Introduction
Diabetes mellitus (DM) is characterized by
persistent hyperglycemia with an inability to fully utilize glucose
and maintain insulin homeostasis, which leads to long-term damage
and dysfunction of the eyes, kidneys, and nervous and
cardiovascular systems in particular (1). Approximately 366,000,000 individuals
are expected to suffer from DM by the year 2030, and almost
592,000,000 individuals, or 1/10, are anticipated to have diabetes
by 2035 (1,2). It is estimated that >40% of diabetic
patients require regular, painful and expensive dialysis due to
end-stage renal disease (ESRD), and the mortality rate remains high
(3).
Diabetic nephropathy (DN) is an emerging clinical
and public health challenge, which is also a life-threatening
long-term microvascular complication of type 1 DM (T1DM) and type 2
DM (T2DM) (1,4,5).
Patients with DN are generally hypertensive, which increases the
risk of cardiovascular disease and mortality (6); hypertension is interconnected with the
initiation and progression of DN. The central pathological
alterations during the initiation and progression of DN comprise
mesangial extension, extracellular matrix alterations,
tubulointerstitial fibrosis and glomerular sclerosis (7). In particular, alterations in the
glomeruli, including fibrosis and apoptosis of mesangial cells, are
relevant to the progression of DN. Evidence suggests that certain
microRNAs (miRNAs) or transcription factors with abnormal
expression may serve a vital role in the development of DN. Wu
et al (8) reported that miRNA
(miR)-455-3p suppresses renal fibrosis through repression of the
expression of Rho-associated kinase 2 (ROCK2) in DN. miR-135a
promotes renal fibrosis in DN by regulating transient receptor
potential cation channel, subfamily C, member 1 (9), and inhibiting miR-192 ameliorates renal
fibrosis in DN (10). Chen et
al (11) reported that high
glucose (HG) promotes the expression of pro-inflammatory cytokines
in vivo or in vitro, by upregulating high mobility
group box 1 (HMGB1) and activating nuclear factor (NF)-κB signaling
in SV40 MES 13 cells. The condition of hyperglycemia also induces
the expression of transforming growth factor-β (TGF-β),
fibronectin, collagen type IV and plasminogen activator inhibitor-1
(PAI-1), which are associated with fibrosis (12). Furthermore, increased renal mRNA and
protein expression levels of TGF-β1 have been observed in various
animal models and in human DN (13).
TGF-β, an important cytokine, is associated with the development of
fibrosis and glomerulosclerosis, particularly mesangial cell
phenotypic transformation in DN (14). Although considerable progress has
been made towards elucidating the potential therapeutic targets and
molecular mechanisms involved in DN, sufficient biomolecular target
information and novel diagnostic and prognostic biomarkers that may
be beneficial for improving the clinical management of DN appear to
be lacking.
Gene expression profiling has been widely applied in
clinical research as a potentially rapid and cost-effective
approach, which is considered useful for identifying the expression
levels of thousands of genes simultaneously. Furthermore, this
method may provide a complete, systematic and reliable comparison
of gene expression profiles among different tissue types.
Accumulated core data of DN have been produced by gene chip
analysis, and have been deposited into the Gene Expression Omnibus
(GEO) database of the National Center for Biotechnology
Information, a public database (http://www.ncbi.nlm.nih.gov/geo/). Therefore, the
present study focused on extracting and reanalyzing the
transcriptome profile of TGF-β1-induced mesangial gene expression,
and the difference between HG and glucosamine (GlcN)-induced mouse
mesangial cells. These data may assist in revealing the molecular
mechanisms of DN in detail.
The main purpose of the present study was to
identify the expression of differently expressed genes (DEGs) in a
TGF-β1-, HG-, and GlcN-induced mouse mesangial cell line (MES-13).
A network pharmacology and bioinformatics reanalysis was performed
of the original data (GSE2558 and GSE2557) of MES-13 cells from a
study by Cheng et al (15),
which were deposited in the GEO database. Subsequently, the DEGs
between the TGF-β1, HG, GlcN and low glucose (LG) groups were
screened via the application of GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), with a
cut-off value of P<0.05 and |log2fold change| ≥1 for
statistical significance, followed by gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses. Subsequently, the protein-protein interaction (PPI)
network of the DEGs between the TGF-β1, HG, GlcN and LG groups were
respectively constructed via the Search Tool for the Retrieval of
Interacting Genes (STRING; http://string-db.org/) database and visualized through
Cytoscape software (http://www.cytoscape.org/), subsequently identifying
hub genes in the PPI network with the highest values for degree,
node betweenness and closeness via network topology calculation.
Furthermore, computational docking was conducted to mimic the
characteristics of anti-DN drug (captopril, enalapril, fosinopril,
irbesartan, lisinopril, losartan, ramipril, telmisartan and
valsartan in the DrugBank database)-target binding, using a web
server for network pharmacology-based prediction and analysis,
SystemsDock (http://systemsdock.unit.oist.jp/iddp/home/index). This
present study applies network pharmacology and bioinformatics
reanalysis to investigate the potential therapeutic target genes
and pathways in TGF-β1, HG and GlcN-induced mouse mesangial cells
(MES-13). The results provide a comprehensive view of the
associations and mechanisms underlying TGF-β1, HG and GlcN-induced
gene expression in renal mesangial cells at the molecular level,
and identifies potential targets with the most appropriate drug for
individualized anti-DN treatment.
Materials and methods
Microarray data
The gene expression profiles of GSE2558 and GSE2557
based on the platform of GPL1261 (Affymetrix Mouse Genome 430 2.0
Array; Affymetrix, Inc., Santa Clara, CA, USA), and deposited by
Cheng et al (15) from Wayne
State University School of Medicine, which were downloaded from the
GEO database of the National Center for Biotechnology Information
(NCBI; http://www.ncbi.nlm.nih.gov/geo/). Renal mesangial
cells serve an important role in the development of diabetic
glomerulosclerosis and renal failure. The following procedures were
deposited with the associated data sets and were previously
published (15). The stable MES-13
murine mesangial cells transformed with non-capsid-forming SV-40
virus were obtained from the ATCC (Manassas, VA, USA) and exhibited
a typical spindle-like appearance, positive staining for vimentin
and desmin, and contraction in response to angiotensin II and
expression of the AT1 receptor. The cells were maintained in DMEM
and Ham's F-12 Nutrient Mixture (4:1 ratio; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing a normal D-glucose
concentration of 5.5 mmol/l, 2% FCS (Gibco; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin, 100 U/ml penicillin and
2 mmol/l glutamine, incubated in a humidified incubator with 5%
CO2 at 37°C, and routinely passaged at confluence every
3 days by trypsinization using 10-cm culture dishes. Monolayers at
~50% confluence were starved in the above medium without FCS for 1
day and then incubated in starvation medium with 100 ng/ml TGF-β1
for 24 h in GSE2558, containing four cell samples (GSM48386,
GSM48392, GSM48394 and GSM4839), including the TGF-β1-treated
MES-13 mouse mesangial cell line and LG-treated cells as a control
sample. For GSE2557, 50% confluent monolayers were starved in the
above medium without FCS for 1 day and incubated in the starvation
medium with the desired concentrations of glucose and GlcN for 48 h
(LG, 5.5 mM; HG, 25 mM; and GlcN, 1.5 mM + LG) in a humidified
incubator containing 5% CO2 at 37°C, including two
HG-treated cell samples (GSM48387 and GSM48388), two GlcN-treated
cell samples (GSM48389 and GSM48390) and two LG-treated cell
samples (GSM48385 and GSM48393) as the control cell samples.
Data processing and DEG
identification
Comparisons were performed between the differently
treated groups of samples (TGF-β1-treated, vs. LG-treated cell
samples in the GSE2558 dataset; HG-treated, vs. LG-treated cell
samples and GlcN-treated, vs. LG-treated cell samples in the
GSE2557 dataset) to identify DEGs. The comparison was performed
using the limma R package-based online program GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/).
GEO2R allows the calculation and graphical representation as a box
plot of the distribution of values for selected samples, and it
applies an adjusted P-value using the Benjamini-Hochberg method to
correct false-positives. Fold-changes (FC) in gene expression were
calculated with threshold criteria of
|log2FC| ≥1, and adjusted P<0.05 was used
as the cut-off criterion for statistically significant DEGs. Venny
software (version 2.1; http://bioinfogp.cnb.csic.es/tools/venny/index.html)
was used to analyze the overlapping DEGs in the three datasets.
Functional enrichment analysis
Gene Ontology (GO) analysis is a bioinformatics
method for annotating genes and proteins to identify characteristic
biological attributes, including biological process, cellular
component, molecular function and biological functions, from
high-throughput genome data (16,17). The
KEGG pathway database is a resource for understanding the
high-level functions and utilities of a biological system, includes
a variety of biochemical pathways (18). The Database for Annotation,
Visualization, and Integrated Discovery (DAVID; version 6.7;
http://david.ncifcrf.gov/), an online
program that offers functional annotation of an enormous quantity
of genes derived from various genomic resources, was used to
perform GO and KEGG pathway analyses on the significant DEGs. The
species was limited to Homo sapiens and P<0.05 was
considered to indicate a statistically significant difference.
PPI network construction and
regulatory network analyses
The STRING database (version 10.0; http://string-db.org), which collects and predicts
interaction information from genomic context predictions,
high-throughput lab experiments, co-expression, automated
text-mining and previous knowledge from databases (19) was used to predict the potential
interactions between gene candidates at the protein level.
Confidence scores >0.4 (median confidence score) were considered
significant. The first three nodes and the overlapping DEGs in
TGF-β1-treated, HG-treated and GlcN-treated PPI networks were
identified for further discussion and molecular docking.
Additionally, Cytoscape software (version 3.4.0; http://www.cytoscape.org/) was utilized to construct
the PPI network.
Molecular docking experiments based on
SystemsDock
The DrugBank (www.drugbank.ca) is a unique, freely available
bioinformatics and chemoinformatics resource containing information
on drugs, drug-target interactions, drug actions and drug
interactions, including Food and Drug Administration (FDA)-approved
drugs in addition to experimental drugs undergoing the FDA approval
process (20). The keyword ‘Diabetic
nephropathy’ was input into the search field. Captopril, enalapril,
fosinopril, irbesartan, lisinopril, losartan, ramipril, telmisartan
and valsartan were selected as anti-DN drugs with different
mechanisms for the molecular docking experiments. The overlapping
DEGs for the crystal structures of Ras homolog gene family, member
B (RHOB; PDB ID: 2FV8) and Krüppel-like factor 15 (KLF15; PDB ID:
4BN2) were obtained from the Protein Data Bank (http://www.rcsb.org/). The structure of complement
factor H (CFH; PDB ID:3R62) was also obtained from the Protein Data
Bank. SystemsDock (http://systemsdock.unit.oist.jp/iddp/home/index), a
web server for network pharmacology-based prediction with
high-precision docking simulation, was used to comprehensively
characterize the ligand selectivity and to illustrate how a ligand
acts on a complex molecular network (21). A docking score of >4.25 is
considered fair, 5≤ docking score <7 is considered good, and
docking score ≥7 is considered excellent; this scoring is
conventionally used to classify ligand binding activity (22).
Results
Data processing and screening of
differentially expressed mRNAs
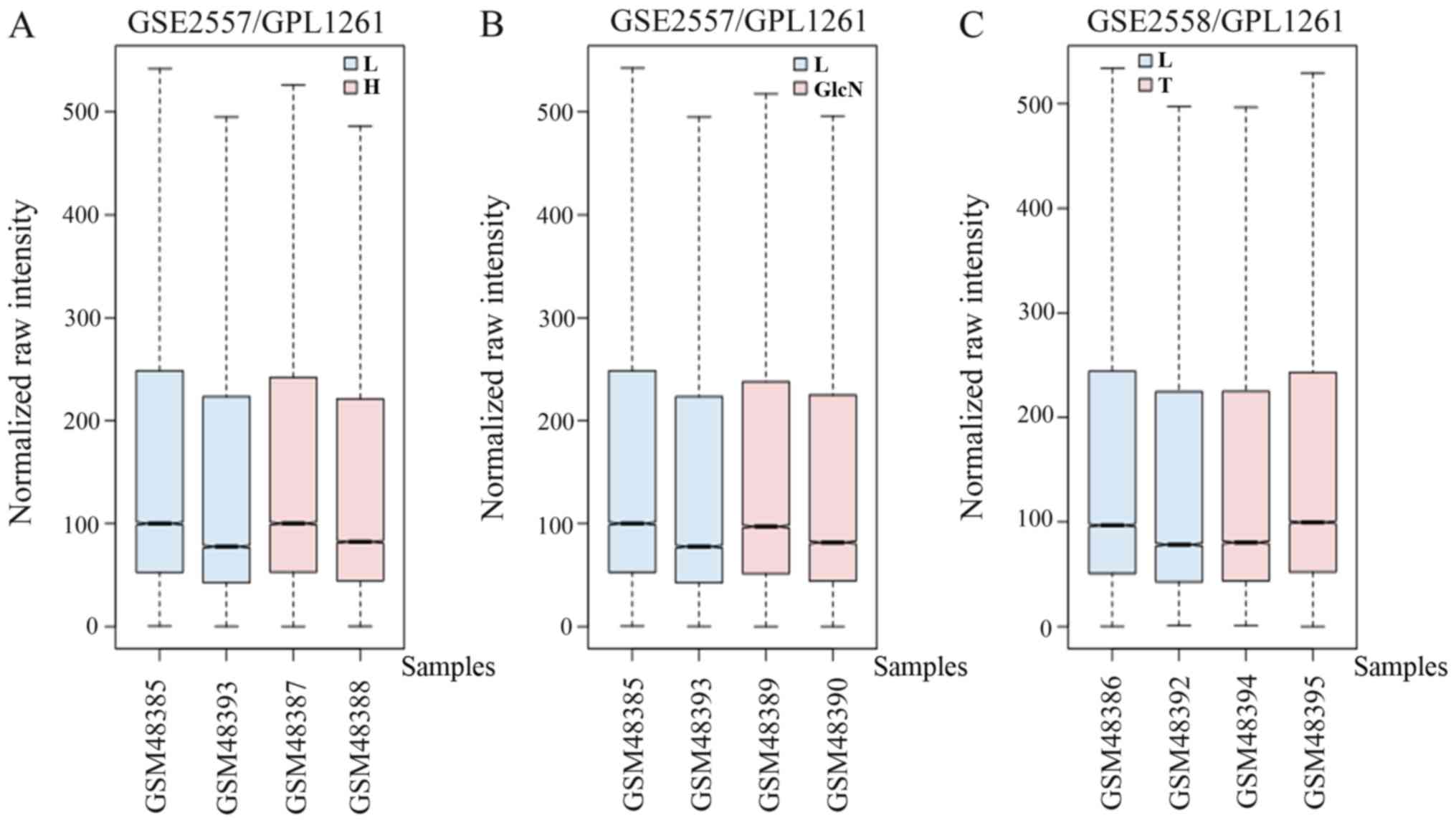
The box plots of the mRNA expression levels were
normalized to the same order of magnitude prior to the statistical
analysis for GSE2557 and GSE2558, as presented in Fig. 1, which clearly demonstrated that the
medians of the different samples were almost equivalent following
normalization, indicating that the data were cross-comparable.
According to the cut-off criteria of P<0.05 and |log2
FC|>1.0, a total of 360 DEGs were obtained, including
202 upregulated and 158 downregulated DEGs from the HG-treated
groups compared with the LG-treated groups, and a total of 241 DEGs
were obtained, including 138 upregulated and 103 downregulated DEGs
from the GlcN-treated groups compared with the LG-treated groups,
when redundant duplicate content had been removed from GSE2557.
Furthermore, via integrated analysis, a set of 125 DEGs were
identified in the TGF-β1-treated groups compared with the
LG-treated groups, including 81 upregulated and 44 downregulated
DEGs in GSE2558.
Functional enrichment analysis
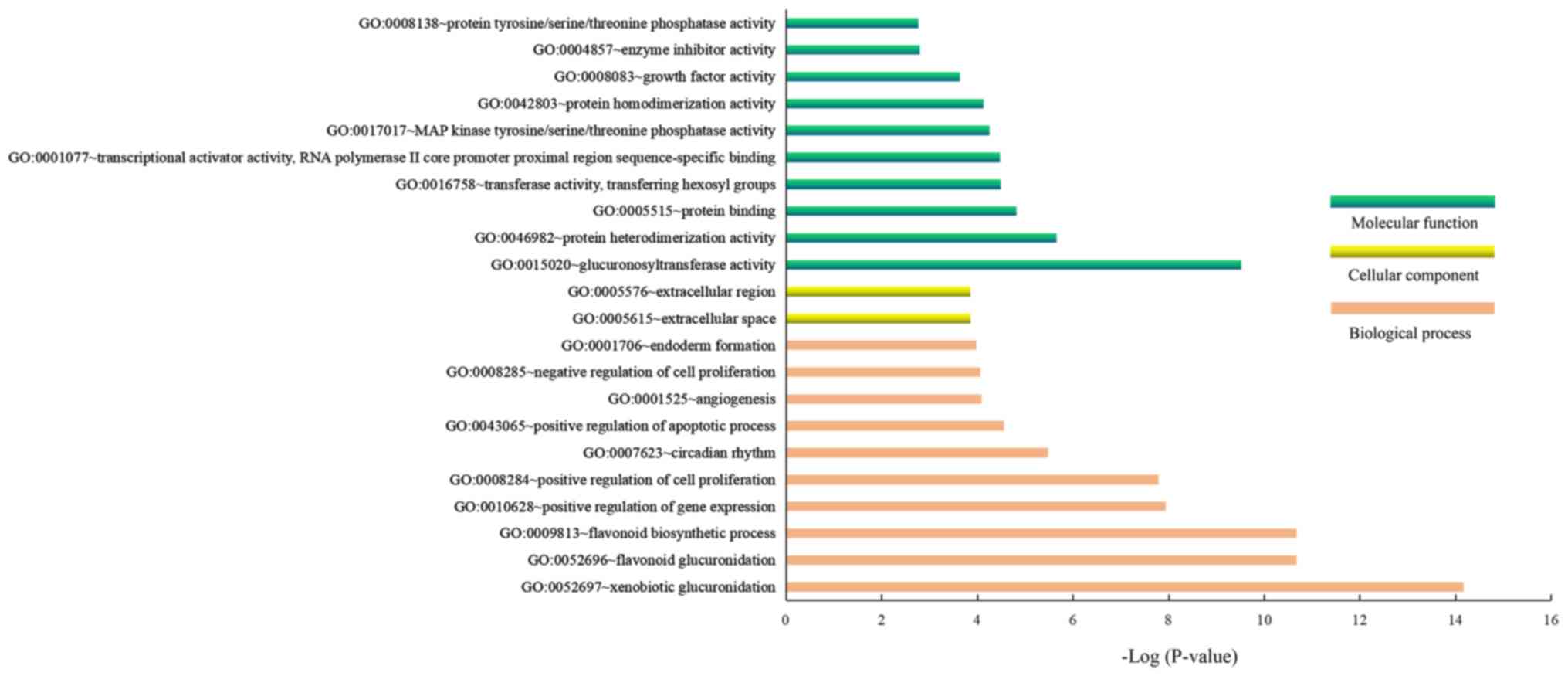
The GO enrichment results demonstrated that the
significantly enriched GO terms of the DEGs in GSE2558 for
biological process were ‘xenobiotic glucuronidation’, ‘flavonoid
glucuronidation’, ‘flavonoid biosynthetic process’ and ‘positive
regulation of cell proliferation’, whereas the significantly
enriched GO terms for cellular component were ‘extracellular space’
and ‘extracellular region’. Furthermore, the significantly enriched
GO terms for molecular functions were ‘glucuronosyltransferase
activity’, ‘protein heterodimerization activity’ and ‘protein
binding’ (Fig. 2). In addition, the
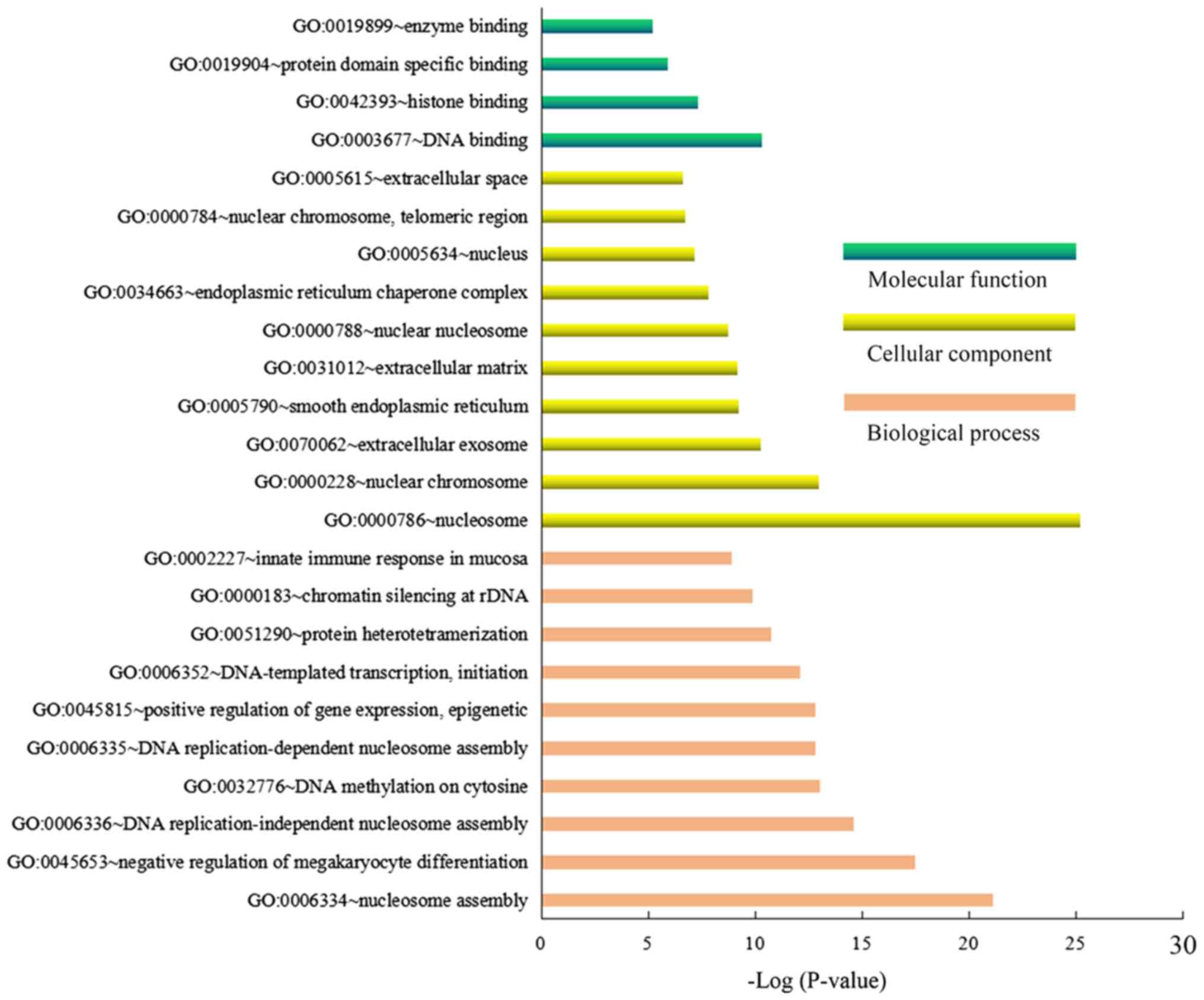
DEGs of the HG-treated groups compared with the LG-treated groups
were mainly enriched in the GO biological process terms of
‘nucleosome assembly’, ‘negative regulation of megakaryocyte
differentiation’ and ‘DNA replication-independent nucleosome
assembly’, ‘cellular component terms of nucleosome’, ‘nuclear
chromosome’ and ‘extracellular exosome’, and molecular functions
terms of ‘DNA binding’, ‘histone binding’ and ‘protein domain
specific binding’ (Fig. 3). The
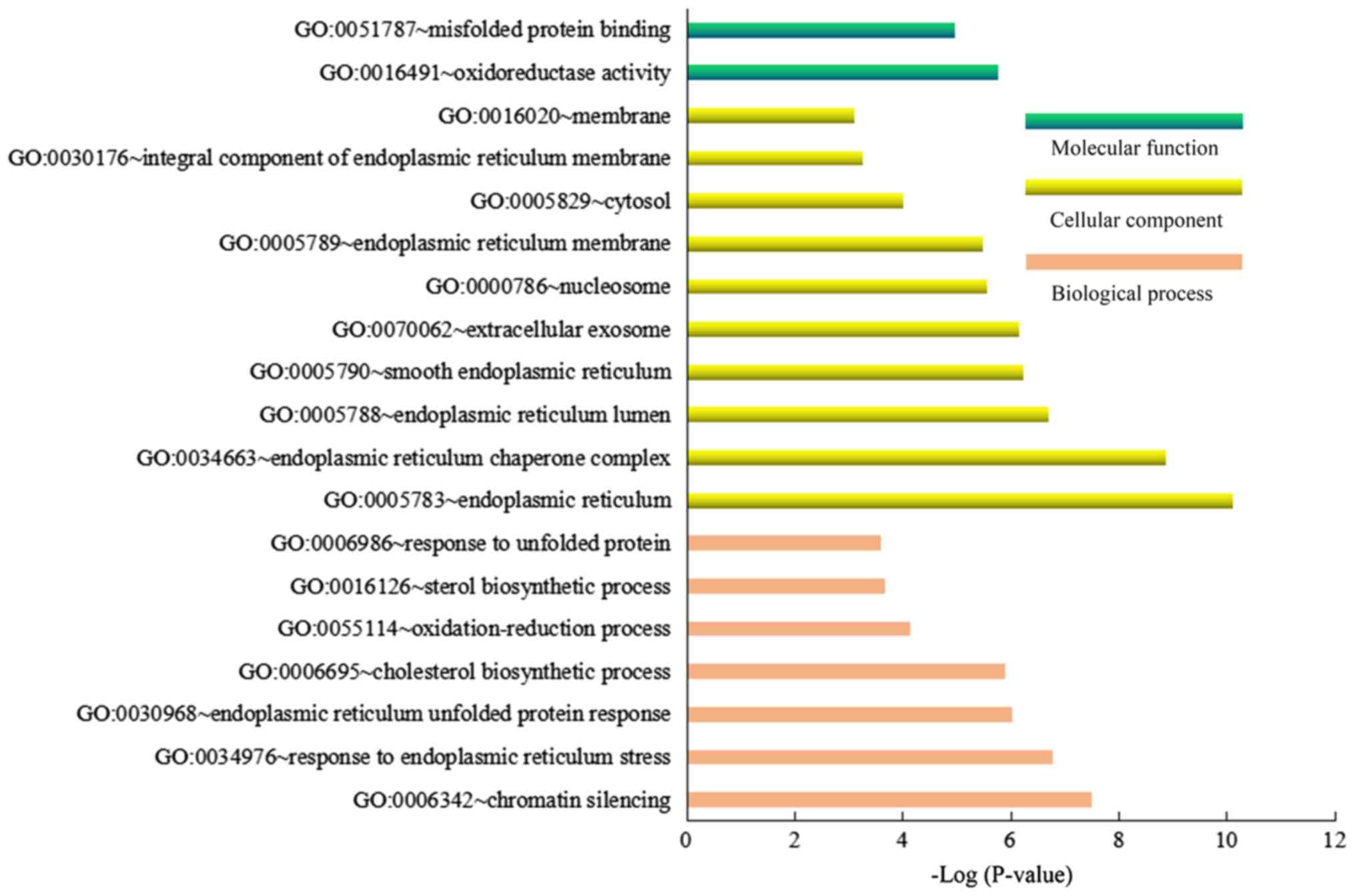
significantly enriched GO biological process terms of ‘chromatin
silencing’, ‘response to endoplasmic reticulum stress’ and
‘endoplasmic reticulum unfolded protein response’, the GO terms of
cellular component ‘endoplasmic reticulum’, ‘endoplasmic reticulum
chaperone complex’ and ‘endoplasmic reticulum lumen’, and the GO
terms of molecular function ‘oxidoreductase activity’ and
‘misfolded protein binding’ were noted for DEGs of the GlcN-treated
groups compared with the LG-treated groups (Fig. 4).
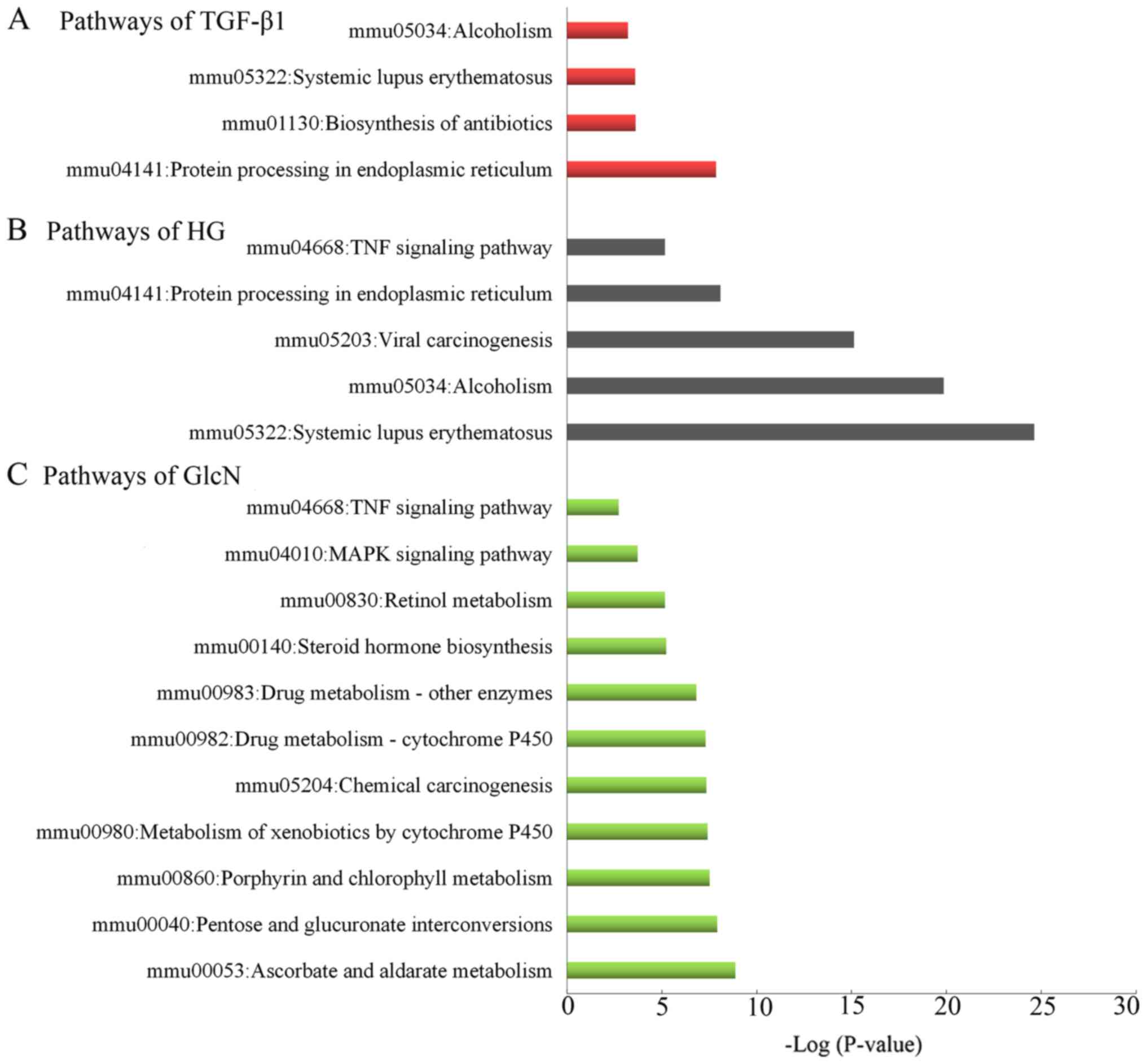
Furthermore, the KEGG pathway analysis indicated
that the ‘protein processing in endoplasmic reticulum’,
‘biosynthesis of antibiotics’ and ‘systemic lupus erythematosus’
pathways served an essential role in the GlcN-treated groups
compared with the LG-treated groups (Fig. 5A). ‘Systemic lupus erythematosus’,
‘alcoholism’ and ‘Viral carcinogenesis’ were the top significantly
enriched pathways in the HG-treated groups compared with the
LG-treated groups (Fig. 5B). As
presented in Fig. 5C. The DEGs of
the TGF-β1-treated groups compared with the LG-treated groups were
primarily enriched in ‘ascorbate and aldarate metabolism’, ‘pentose
and glucuronate interconversions’ and ‘porphyrin and chlorophyll
metabolism’. Notably, the intersecting pathway was ‘TNF signaling
pathway’ in the TGF-β1-treated and HG-treated groups, compared with
the LG-treated groups.
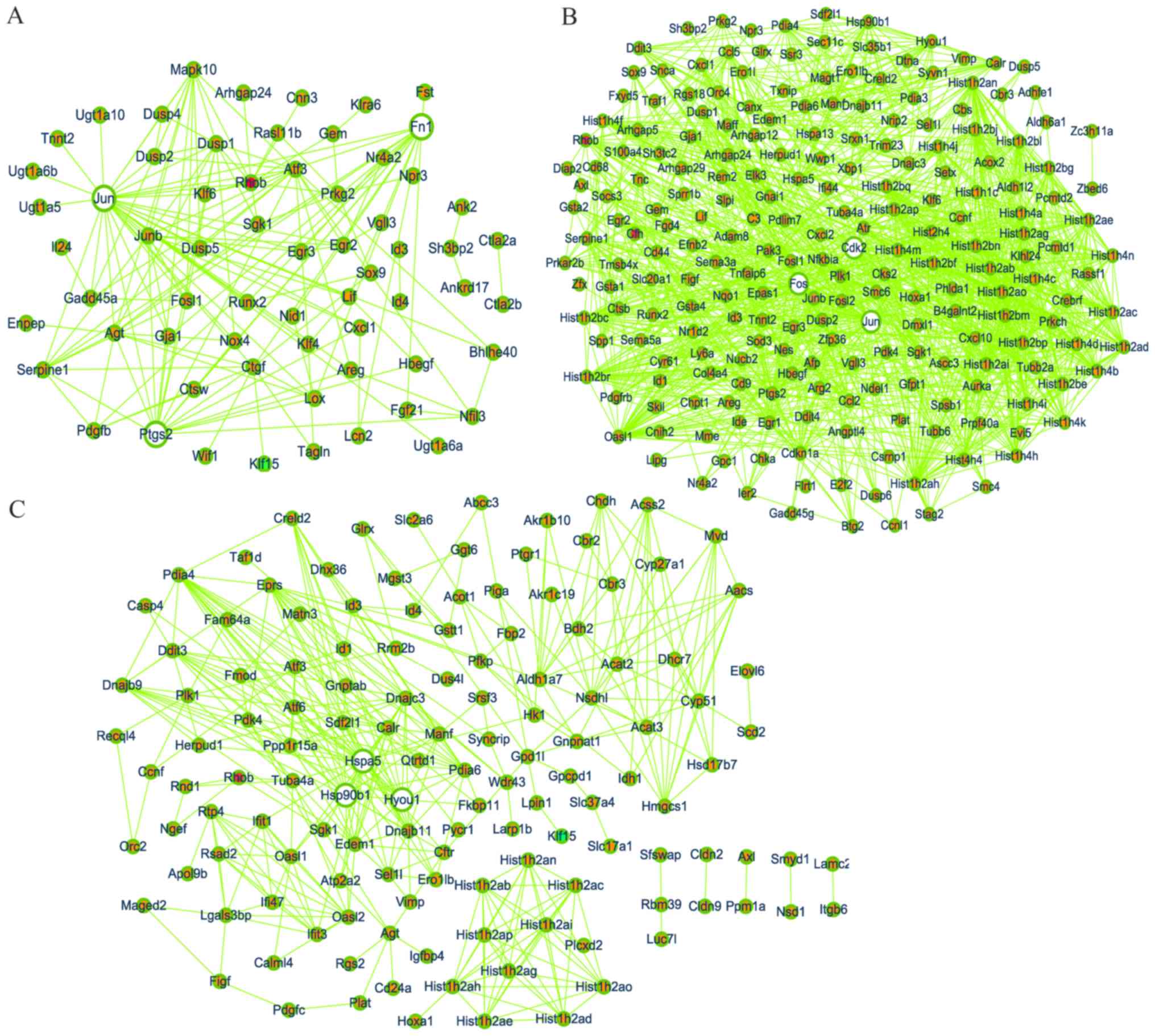
PPI network
Based on the information obtained from the STRING
database, a network diagram was produced separately for significant
DEGs in GSE2557 and GSE2558; certain DEGs were on the margins and
isolated, suggesting no association with other genes, and were
removed from the PPI network diagram using Cytoscape. There were 64
nodes and 149 edges in the network for the TGF-β1-treated groups
compared with the LG-treated groups (Fig. 6A), 223 nodes with 1,245 edges in the
network for the HG-treated groups compared with the LG-treated
groups (Fig. 6B), and 134 nodes with
366 edges in the network for the GlcN-treated groups compared with
the LG-treated groups (Fig. 6C). In
a PPI network, the greater the characteristic properties of degree,
betweenness and closeness of a node, the more important its role.
The parameters ‘degree’, ‘betweenness’ and ‘closeness’ were used to
select the first three nodes, Jun (degree=32,
betweenness=0.49093999, closeness=0.68292683),
prostaglandin-endoperoxide synthase 2 (Ptgs2; degree=17,
betweenness=0.0727469, closeness=0.52336449) and fibronectin 1
(Fn1; degree=16, betweenness=0.18112793, closeness=0.55445545) in
Fig. 4A; Jun (degree=56,
betweenness=0.25066792, closeness=0.51318945), cyclin-dependent
kinase (Cdk2; degree=43, betweenness=0.14884068,
closeness=0.49195402) and Fos (degree=41, betweenness=0.08047788,
closeness=0.47240618) in Fig. 4B;
heat shock protein family A (Hsp70) member (Hspa)5 (degree=30,
betweenness=0.38665873, closeness=0.42635659), Hsp90b1 (degree=21,
betweenness=0.07358151, closeness=0.38327526) and Hyou1 (degree=17,
betweenness=0.01014303, closeness=0.34920635) in Fig. 4C. These genes were assessed as hub
genes.
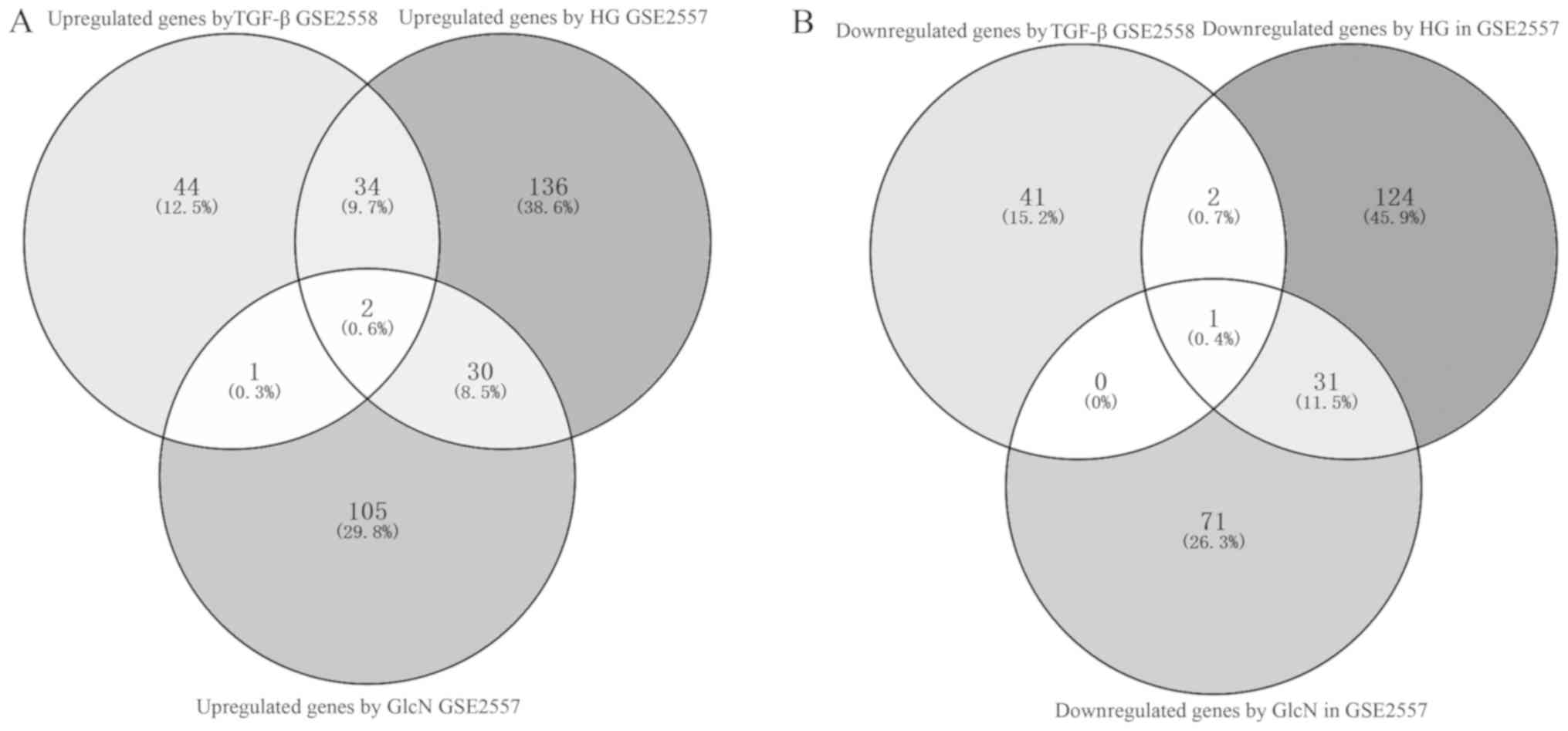
Finally, to improve the specificity, a total of two
upregulated potentially interacting genes (Rhob and Cfh) in the
Venn diagram (Fig. 7A), and one
downregulated potential therapeutic target gene (Klf15) (Fig. 7B) in the three sets of results were
identified and selected for further investigation.
Molecular docking analysis of binding
to RHOB, CFH and KLF15
Computational molecular docking experiments were
conducted to mimic the characteristics of anti-hypertension or
DN-target binding. The crystal structure of RHOB (PDB ID: 2FV8),
KLF15 (PDB ID: 4BN2) and the structure of CFH (PDB ID:3R62) were
obtained from the Protein Data Bank (http://www.rcsb.org/). The docking exercise was
conducted using systemsDock (http://systemsdock.unit.oist.jp/iddp/home/index)
(21). The results indicated that a
number of the selected drugs, lisinopril, enalapril, ramipril,
fosinopril and valsartan, in the drugbank database had higher
docking scores, of 5.727, 5.936, 6.19, 6.415 and 6.692
respectively, compared with the native ligand (5.7) of RHOB
(Fig. 8A). As presented in Fig. 8B, the binding scores of irbesartan,
captopril, losartan, lisinopril, valsartan, enalapril, ramipril and
fosinopril were 3.904, 4.409, 4.836, 5.749, 5.924, 5.957, 6.231 and
6.464, respectively, which were higher than that of the native
ligand (3.7) of CFH. Similarly, the binding scores of lisinopril,
enalapril, ramipril, valsartan and fosinopril were 5.717, 5.760,
5.844, 6.248 and 6.614, respectively, which were higher than the
score of 5.70 of the natural ligand of KLF15 (Fig. 8C). The results of the molecular
docking of binding to RHOB, CFH and KLF15 indicted that valsartan,
fosinopril, or a combination of the two may have a superior
therapeutic effect via the putative targets.
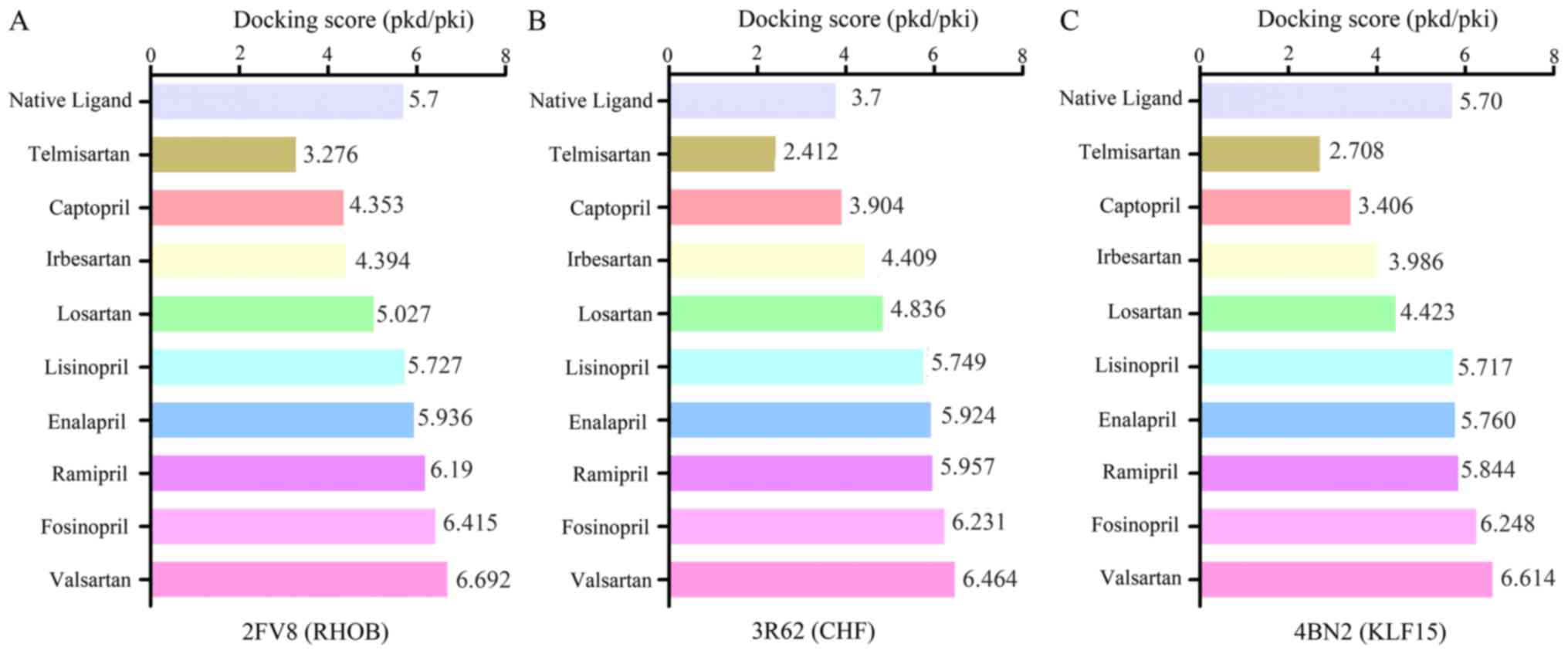 | Figure 8.Molecular docking of binding to RHOB,
CFH and KLF15. The y-axis represents captopril, enalapril,
fosinopril, irbesartan, lisinopril, losartan, ramipril, telmisartan
and valsartan as antidiabetic nephropathy drugs, and the x-axis
represents the docking score values. The docking score of these
drugs binding to (A) RHOB, (B) CFH and (C) KLF15 are presented.
Selected drugs, lisinopril, enalapril, ramipril, fosinopril and
valsartan, exhibited similar or superior docking than the native
ligands. RHOB, Ras homolog gene family, member B; CFH, complement
factor H; KLF15, Krüppel-like factor 1. |
Discussion
DN, a common microvascular complication of diabetes,
and a major cause of ESRD, a leading cause of mortality among
diabetic patients (23,24). It is characterized by proteinuria,
albuminuria, renal glomerular hypertrophy, basement membrane
thickening, podocyte dysfunction, and mesangial and
tubulointerstitial fibrosis due to accumulation of extracellular
matrix (ECM) proteins (25–27). Previous investigations have
demonstrated that one of the principal classical pathways leading
to renal fibrosis is the TGF-β1/Smad signaling pathway, and
culturing mesangial cells, the culprit cells in DN, in HG media
stimulates the synthesis of ECM proteins, inhibits cell growth and
alters the production of growth factors (15). TGF-β1 is one of the most important
mediators of fibrosis, in addition to cellular hypertrophy and
survival (26,28). Following TGF-β1/Smad signaling
pathway activation, the expression of TGF-β1 is increased, inducing
excessive deposition of ECM and increased epithelial-mesenchymal
transition in renal cells, including mesangial cells and tubular
cells in DN, indicating that TGF-β1 serves a critical role in the
development and progression of fibrosis in DN. Comprehensive
investigations to improve current understanding of the molecular
mechanisms of the pathogenesis of DN are thus important and
necessary.
In the present study, putative hub candidate genes
and enriched pathways of TGF-β1, HG-, and GlcN-stimulated murine
mesangial (MES-13) cells were identified by bioinformatics analysis
and the verification of hub genes was performed via molecular
docking. The gene expression data from GSE2557 and GSE2558 were
reanalyzed and identified. A total of 360 DEGs were screened,
consisting of 202 upregulated and 158 downregulated DEGs from the
HG-treated groups compared with the LG-treated groups; in addition,
241 DEGs were obtained, including 138 upregulated and 103
downregulated DEGs from the GlcN-treated groups compared with the
LG-treated groups in GSE2557, and 125 DEGs were identified,
including 81 upregulated and 44 downregulated DEGs, in GSE2558. The
present study identified three overlapping DEGs, two upregulated
and one downregulated, across the three conditions. Furthermore,
the results of the GO analysis indicated that the DEGs of the
TGF-β1-treated groups compared with those in the LG-treated groups
were primarily involved in the ‘xenobiotic glucuronidation’,
‘flavonoid glucuronidation’, ‘flavonoid biosynthetic process’ and
‘positive regulation of cell proliferation’, and the ‘ascorbate and
aldarate metabolism’ pathways; the DEGs of the HG-treated groups
compared with the LG-treated groups were principally enriched in
the ‘nucleosome assembly’, ‘negative regulation of megakaryocyte
differentiation’ and the ‘systemic lupus erythematosus’ pathways;
and the DEGs of the GlcN-treated groups compared with the
LG-treated groups were involved in the ‘chromatin silencing’,
‘response to endoplasmic reticulum stress’, ‘endoplasmic reticulum
unfolded protein response’ and the ‘protein processing in
endoplasmic reticulum’ pathways. In addition, KEGG pathway
enrichment analysis demonstrated that the overlapping pathway was
the ‘TNF signaling pathway’ in the TGF-β1-treated and HG-treated
groups compared with the LG-treated groups. There is evidence to
suggest that oxidative stress, inflammation and fibrosis serve a
pivotal role in the progression of DN. Based on a number of human
and animal studies, tumor necrosis factor-α (TNF-α), interleukin-1β
(IL-1β) and interleukin-6 (IL-6), in addition to the expression of
numerous pro-inflammatory cytokines and chemokines, are increased
in DN with the activation of the NF-κB pathway in the kidneys of
diabetic patients, which in turn activates the TGF-β1/Smad pathway
to promote the progression of fibrosis (29–31).
Notably, TGF-β1 may be acting through the TNF signaling pathway and
worsening inflammatory responses in DN. These enriched GO terms and
pathways provide insights into the molecular mechanism of DN, and
may therefore be useful for the development of novel therapeutic
strategies.
In the present study, DEGs were screened from the
GES2557 and GSE2558 microarray datasets. Jun, Ptgs2 and Fn1 were
identified as hub genes under treatment with TGF-β1, and Cdk2, Fos,
Hspa5, Hsp90b1 and Hyou1 also were selected as hub genes. A number
of these genes have been linked to DN in previous studies. Jun is
one of the components of the nuclear transcription factor activator
protein-1 (AP-1), and binds to TRE elements in target gene promoter
regions in order to activate transcription (32). Weigert et al (33) revealed that the mutation of Jun
binding sites in the TGFβ1 promoter or treatment with a Jun
inhibitor eliminates HG-induced TGFβ1 promoter activity in
mesangial cells. Similarly, Gao et al (34) suggested that the increase in the
expression of Jun and SP1 is synergistically activated, and is
positively correlated with profibrotic expression of TGFβ1 in
HG-treated human renal mesangial cells and in the development of DN
by autoregulation, cross-activation and phospho-modification. PTGS,
also known as cyclooxygenase (COX), consists of two major isoforms,
Ptgs1 (COX-1) and Ptgs2 (COX-2). The cyclooxygenase/prostaglandin
system contributes to the development of DN (35). COX-1 is expressed constitutively in
numerous tissues, whereas COX-2 is expressed in various organs
during inflammation and is expressed at high levels in podocytes,
mesangial cells and macula densa cells in diabetic rats (36). Cheng et al (37) and Cheng et al (38) reported that DN increases podocyte
expression of COX-2, and the inhibition of COX-2 reduces
proteinuria and glomerular injury and delays DN progression in
animal models of diabetes. ECM accumulation may induce further
tubular endothelium fibrosis and glomerular sclerosis and cause the
glomerular filtration rate to progressively decrease, leading to
renal failure in DN. Fn1, a noncollagenous glycoprotein and the
principal component of ECM, is increased substantially in DN and
leads to glomerular sclerosis and eventual fibrosis (39). CDKs serve key roles in cell
proliferation, and Cdk2 is a member of the CDK family, which is
considered to be essential in the cell cycle and associates with
cyclin A, driving cells through the S phase (40). Saurus et al (41) provided a novel strategy to prevent
podocyte apoptosis and the progression of DN but preventing the
downregulation of Cdk2, by inhibiting the Toll-like receptor (TLR)
pathway with 4,5-dihydro-3-phenyl-5-isoxazoleacetic acid. The Fos
(c-Fos, FosB, Fra-1 and Fra-2) and Jun (c-Jun, JunB and JunD)
proteins form dimers with AP-1, which serve an important role in DN
(42). In the present study, the
expression of Fos was significantly upregulated, which was
consistent with the activation of AP-1 by HG (42). The activation of AP-1 induced by
hyperglycemia, oxidative stress, advanced glycation-end products,
low-density lipoprotein, angiotensin II and proinflammatory
cytokines under diabetic conditions may regulate the expression of
FN, TGF-β1 and intercellular adhesion molecule, indicating that
AP-1 is closely correlated with inflammation, cell proliferation
and ECM accumulation during the progression of DN (42–44).
Endoplasmic reticulum (ER) stress, resulting from the accumulation
of misfolded and/or unfolded proteins in ER membranes, and induced
podocyte apoptosis serve a critical role in the development of DN
(45,46). Hspa5, a member of the HSP70 family,
is also known as glucose-regulated protein 78 (GRP78/Bip), an
important molecular chaperone localized to the ER, and has been
extensively considered as an important indicator for the induction
of ER stress; the upregulated expression of GRP78 also indicates
the activation of ER stress (46).
Hyou1 also belongs to the ER chaperone family. Lindenmeyer et
al (47) reported that patients
with established DN had a marked increase in the mRNA expression of
the genes Hspa5 and Hyou1. Cao et al (45) and Chen et al (48) respectively reported that the ER
stress inhibitors ursodeoxycholic acid and 4-phenylbutyrate and
terpene glycoside component from Moutan Cortex ameliorate DN by
regulating ER stress-associated inflammatory responses, as
evidenced by the decreased expression of GRP78/Bip. Hsp90b1 encodes
a member of a family of adenosine triphosphate-metabolizing
molecular chaperones with roles in stabilizing and folding other
proteins, also known as GRP94. GRP94 is the most abundant protein
contributing to ER quality control by chaperoning the folding of
proteins, participating in calcium storage and assisting in the
targeting of misfolded proteins for ER-associated degradation in
the ER lumen (49). Based on the
results from GSE2557 in the present study, GlcN may downregulate
the expression of GRP94 in MES-13 cells.
The results of the molecular docking demonstrated
that valsartan with the overlapping gene RHOB, and fosinopril with
CFH and KLF15 had preferential binding activity. RHOB is an
early-response gene whose expression is elevated by cellular
stresses, and which serves an important role in the onset of DM and
its associated complications (50).
Bravo-Nuevo et al (51)
revealed that RHOB loss prevents streptozotocin-induced diabetes
and ameliorates diabetic complications in mice. The complement
system is a key component of innate immunity. CFH is a 155-kDa
glycoprotein and consists of 20 short census repeat domains. It is
located on chromosome 1q32 and encodes the factor H protein, a
critical inhibitor of the alternative pathway of complement in the
fluid phase and on the surface of host cells (52,53).
Bonomo et al (53) indicated
that a subset of cases with end-stage kidney disease clinically
ascribed to the effects of hypertension or glomerulosclerosis
actually have CFH-associated forms of mesangial proliferative
glomerulonephritis. Podocyte injury is the hallmark of proteinuric
kidney disease. The kidney-enriched zinc finger transcription
factor KLF15 belongs to a 17-member family of DNA-binding zinc
finger transcription factors associated with differentiation,
mitochondrial biogenesis, and cell cycle and DNA repair, a diverse
set of cellular processes (54).
Furthermore, previous reports have suggested that inducing
podocyte-specific KLF15 attenuates kidney injury by directly and
indirectly upregulating genes critical for podocyte
differentiation, suggesting that KLF15 induction may be a potential
strategy for treating proteinuric kidney disease (55).
In conclusion, this comprehensive bioinformatics
analysis identified that a total of 11 hub genes, including Jun,
Ptgs2, Fn1, Cdk2, Fos, Hspa5, Hsp90b1, Hyou1, RHOB, CFH and KLF15,
and the TNF signaling pathway in the MES-13 mouse mesangial cell
line may serve a vital role in the progression of DN. These
findings provide a set of useful molecular targets for future
investigation of the mechanisms and selection of biotargets for DN.
Further experimental investigations or the use of other datasets
are to be performed to confirm the function of the identified genes
in the present observations.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81774217 and
81273623), a grant from Zhejiang Administration of Traditional
Chinese Medicine (grant no. 2017ZKL016), and a grant from the
Science and Technology Commission of Hangzhou (grant no.
20150733Q42).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XM, DiYZ and DaYZ collected and analyzed the
majority of the data and drafted the initial manuscript. YHL and
KYL collected important background information. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sathibabu Uddandrao VV, Brahmanaidu P,
Ravindarnaik R, Suresh P, Vadivukkarasi S and Saravanan G:
Restorative potentiality of S-allylcysteine against diabetic
nephropathy through attenuation of oxidative stress and
inflammation in streptozotocin-nicotinamide-induced diabetic rats.
Eur J Nutr. July 30–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wagnew F, Eshetie S, Kibret GD, Zegeye A,
Dessie G, Mulugeta H and Alemu A: Diabetic nephropathy and
hypertension in diabetes patients of sub-Saharan countries: A
systematic review and meta-analysis. BMC Res Notes. 11:5652018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mafi A, Aghadavod E, Mirhosseini N, Mobini
M and Asemi Z: The effects of expression of different microRNAs on
insulin secretion and diabetic nephropathy progression. J Cell
Physiol. 234:42–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song X, Gong M, Chen Y, Liu H and Zhang J:
Nine hub genes as the potential indicator for the clinical outcome
of diabetic nephropathy. J Cell Physiol. 234:1461–1468. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu P, Peng L, Zhang H, Tang PM, Zhao T,
Yan M, Zhao H, Huang X, Lan H and Li P: Tangshen formula attenuates
diabetic nephropathy by promoting ABCA1-mediated renal cholesterol
efflux in db/db Mice. Front Physiol. 9:3432018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu T, Li Q, Wu T and Liu HY:
Identification of biological targets of therapeutic intervention
for diabetic nephropathy with bioinformatics approach. Exp Clin
Endocrinol Diabetes. 122:587–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ziyadeh FN and Wolf G: Pathogenesis of the
podocytopathy and proteinuria in diabetic glomerulopathy. Curr
Diabetes Rev. 4:39–45. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu J, Liu J, Ding Y, Zhu M, Lu K, Zhou J,
Xie X, Xu Y, Shen X, Chen Y, et al: MiR-455-3p suppresses renal
fibrosis through repression of ROCK2 expression in diabetic
nephropathy. Biochem Biophys Res Commun. 503:977–983. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He F, Peng F, Xia X, Zhao C, Luo Q, Guan
W, Li Z, Yu X and Huang F: MiR-135a promotes renal fibrosis in
diabetic nephropathy by regulating TRPC1. Diabetologia.
57:1726–1736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Putta S, Lanting L, Sun G, Lawson G, Kato
M and Natarajan R: Inhibiting MicroRNA-192 Ameliorates renal
fibrosis in diabetic nephropathy. J Am Soc Nephrol. 23:458–469.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Qiao F, Zhao Y, Wang Y and Liu G:
HMGB1 is activated in type 2 diabetes mellitus patients and in
mesangial cells in response to high glucose. Int J Clin Exp Pathol.
8:6683–6691. 2015.PubMed/NCBI
|
|
12
|
Seo E, Kang H, Oh YS and Jun HS: Psoralea
corylifolia L. Seed extract attenuates diabetic nephropathy by
inhibiting renal fibrosis and apoptosis in streptozotocin-induced
diabetic mice. Nutrients. 9(pii): E8282017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Braga Gomes K, Fontana Rodrigues K and
Fernandes AP: The role of transforming growth factor-beta in
diabetic nephropathy. Int J Med Genet. 2014:1–6. 2014. View Article : Google Scholar
|
|
14
|
Hills CE and Squires PE: The role of TGF-β
and epithelial-to mesenchymal transition in diabetic nephropathy.
Cytokine Growth Factor Rev. 22:131–139. 2011.PubMed/NCBI
|
|
15
|
Cheng DW, Jiang Y, Shalev A, Kowluru R,
Crook ED and Singh LP: An analysis of high glucose and
glucosamine-induced gene expression and oxidative stress in renal
mesangial cells. Arch Physiol Biochem. 112:189–218. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16:162016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Y, Zhang Z, Tang Y, Chen X and Zhou
J: Identification of potential target genes in pancreatic ductal
adenocarcinoma by bioinformatics analysis. Oncol Lett.
16:2453–2461. 2018.PubMed/NCBI
|
|
18
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res 40 (Database
Issue). D109–D114. 2012. View Article : Google Scholar
|
|
19
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46:D1074–D1082. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K,
Watanabe T, Kawaoka Y and Kitano H: SystemsDock: A web server for
network pharmacology-based prediction and analysis. Nucleic Acids
Res. 44:W507–W513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsin KY, Ghosh S and Kitano H: Combining
machine learning systems and multiple docking simulation packages
to improve docking prediction reliability for network pharmacology.
PLoS One. 8:e839222013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma J, Zhang L, Hao J, Li N, Tang J and Hao
L: Up-regulation of microRNA-93 inhibits TGF-β1-induced EMT and
renal fibrogenesis by down-regulation of Orai1. J Pharmacol Sci.
136:218–227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deshpande S, Abdollahi M, Wang M, Lanting
L, Kato M and Natarajan R: Reduced autophagy by a microRNA-mediated
signaling cascade in diabetes-induced renal glomerular hypertrophy.
Sci Rep. 8:69542018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song KH, Park J, Park JH, Natarajan R and
Ha H: Fractalkine and its receptor mediate extracellular matrix
accumulation in diabetic nephropathy in mice. Diabetologia.
56:1661–1669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanwar YS, Sun L, Xie P, Liu FY and Chen
S: A glimpse of various pathogenetic mechanisms of diabetic
nephropathy. Annu Rev Pathol. 6:395–423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reidy K, Kang HM, Hostetter T and Susztak
K: Molecular mechanisms of diabetic kidney disease. J Clin Invest.
124:2333–2340. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng XM, Nikolic-Paterson DJ and Lan HY:
TGF-β: The master regulator of fibrosis. Nat Rev Nephrol.
12:325–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim AK and Tesch GH: Inflammation in
diabetic nephropathy. Mediators Inflamm. 2012:1461542012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lan HY: Transforming growth factor-β/Smad
signalling in diabetic nephropathy. Clin Exp Pharmacol Physiol.
39:731–738. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu L, Shen P, Bi Y, Chen J, Xiao Z, Zhang
X and Wang Z: Danshen injection ameliorates STZ-induced diabetic
nephropathy in association with suppression of oxidative stress,
pro-inflammatory factors and fibrosis. Int Immunopharmacol.
38:385–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jochum W, Passegué E and Wagner EF: AP-1
in mouse development and tumorigenesis. Oncogene. 20:2401–2412.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weigert C, Sauer U, Brodbeck K, Pfeiffer
A, Häring HU and Schleicher ED: AP-1 proteins mediate
hyperglycemia-induced activation of the human TGF-beta1 promoter in
mesangial cells. J Am Soc Nephrol. 11:2007–2016. 2000.PubMed/NCBI
|
|
34
|
Gao P, Wei Y, Zhang Z, Zeng W, Sun D, Liu
D, Hou B, Zhang C, Zhang N, Li H and Li L: Synergistic effects of
c-Jun and SP1 in the promotion of TGFβ1-mediated diabetic
nephropathy progression. Exp Mol Pathol. 100:441–450. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Yao B, Wang Y, Fan X, Wang S, Niu
A, Yang H, Fogo A, Zhang MZ and Harris RC: Macrophage
Cyclooxygenase-2 protects against development of diabetic
nephropathy. Diabetes. 66:494–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Watanabe Y, Yamaguchi T, Ishihara N,
Nakamura S, Tanaka S, Oka R, Imamura H, Sato Y, Ban N, Kawana H, et
al: 7-Ketocholesterol induces ROS-mediated mRNA expression of
12-lipoxygenase, cyclooxygenase-2 and pro-inflammatory cytokines in
human mesangial cells: Potential role in diabetic nephropathy.
Prostaglandins Other Lipid Mediat. 134:16–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng HF, Wang CJ, Moeckel GW, Zhang MZ,
McKanna JA and Harris RC: Cyclooxygenase-2 inhibitor blocks
expression of mediators of renal injury in a model of diabetes and
hypertension1. Kidney Int. 62:929–939. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng H, Fan X, Moeckel GW and Harris RC:
Podocyte COX-2 exacerbates diabetic nephropathy by increasing
podocyte (pro)renin receptor expression. J Am Soc Nephrol.
22:1240–1251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma X, Lu C, Lv C and Wang Q: The
expression of miR-192 and its significance in diabetic nephropathy
patients with different urine albumin creatinine ratio. J Diabetes
Res. 2016:67894022016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q,
Li YX, Liu M, Li X and Tang H: MicroRNA-372 is down-regulated and
targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human
cervical cancer, which may contribute to tumorigene. J Biol Chem.
286:25556–25563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saurus P, Kuusela S, Dumont V, Lehtonen E,
Fogarty CL, Lassenius MI, Forsblom C, Lehto M, Saleem MA, Groop PH
and Lehtonen S: Cyclin-dependent kinase 2 protects podocytes from
apoptosis. Sci Rep. 6:216642016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang K, Huang J, Chen C, Hao J, Wang S,
Huang J, Liu P and Huang H: AP-1 regulates sphingosine kinase 1
expression in a positive feedback manner in glomerular mesangial
cells exposed to high glucose. Cell Signal. 26:629–638. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen S, Mukherjee S, Chakraborty C and
Chakrabarti S: High glucose-induced, endothelin-dependent
fibronectin synthesis is mediated via NF-kappa B and AP-1. Am J
Physiol Cell Physiol. 284:C263–C272. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Haneda M, Koya D, Isono M and Kikkawa R:
Overview of glucose signaling in mesangial cells in diabetic
nephropathy. J Am Soc Nephrol. 14:1374–1382. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cao AL, Wang L, Chen X, Wang YM, Guo HJ,
Chu S, Liu C, Zhang XM and Peng W: Ursodeoxycholic acid and
4-phenylbutyrate prevent endoplasmic reticulum stress-induced
podocyte apoptosis in diabetic nephropathy. Lab Invest. 96:610–622.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Y, Gui D, Chen J, He D, Luo Y and
Wang N: Down-regulation of PERK-ATF4-CHOP pathway by Astragaloside
IV is associated with the inhibition of endoplasmic reticulum
stress-induced podocyte apoptosis in diabetic rats. Cell Physiol
Biochem. 33:1975–1987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lindenmeyer MT, Rastaldi MP, Ikehata M,
Neusser MA, Kretzler M, Cohen CD and Schlöndorff D: Proteinuria and
hyperglycemia induce endoplasmic reticulum stress. J Am Soc
Nephrol. 19:2225–2236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen J, Hou XF, Wang G, Zhong QX, Liu Y,
Qiu HH, Yang N, Gu JF, Wang CF, Zhang L, et al: Terpene glycoside
component from Moutan Cortex ameliorates diabetic nephropathy by
regulating endoplasmic reticulum stress-related inflammatory
responses. J Ethnopharmacol. 193:433–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Eletto D, Dersh D and Argon Y: GRP94 in ER
quality control and stress responses. Semin Cell Dev Biol.
21:479–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang M and Prendergast GC: RhoB in cancer
suppression. Histol Histopathol. 21:213–218. 2006.PubMed/NCBI
|
|
51
|
Bravo-Nuevo A, Sugimoto H, Iyer S, Fallon
Z, Lucas JM, Kazerounian S, Prendergast GC, Kalluri R, Shapiro NI
and Benjamin LE: RhoB loss prevents streptozotocin-induced diabetes
and ameliorates diabetic complications in mice. Am J Pathol.
178:245–252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mehta G, Ferreira VP, Skerka C, Zipfel PF
and Banda NK: New insights into disease-specific absence of
complement factor H related protein C in mouse models of
spontaneous autoimmune diseases. Mol Immunol. 62:235–248. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bonomo JA, Palmer ND, Hicks PJ, Lea JP,
Okusa MD, Langefeld CD, Bowden DW and Freedman BI: Complement
factor H gene associations with end-stage kidney disease in African
Americans. Nephrol Dial Transplant. 29:1409–1414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mallipattu SK, Estrada CC and He JC: The
critical role of Krüppel-like factors in kidney disease. Am J
Physiol Renal Physiol. 312:F259–F265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guo Y, Pace J, Li Z, Ma'ayan A, Wang Z,
Revelo MP, Chen E, Gu X, Attalah A, Yang Y, Estrada C, et al:
Podocyte-specific induction of Krüppel-like factor 15 restores
differentiation markers and attenuates kidney injury in proteinuric
kidney disease. J Am Soc Nephrol. 29:2529–2545. 2018. View Article : Google Scholar : PubMed/NCBI
|