig. 5B-D).
However, simultaneous knockdown of miR-552-5p and WIF1 rescued the
proliferation, migration, and invasion of MG63 and U2OS cells to
the control level (Fig. 5B-D). It
may be concluded that miR-552-5p regulated the proliferation,
migration, and invasion of OS cells by targeting WIF1.
Discussion
OS is the most malignant bone cancer worldwide and
is associated with poor clinical outcomes due to tumor metastasis
and recurrence (21). Therefore,
research into the molecular mechanism that regulates OS occurrence
and progression is required in order to screen new effective
therapeutic targets. In the current study, miR-552-5p was
overexpressed in OS tissues and cell lines compared with normal
tissues and cells. In addition, miR-552-5p promoted the
proliferation, migration and invasion of OS cells, suggesting that
miR-552-5p may be a novel target for OS therapy.
A previous study has demonstrated that the
dysregulation of miRNAs, which target specific genes to regulate
their expression, was associated with different types of human
cancer (22). Increasing number of
studies indicate that miRNAs regulate the proliferation, apoptosis
and metastasis of various types of cancer cells (23,24).
miR-124 suppresses the invasion and epithelial-mesenchymal
transition of gastric cancer cells by inhibiting snail family
transcriptional repressor 2 (23).
Tang et al (25) reported
that miR-29a inhibited cell proliferation, migration and invasion
in esophageal squamous cell carcinoma. Liu et al (26) demonstrated that miR-1297 promoted
breast cancer growth by activating the phosphatase and tensin
homolog/phosphoinositide 3-kinase/protein kinase B signaling
pathway. miRNA-552 promotes growth and metastasis of colorectal
cancer cells (15). Cao et al
(15) revealed that miR-552 targeted
DACH1 and regulate the Wnt/β-catenin pathway in colorectal cancer.
Wang et al (16) demonstrated
that miR-552 targeted ADAM metallopeptidase domain 28 to enhance
metastasis in colorectal cancer. In the present study, CCK-8 and
Transwell assays revealed that miR-552-5p promoted proliferation,
migration and invasion of tumor cells in vitro.
The Wnt/β-catenin pathway regulates numerous
cellular processes, including cell proliferation, metastasis and
differentiation (27,28). Abnormal activation of the
Wnt/β-catenin pathway promotes OS development and progression
(29). WIF1 serves as a regulator of
Wnt/β-catenin signaling (30). WIF1
inhibits activation of the Wnt signaling pathway by associating
with Wnt proteins (31).
Furthermore, WIF1 inhibits the progression of various types of
cancer, including colon cancer, non-small cell lung cancer and
endometrial adenocarcinoma (20,31,32). A
previous report indicated that WIF1 suppresses OS development and
metastasis (33). In the present
study, miR-552-5p directly targeted the 3′-UTR of WIF1 mRNA in OS
cells. RT-qPCR results revealed an inverse correlation between the
expression levels of miR-552-5p and WIF1 in OS tissues. The WIF1
expression was downregulated in OS tissues and cells. To determine
whether the promotion of OS progression by miR-552-5p was dependent
on WIF1, WIF1 and miR-552-5p were knocked down in MG63 and U2OS
cells. The results of the CCK-8 and Transwell assays revealed that
WIF1 knockdown rescued proliferation, migration, and invasion of OS
cells with downregulated miR-552-5p expression. However, further
investigation is required to confirm whether miR-552-5p
overexpression promotes activation of the Wnt/β-catenin signaling
pathway in OS cells.
In conclusion, the present study demonstrated that
miR-552-5p promoted proliferation, migration and invasion of OS
cells by targeting WIF1. Therefore, the miR-552-5p/WIF1 signaling
may serve as a therapeutic target for OS treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZS and WC initiated and designed this study,
analyzed and interpreted the results and wrote the manuscript. WC
participated in all the experiments. YX, JY and WZ performed
western blotting. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Nanjing Medical University (Nanjing, China) and
all enrolled patients signed a written informed consent
document.
Patient consent for publication
All patients included in this study provided consent
for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu JJ, Pi WS, Cao Y, Peng AF, Cao ZY, Liu
JM, Huang SH, Liu ZL and Zhang W: Let-7a inhibits osteosarcoma cell
growth and lung metastasis by targeting Aurora-B. Cancer Manag Res.
10:6305–6315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng WP, Huang YP, Chen HY, Wang N, Xiao
W, Liang Y, Jiang X, Su W and Wen S: Nomogram application to
predict overall and cancer-specific survival in osteosarcoma.
Cancer Manag Res. 10:5439–5450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pan R, He Z, Ruan W, Li S, Chen H, Chen Z,
Liu F, Tian X and Nie Y: lncRNA FBXL19-AS1 regulates osteosarcoma
cell proliferation, migration and invasion by sponging miR-346.
Onco Targets Ther. 11:8409–8420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q,
Chen J, Chen X, Zhang S, Liu Z, et al: miR-137 targets Cdc42
expression, induces cell cycle G1 arrest and inhibits invasion in
colorectal cancer cells. Int J Cancer. 128:1269–1279. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mei Q, Li X, Guo MZ, Fu XB and Han WD: The
miRNA network: micro-regulator of cell signaling in cancer. Expert
Rev Anticancer Ther. 14:1515–1527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry (Mosc). 81:315–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nature Reviews Cancer. 6:857–866. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: Microrna expression profiles classify human cancers. Nature.
435:834–8. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim KJ and Cho SB: Exploring features and
classifiers to classify microrna expression profiles of human
cancer. Neural Inf Process Models Appl. 6444:234–241. 2010.
|
|
11
|
Bai X, Meng L, Sun H, Li Z, Zhang X and
Hua S: MicroRNA-196b inhibits cell growth and metastasis of lung
cancer cells by targeting Runx2. Cell Physiol Biochem. 43:757–767.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Zeng H, Li H, Chen T, Wang L,
Zhang K, Chen J, Wang R, Li Q and Wang S: MicroRNA-101 inhibits
growth, proliferation and migration and induces apoptosis of breast
cancer cells by targeting sex-determining region Y-Box 2. Cell
Physiol Biochem. 43:717–732. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang W, Liu J, Xu T and Yu X: MiR-329
suppresses osteosarcoma development by downregulating Rab10. FEBS
Lett. 590:2973–2981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao J, Qin L, Miao S, Wang X and Wu X:
Overexpression of miR-506 suppresses proliferation and promotes
apoptosis of osteosarcoma cells by targeting astrocyte elevated
gene-1. Oncol Lett. 12:1840–1848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao J, Yan XR, Liu T, Han XB, Yu JJ, Liu
SH and Wang LB: MicroRNA-552 promotes tumor cell proliferation and
migration by directly targeting DACH1 via the Wnt/β-catenin
signaling pathway in colorectal cancer. Oncol Lett. 14:3795–3802.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang
D, Ma X, Du Y, Liu X and Yang Y: MicroRNA-552 enhances metastatic
capacity of colorectal cancer cells by targeting a disintegrin and
metalloprotease 28. Oncotarget. 7:70194–70210. 2016.PubMed/NCBI
|
|
17
|
Luo X, Ye S, Jiang Q, Gong Y, Yuan Y, Hu
X, Su X and Zhu W: Wnt inhibitory factor-1-mediated autophagy
inhibits Wnt/β-catenin signaling by downregulating dishevelled-2
expression in non-small cell lung cancer cells. Int J Oncol.
53:904–914. 2018.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou Z, Guo K, Sun X, Hu F, Chen Q, Luo X,
Wang G, Hu J and Sun L: TRIB2 functions as novel oncogene in
colorectal cancer by blocking cellular senescence through AP4/p21
signaling. Mol Cancer. 17:1722018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng ZY, Xu XH, Cen DZ, Luo CY and Wu SB:
miR-590-3p promotes colon cancer cell proliferation via
Wnt/β-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur
Rev Med Pharmacol Sci. 21:4844–4852. 2017.PubMed/NCBI
|
|
21
|
Zhang Y, Zhang L, Zhang G, Li S, Duan J,
Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al: Osteosarcoma
metastasis: Prospective role of ezrin. Tumour Biol. 35:5055–5059.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li SL, Gao HL, Lv XK, Hei YR, Li PZ, Zhang
JX and Lu N: MicroRNA-124 inhibits cell invasion and
epithelial-mesenchymal transition by directly repressing Snail2 in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:3389–3396.
2017.PubMed/NCBI
|
|
24
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang L, Hao ZP, Deng Y, Zhang P, Fu XN and
Zhang N: MiR-29a-3p suppresses the proliferation, migration and
invasion of esophageal squamous cell carcinoma by targeting IGF-1.
Int J Clin Exp Pathol. 10:1293–1302. 2017.
|
|
26
|
Liu C, Liu Z, Li X, Tang X, He J and Lu S:
MicroRNA-1297 contributes to tumor growth of human breast cancer by
targeting PTEN/PI3K/AKT signaling. Oncol Rep. 38:2435–2443. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ring A, Kim YM and Kahn M: Wnt/catenin
signaling in adult stem cell physiology and disease. Stem Cell Rev.
10:512–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stewart DJ, Chang DW, Ye Y, Spitz M, Lu C,
Shu X, Wampfler JA, Marks RS, Garces YI, Yang P and Wu X: Wnt
signaling pathway pharmacogenetics in non-small cell lung cancer.
Pharmacogenom J. 14:509–522. 2014. View Article : Google Scholar
|
|
29
|
Dai G, Zheng D, Wang Q, Yang J, Liu G,
Song Q, Sun X, Tao C, Hu Q, Gao T, et al: Baicalein inhibits
progression of osteosarcoma cells through inactivation of the
Wnt/β-catenin signaling pathway. Oncotarget. 8:86098–86116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Hu B, Wang Z, Zhang F, Wei H and
Li L: miR-181c contributes to cisplatin resistance in non-small
cell lung cancer cells by targeting Wnt inhibition factor 1. Cancer
Chemother Pharmacol. 80:973–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Deng X, Hou C, Wang H, Liang T and Zhu L:
Hypermethylation of WIF1 and its inhibitory role in the tumor
growth of endometrial adenocarcinoma. Mol Med Rep. 16:7497–7503.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rubin EM, Guo Y, Tu K, Xie J, Zi X and
Hoang BH: Wnt inhibitory factor 1 decreases tumorigenesis and
metastasis in osteosarcoma. Mol Cancer Ther. 9:731–741. 2010.
View Article : Google Scholar : PubMed/NCBI
|