Introduction
Epilepsy is the most common chronic neurological
disorders, affecting ~70 million individuals worldwide (1). Furthermore, ~0.5–1% of the pediatric
population suffer from epilepsy (2)
and approximately one-third of these patients are refractory to
epilepsy medication (3). Valproic
acid (VPA) is an anti-epileptic drug recommended by the National
Institute for Health and Care Excellence guidelines as the
first-line therapy for absence epilepsy (4,5), and has
been used for 50 years due to its efficacy and high tolerability
(6,7). Previous studies have indicated that
approximately one-third of patients are non-responsive to VPA
(8,9), and the reason for this phenomenon
remains elusive. Recently, Gesche et al (10) have demonstrated that resistance to
VPA has a specificity of 100% regarding the identification of
genetic generalized epileptic patients. Consequently, it is
important to elucidate the mechanism underlying VPA efficacy and
identify biomarkers predictive of VPA responses.
At present, two hypotheses for pharmaco-resistant
epilepsy are commonly accepted, namely the multi-transporter
hypothesis and the drug targets hypothesis. However, the mechanism
of VPA resistance is distinct from that of other anti-epileptic
drugs (AEDs). In fact, VPA is neither the substrate of
multi-transporters, including P-glycoprotein, multi-drug resistant
protein and breast cancer resistance protein, nor does it induce
the expression of the multi-transporters in the brain (11–14).
However, reported targets of VPA, including γ-aminobutyric acid
receptor (15,16), sodium channels and calcium channels,
appear to not be involved in VPA resistance (17–19).
These results suggest that the aforementioned hypotheses hardly
explain the mechanisms of VPA resistance or efficacy.
Genome-wide gene expression profiling has been
increasingly used to investigate pathogenetic mechanism and
identify potential biomarkers for various human diseases (20,21). RNA
sequencing (RNA-seq) is a widely used method to study overall
transcriptional activity and has a broad coverage. Previous studies
employing the RNA-seq method led to the discovery of potential
biomarkers for Alzheimer's disease and malignant glioma, including
NeuroD6 and F11R, in brain tissues (22,23).
Fibronectin 1 and 12 other genes implicated in oxidative
phosphorylation and glycolysis/gluconeogenesis pathways in the
brain were identified as candidate critical factors for temporal
lobe epilepsy, with and without hippocampal sclerosis (24,25).
However, few studies have addressed VPA efficacy and resistance,
also due to the limited accessibility of brain tissue from
VPA-treated patients.
Peripheral blood may be obtained non-invasively and
is commonly used for studying biomarkers. Liew et al
(26) have indicated that ~81.9% of
the genes expressed in the brain were also expressed in the
whole-blood microarray dataset. Borovecki et al (27) reported that the blood mRNA levels of
specific target genes are associated with Huntington's disease
severity and response to a histone deacetylase inhibitor. VPA is a
histone deacetylase inhibitor (28,29),
potentially affecting, directly or indirectly, between 2 and 5% of
all genes (30). A previous study
reported that 11 genes were differentially expressed after a
3-month VPA treatment (31). In the
present study, the mRNA expression profile in the blood of VPA
responders and non-responders was analyzed after a treatment period
of ≥1 year, to identify possible biomarkers for the prediction of
VPA efficacy.
Materials and methods
Patients
Subjects aged from 0 to 18 years were enrolled at
the Children's Hospital of Fudan University (Shanghai, China)
between July 2016 and May 2018. Each patient was evaluated
according to the inclusion and exclusion criteria (32). The inclusion criteria were as
follows: Pediatric patients diagnosed with epilepsy or an epileptic
syndrome and administration of VPA for at least one year. The
exclusion criteria were as follows: Patients with abnormal liver
and kidney function, and patients who had developed infectious
diseases, including upper respiratory infection and urinary tract
infection, in the last three months.
Patients with a complete disappearance of seizures
and a normal electroencephalogram were considered as VPA
responders, while patients who continued to experience seizures
were assigned to the non-responsive groups. Seizure types were
identified according to the International League Against Epilepsy
definition (33). Focal epileptic
seizures were defined as events originating within networks limited
to one hemisphere. Generalized epileptic seizures were
conceptualized as originating at a certain point within, and
rapidly engaging, bilaterally distributed networks (34).
RNA preparation
Blood from three VPA responders and five
non-responders without seizure for 12 h was collected in PAXgene
blood RNA tubes and stored at −80°C until use. PAXgene tubes were
stabilized for 2 h at room temperature. After centrifugation for 10
min at 3,000–5,000 × g at 4°C by using a swing-out rotor, the
supernatant was removed by pipetting. Subsequently, 4 ml of
RNase-free water were added to the pellet. Total RNA was extracted
according to the instructions of the PAXgene Blood RNA kit
(Qiagen), and the quality and quantity of total RNA were determined
using a Qubit 2.0 (Thermo Fisher Scientific, Inc.) and a
Bioanalyzer 2100 (Agilent Technologies).
RNA-seq
Transcriptome sequencing libraries were prepared by
using the TruSeq RNA LT V2 Sample Prep lit (Illumina, Inc.) and
were qualified using the Qubit 2.0. Paired-end sequencing for 150
base pair was performed by an Illumina HiSeq 2500 instrument
(Illumina, Inc.). All of the paired-end raw reads were
quality-checked for low-quality bases and adapter sequences.
Analysis of differentially expressed
genes (DEGs)
Low-quality fractions and Illumina universal
adapters were deleted by using Trim Galore v0.4.2 with the
threshold of Q<30. Fasta QC (version 0.11.5; Illumina, Inc.) was
employed to assess the quality of the data. The paired-end
sequencing reads were aligned to the reference genome (hg19),
downloaded from the University of California Santa Cruz (UCSC)
website (http://hgdownload.soe.ucsc.edu/downloads.html).
Statistically significant expression changes between responders and
non-responders were estimated using the Cuffilinks 2.2.1 software
(http://cole-trapnell-lab.github.io/cufflinks/install/)
with a threshold of 10 for NO TEST. Student's t-test was
performed to calculate the P-value, while the false discovery rate
controlled by the Benjamini-Hochberg procedure was used for
determining the Q-value. Genes with a fold change of >1 and
P<0.05 were defined as DEGs.
Validation by reverse
transcription-quantitative (RT-qPCR)
Changes in the mRNA expression of specific DEGs
associated with epilepsy [chemokine (C-C motif) ligand 3 and FOS],
exhibiting high fold changes, were validated by qPCR in 17 samples
(including 6 VPA responders and 11 non-responders). Specific
primers for selected genes were designed using Primer-BLAST
(http://www.ncbi.nlm.nih.gov/tools/primer-blast). All
samples were amplified in triplicate. qPCR amplifications were
performed with the following cycling parameters: An initial hot
start at 95°C for 5 min followed by 45 cycles of 95°C for 15 sec
and 60°C for 30 sec. In order to normalize the qPCR results, GAPDH
was included as the reference gene. The relative expression of
genes was calculated based on the average quantification cycle (Cq)
values across samples. Relative expression=2{-[(Cq gene of
interest-Cq GAPDH of interest) non-responders]-(Cq gene of
interest-Cq GAPDH of interest) responders}} (35).
Gene functions and pathways
Gene Ontology (GO) gene functions and biochemical
pathways enriched by the DEGs were determined by using the
web-based annotation tool DAVID v6.7 (https://david-d.ncifcrf.gov/summary.jsp) (36), providing GO terms in the categories
biological process (BP), cellular component (CC), and molecular
function (MF) and Kyoto Encyclopedia of Genes and Genomes pathways.
P<0.05 was used as the significance threshold.
Construction and visualization of the
protein-protein interaction (PPI) network
The PPI network based on the DEGs identified were
constructed by using the STRING database (https://string-db.org), a pre-computed database
wherein associations between proteins are assigned on the basis of
high-throughput experiments, literature mining, gene fusion,
co-occurrence, co-expression analysis, and also computational
predictions, e.g. genomic meta-analysis. Interactions with a
confidence score of 0.7 were considered for visualization by
Cytoscape v3.4.0 (https://cytoscape.org/).
Network module analysis
The Molecular Complex Deletion (MCODE) plugin for
Cytoscape was used to analyze the network modules (37). Densely connected regions or clusters
in the co-expression network were identified using the following
parameters: Degree cut-off=2, k-core=2 and max. depth=100.
Statistical analysis
DEGs analysis was performed by using Cuffilinks
2.2.1 software (cuffdiff,
ttp://cole-trapnell-lab.github.io/cufflinks/install/). RT-qPCR data
were analyzed using GraphPad Prism version 7.0 software (GraphPad,
Inc.). Values were expressed as the mean ± standard error.
Student's t-test was performed to analyze differences between
VPA-responders and non-responders.
Results
Demographic data
A total of 8 pediatric patients with epilepsy were
recruited, of which 3 were responders, while 5 were non-responders.
All responders were males, while 3 of the non-responders were males
and 2 females. The average age of the responders was 7.0±2.2 years
and was not significantly different from that of the non-responsive
group (4.8±1.8 years, P=0.47). Furthermore, no significant
difference in the plasma VPA concentration was identified between
the responsive and non-responsive groups (107.2±17.7 vs. 80.4±17.4,
P=0.35; Table I).
 | Table I.Demographic data. |
Table I.
Demographic data.
| Demographics | Responders | Non-responders |
|---|
|
|
|
|
|---|
| ID | 1 | 2 | 3 | Median (min,
max) | 1 | 2 | 3 | 4 | 5 | Median (min,
max) | P-value |
|---|
| Age (Years) | 10.7 | 3 | 7.4 | 7.4 (3, 10.7) | 9.6 | 4.8 | 8 | 0.3 | 1.3 | 4.8 (0.3, 9.6) | 0.47 |
| Sex | Male | Male | Male | – | Male | Female | Male | Female | Male | – | – |
| Seizure type | Unknown | Generalized | Generalized | – | Focal | Generalized | Generalized | Generalized | Generalized | – | – |
| AEDs | VPA+TPM | VPA | VPA | – | VPA | VPA+LTG+OXC | VPA | VPA | VPA+LEV | – | – |
| Dose of
VPA(mg/kg) | 33.3 | 21.5 | 18.1 | 21.5 | 29.7 | 28.6 | 31.2 | 29.1 | 36.4 | 29.7 | 0.13 |
|
|
|
|
| (18.1, 33.3) |
|
|
|
|
| (28.6, 36.4) |
|
| Plasma
concentration | 139.1 | 77.9 | 104.6 | 104.6 | 56.7 | 62.0 | 148.2 | 57.3 | 77.7 | 62 (56.7,
148.2) | 0.35 |
| (µg/ml) |
|
|
|
| (77.9, 139.1) |
|
|
|
|
|
|
Comparative transcriptome
profiling
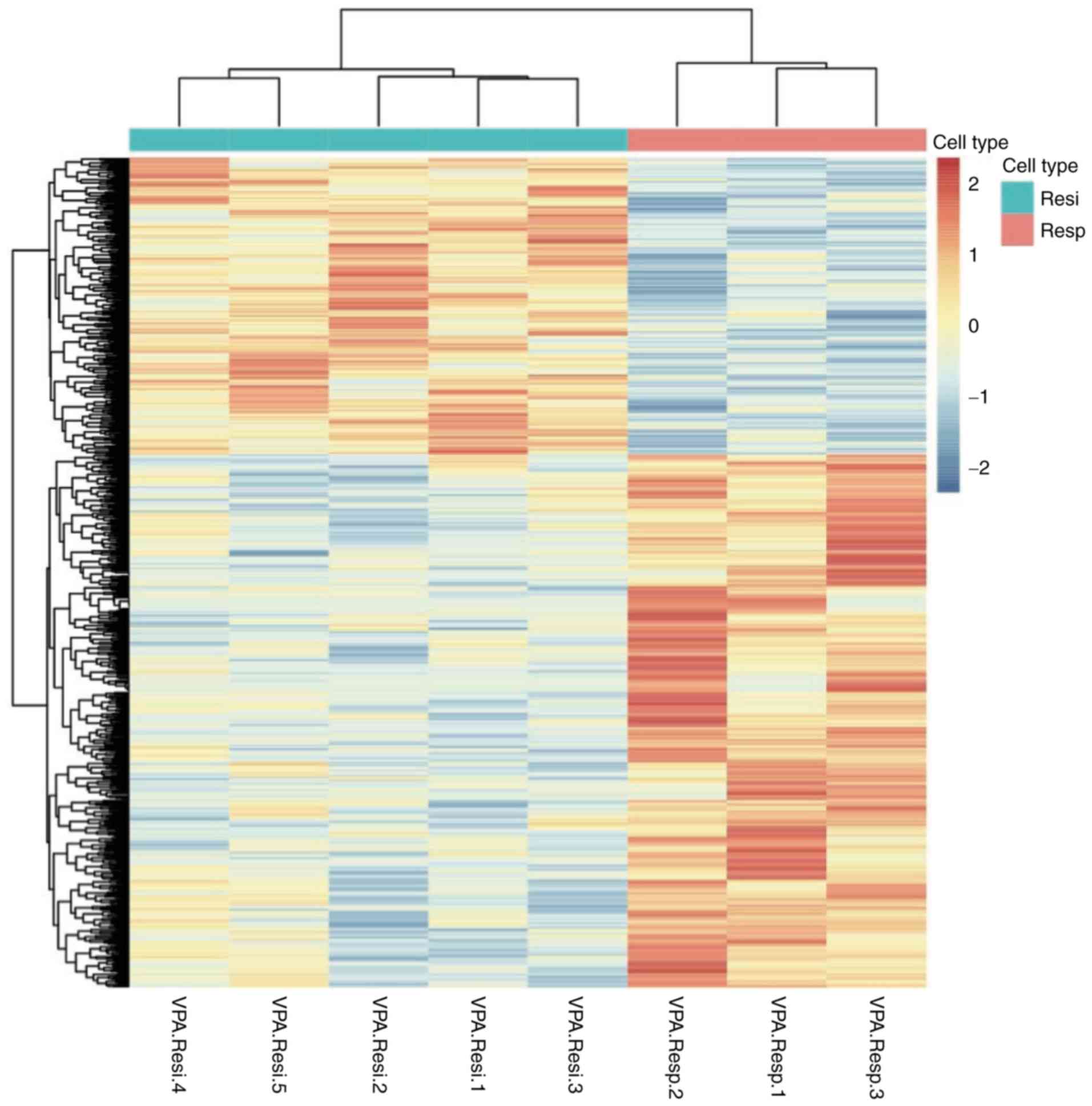
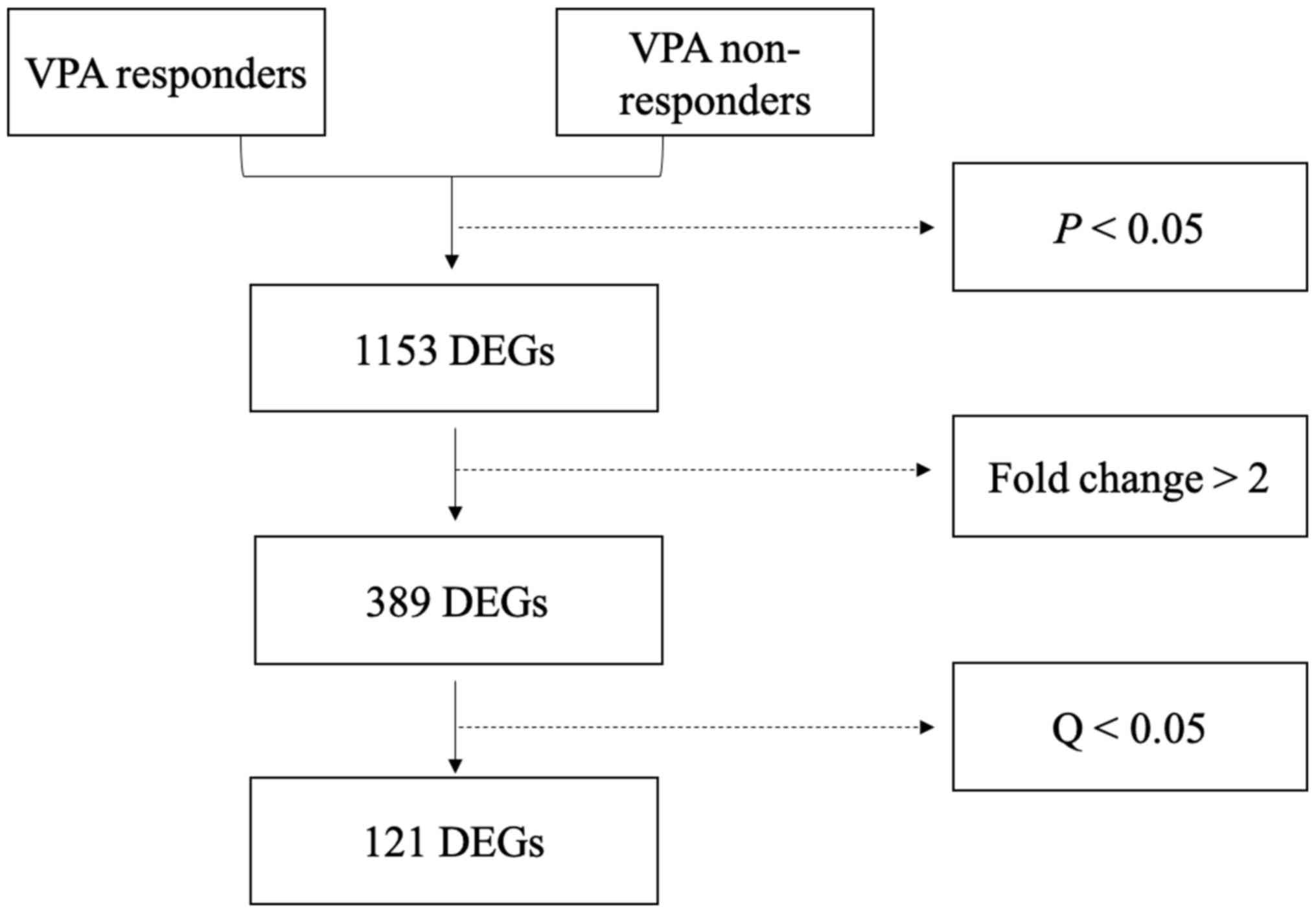
A total of 1,153 genes were differentially expressed
between responders and non-responders (P<0.05; Fig. 1). Of these DEGs, 389 had a |Log 2
fold change|≥1 and comprised of 227 upregulated and 162
downregulated genes. Among the 389 DEGs, 121 variations had a
Q-value of <0.05 and included 84 upregulated and 37
downregulated genes (Fig. 2). The 20
most significantly upregulated and downregulated genes are listed
in Tables II and III, respectively.
 | Table II.Top 20 upregulated differentially
expressed genes. |
Table II.
Top 20 upregulated differentially
expressed genes.
| Gene | Definition | Log2 fold
change | P-value | Q-value |
|---|
| TSIX | TSIX transcript,
XIST antisense RNA | 7.59 |
5.00×10−5 |
9.00×10−3 |
| CXCL10 | Chemokine (C-X-C
motif) ligand 10 | 5.99 |
5.00×10−5 |
9.00×10−3 |
| LILRA3 | Leukocyte
immunoglobulin-like receptor subfamily a (without tm domain) member
3 | 5.96 |
5.00×10−5 |
9.00×10−3 |
| FN1 | Fibronectin 1 | 3.47 |
5.00×10−5 |
9.00×10−3 |
| GPR84 | G protein-coupled
receptor 84 | 3.25 |
5.00×10−5 |
9.00×10−3 |
| PDK4 | Pyruvate
dehydrogenase kinase, isozyme 4 | 2.99 |
5.00×10−5 |
9.00×10−3 |
| SEMA6B | Sema domain.
Transmembrane domain (tm) and cytoplasmic domain (semaphorin)
6b | 2.74 |
5.00×10−5 |
9.00×10−3 |
| HLA-DRB5 | Major
histocompatibility complex class ii dr β 5 | 2.73 |
5.00×10−5 |
9.00×10−3 |
| PTGES | Prostaglandin E
synthase | 2.73 |
5.00×10−5 |
9.00×10−3 |
| MYOM2 | Myomesin 2 | 2.68 |
5.00×10−5 |
9.00×10−3 |
| CCL3 | Chemokine (C-C
motif) ligand 3 | 2.60 |
5.00×10−5 |
9.00×10−3 |
| IL1B | Interleukin 1β | 2.55 |
5.00×10−5 |
9.00×10−3 |
| IFI27 | Interferon
α-inducible protein 27 | 2.32 |
5.00×10−5 |
9.00×10−3 |
| HJURP | Holliday junction
recognition protein | 2.17 |
5.00×10−5 |
9.00×10−3 |
| RRM2 | Ribonucleotide
reductase M2 | 2.06 |
5.00×10−5 |
9.00×10−3 |
| CDCA5 | Cell division cycle
associated 5 | 2.04 |
5.00×10−5 |
9.00×10−3 |
| FOLR3 | Folate receptor 3
(γ) | 1.97 |
5.00×10−5 |
9.00×10−3 |
| TNF | Tumor necrosis
factor | 1.89 |
5.00×10−5 |
9.00×10−3 |
| PLK1 | Polo-like kinase
1 | 1.83 |
5.00×10−5 |
9.00×10−3 |
| SEC14L2 | SEC14-like 2 (S.
cerevisiae) | 1.79 |
5.00×10−5 |
9.00×10−3 |
 | Table III.Top 20 downregulated differentially
expressed genes. |
Table III.
Top 20 downregulated differentially
expressed genes.
| Gene | Definition | Log2 fold
change | P-value | Q-value |
|---|
| ARHGEF10 | Rho guanine
nucleotide exchange factor 10 | −3.33 |
5.00×10−5 |
9.00×10−3 |
| KCNG1 | Potassium
voltage-gated channel subfamily g member 1 | −2.63 |
5.00×10−5 |
9.00×10−3 |
| FOS | FBJ murine
osteosarcoma viral oncogene homolog | −2.20 |
5.00×10−5 |
9.00×10−3 |
| PAQR8 | Progestin and
adipoq receptor family member VIII | −2.06 |
5.00×10−5 |
9.00×10−3 |
| C21orf15 | Chromosome 21 open
reading frame 15 | −2.06 |
5.00×10−5 |
9.00×10−3 |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2
(prostaglandin G/H synthase and cyclooxygenase) | −1.72 |
5.00×10−5 |
9.00×10−3 |
| HCAR2 | Hydroxycarboxylic
acid receptor 2 | −1.71 |
5.00×10−5 |
9.00×10−3 |
| IL8 | Interleukin 8 | −1.67 |
5.00×10−5 |
9.00×10−3 |
| TGFA | Transforming growth
factor α | −1.57 |
5.00×10−5 |
9.00×10−3 |
| HLA-DQA2 | Major
histocompatibility complex class II DQ α 2 | −1.49 |
5.00×10−5 |
9.00×10−3 |
| FOSB | FBJ murine
osteosarcoma viral oncogene homolog B | −1.47 |
5.00×10−5 |
9.00×10−3 |
| TNFRSF10C | Tumor necrosis
factor receptor superfamily member 10c decoy without an
intracellular domain | −1.46 |
5.00×10−5 |
9.00×10−3 |
| DUSP1 | Dual specificity
phosphatase 1 | −1.43 |
5.00×10−5 |
9.00×10−3 |
| RTN1 | Reticulon 1 | −1.40 |
5.00×10−5 |
9.00×10−3 |
| TLR10 | Toll-like receptor
10 | −1.25 |
5.00×10−5 |
9.00×10−3 |
| KCNE3 | Potassium
voltage-gated channel Isk-related family member 3 | −1.20 |
5.00×10−5 |
9.00×10−3 |
| THBD | Thrombomodulin | −1.19 |
5.00×10−5 |
9.00×10−3 |
| MYBL1 | v-myb
myeloblastosis viral oncogene homolog (avian)-like 1 | −1.09 |
5.00×10−5 |
9.00×10−3 |
| ARFIP1 | ADP-ribosylation
factor interacting protein 1 | −1.38 |
1.00×10−4 |
1.60×10−2 |
| FCRL5 | Fc receptor-like
5 | −1.27 |
1.00×10−4 |
1.60×10−2 |
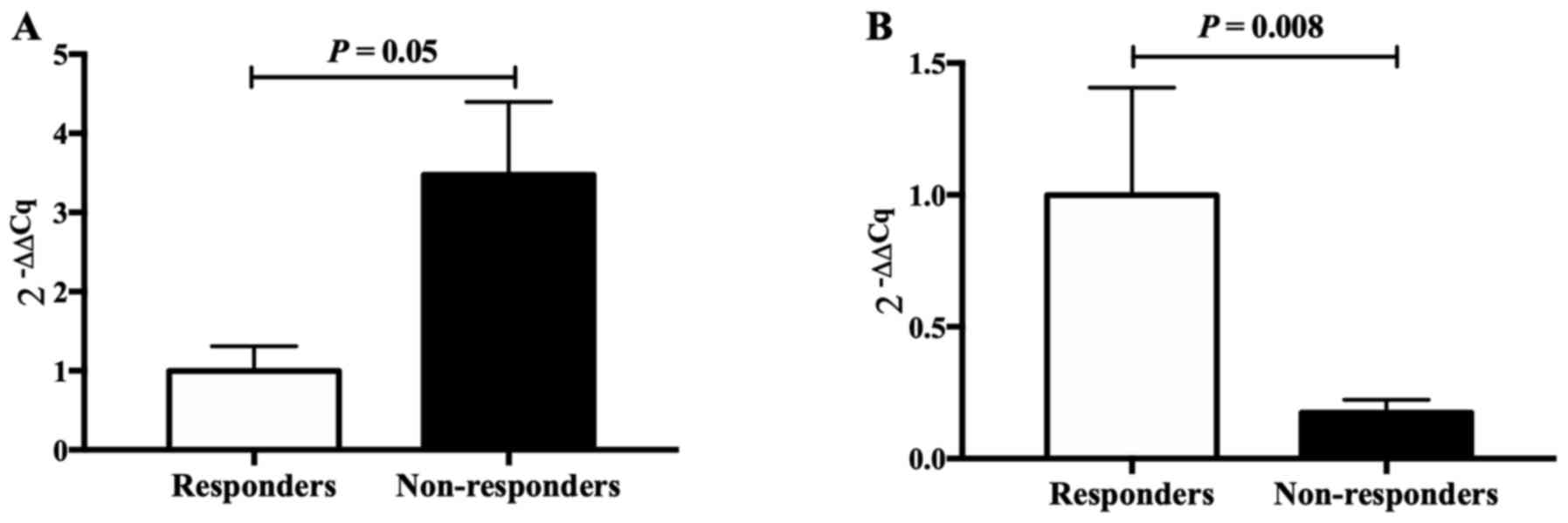
Two genes (CCL3 and FOS), closely associated with
epilepsy, were selected for validation by RT-qPCR, revealing
significant differences in expression between VPA responders and
non-responders, in accordance with the results of the RNA-seq. This
indicated that the results obtained by the RNA-seq analysis were
reliable (Fig. 3).
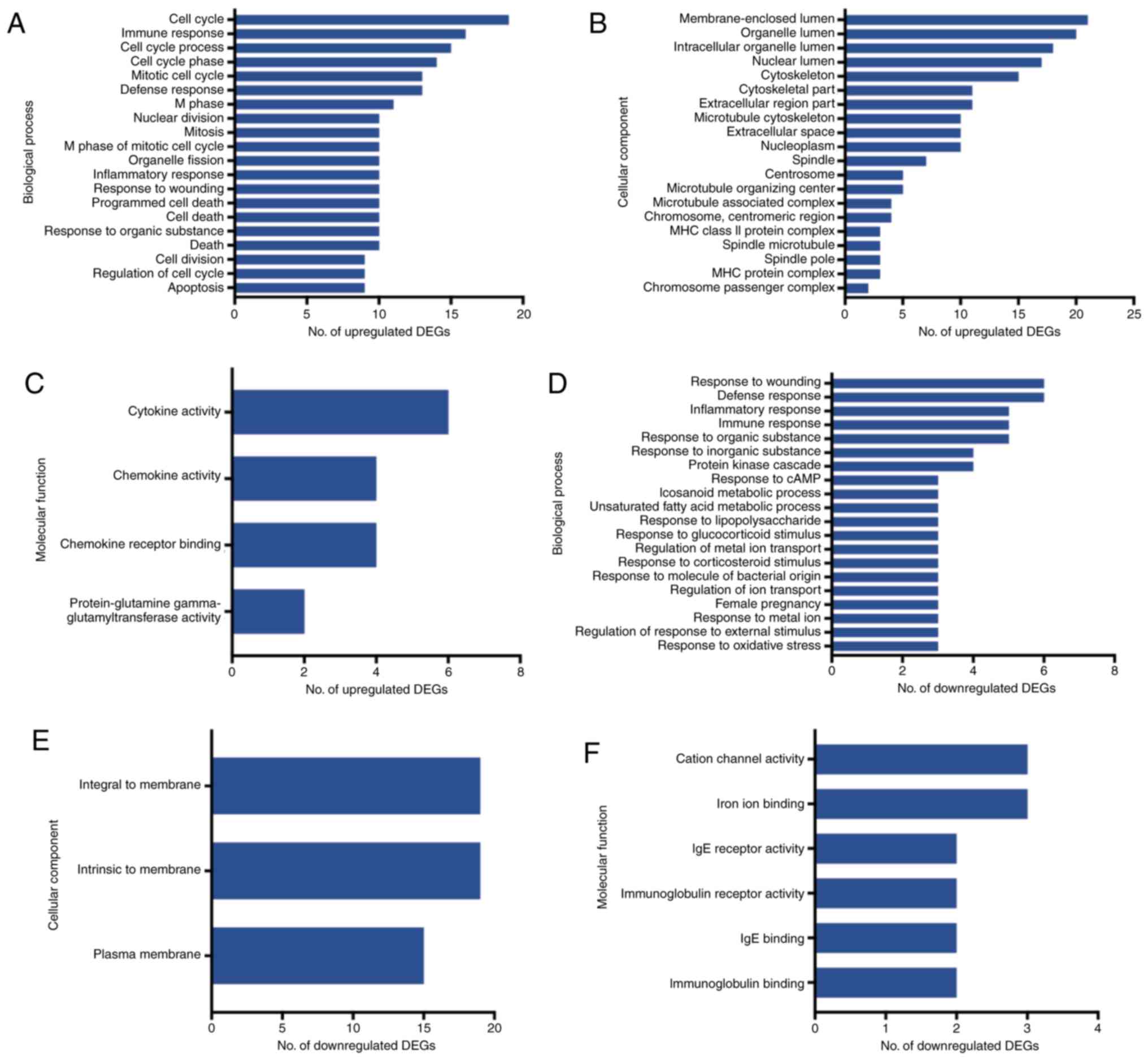
GO and pathway analysis
The GO and pathway enrichment analyses were
performed for the 121 final DEGs, including 84 upregulated and 37
downregulated genes. In the GO category BP, the upregulated DEGs
were significantly enriched in the GO terms ‘cell cycle’, ‘immune
response’ and ‘cell cycle process’ (Fig.
4A). In the category CC, the upregulated DEGs were enriched in
the GO terms ‘membrane-enclosed lumen’, ‘organelle lumen’ and
‘organelle lumen’ (Fig 4B), and in
the category MF, they were enriched in the GO terms ‘cytokine
activity’, ‘chemokine activity’ and ‘chemokine receptor binding’
(Fig 4C). On the other hand, the
group of downregulated DEGs was highly enriched in the GO terms
‘response to wounding’, ‘defense response’ and ‘inflammatory
response’ in the category BP (Fig
4D). In the category CC, the downregulated DEGs were highly
enriched in the GO terms ‘integral to membrane’, ‘intrinsic to
membrane’ and ‘plasma membrane’ (Fig
4E), and in the category MF, they accumulated in the GO terms
‘cation channel’, ‘iron ion binding’ and ‘IgE receptor activity’
(Fig. 4F).
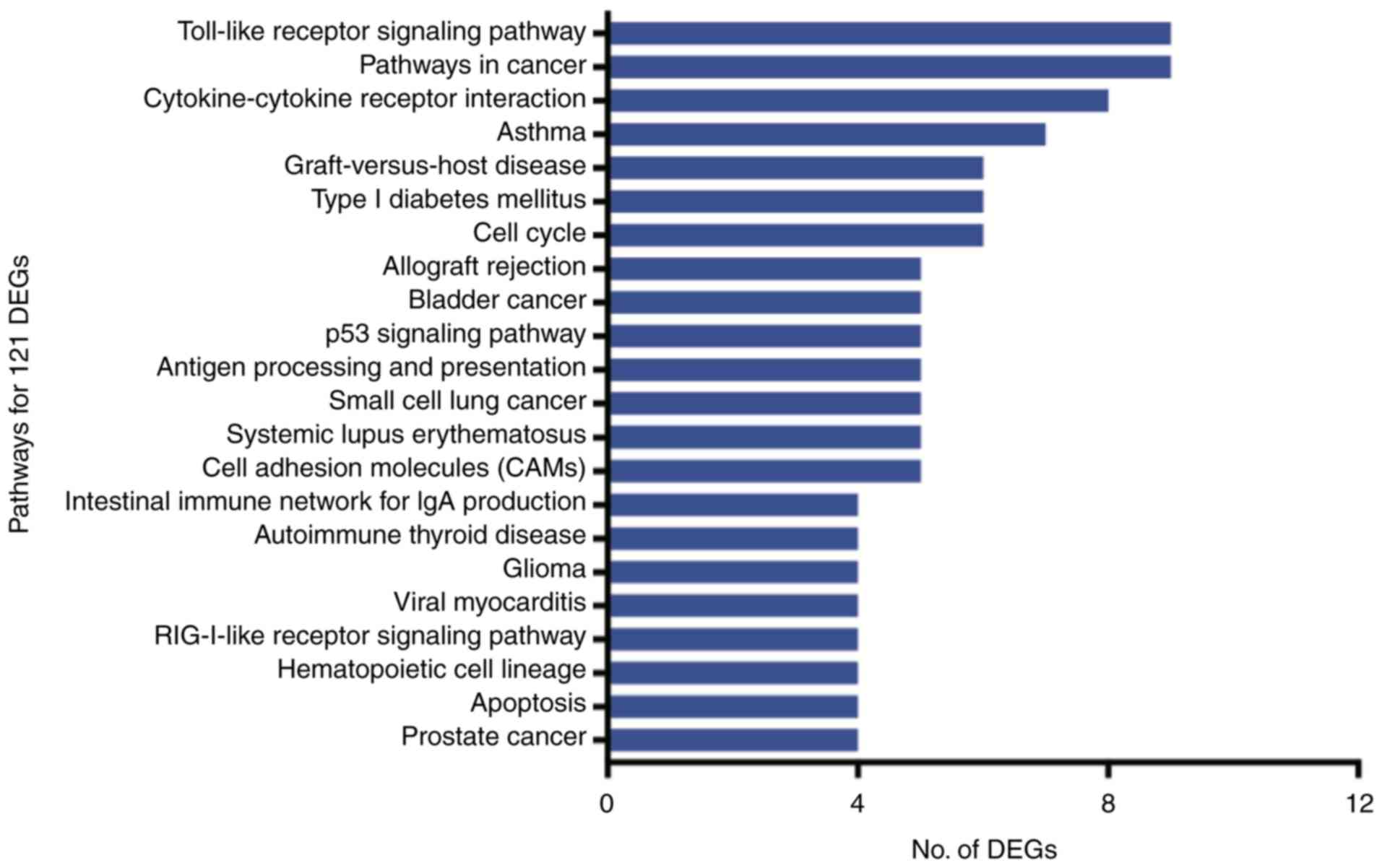
The 121 DEGs were enriched in 22 pathways
(P<0.05), of which the Toll-like receptor signaling pathway,
cancer pathways and cytokine-cytokine receptor interactions were
the most represented (Fig. 5).
PPI network analysis and module
identification
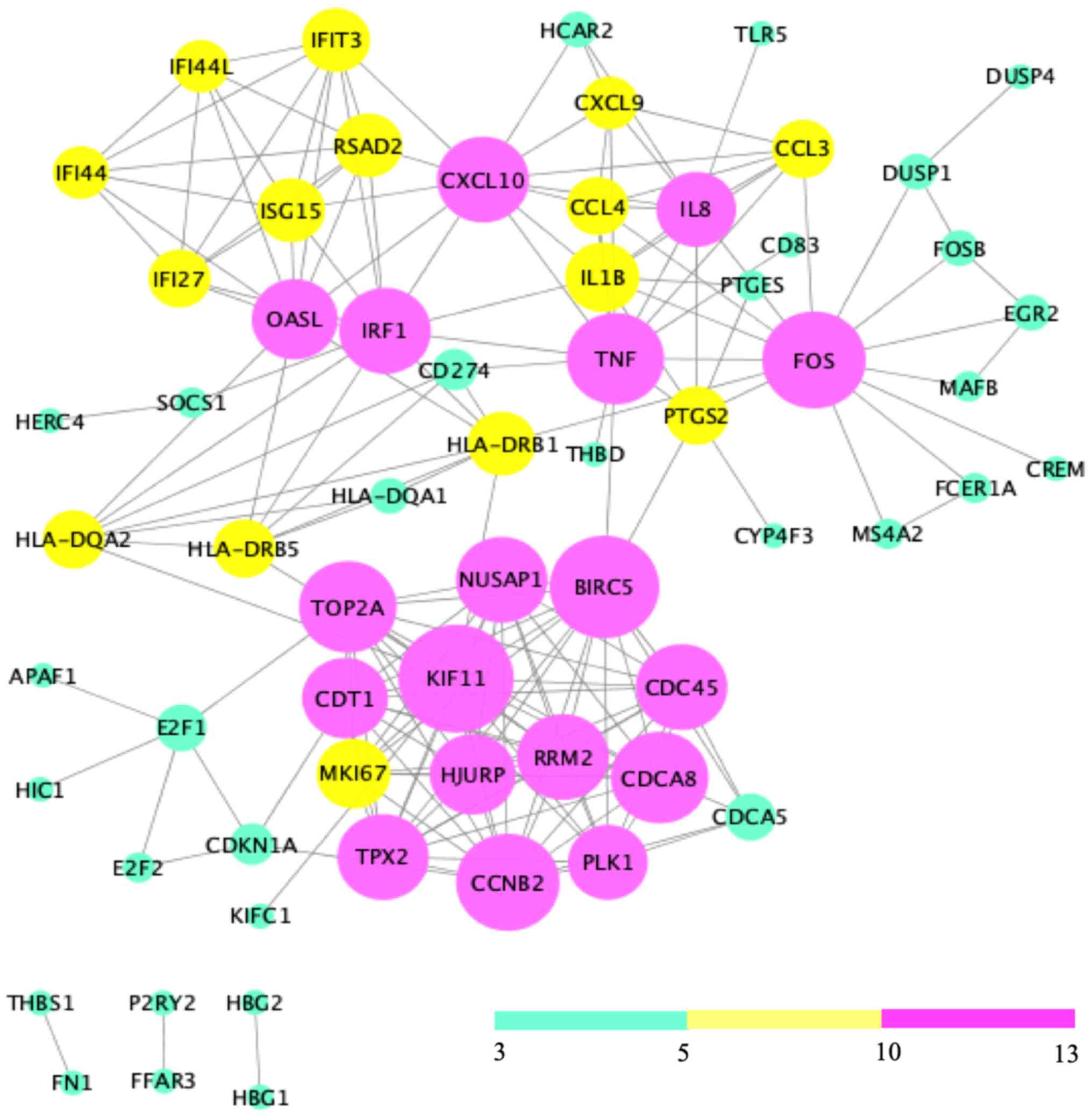
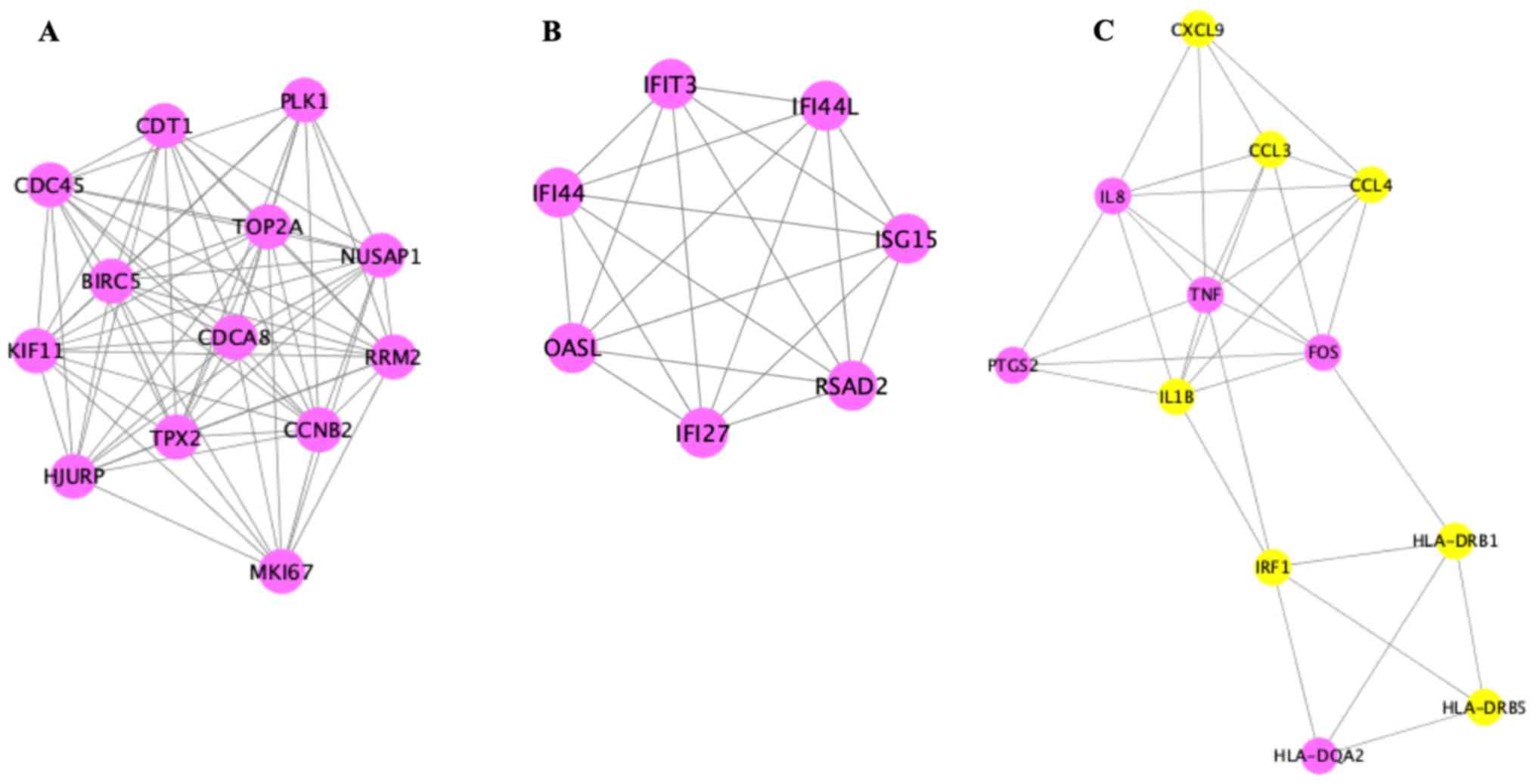
Analysis of the 121 DEGs by STRING and visualization
with the Cytoscape plugin MCODE revealed 197 interactions, covering
three modules (Fig. 6). Of these,
module 1 contained 73 interactions with 13 nodes (CDT1, PLK1,
NUSAP1, RRM2, MKI67, CCNB2, HJURP, TPX2, CDCA8, TOP2A, BIRC5,
KIF11, CDC45), module 2 included 21 interactions with 7 nodes
(IFIT3, IFI44L, ISG15, RSAD2, IFI27, OASL, IFI44L), and module 3
contained 32 interactions with 12 nodes [chemokine (C-X-C motif)
ligand 9 (CXCL9), CCL3, CCL4, tumor necrosis factor-α (TNF-α),
interleukin (IL)-1β, IRF1, HLA-DRB1, HLA-DRB5, IL8, PTGS2, FOS,
HLA-DQA2)] (Fig. 7).
A literature review in PubMed confirmed that the
function of CCL3, CCL4, CXCL9, TNF-α, IL-1β, and FOS is
associated with epilepsy (Table IV)
(38–44).
 | Table IV.Functions of chemokine genes in
epilepsy. |
Table IV.
Functions of chemokine genes in
epilepsy.
| Gene | Function | (Refs.) |
|---|
| CCL3 | Inhibition of
systemic receptor leads to decrease in seizure activity | (38,39) |
| CCL4 | Inhibition of
systemic receptor leads to decrease in seizure activity | (38,39) |
| CXCL9 | Immune-cell
recruitment across the BBB | (38) |
| TNF | Activation of NF-κB
and regulation of the process of post-seizure neurogenesis | (40,41) |
| IL-1β | Induction of
spontaneously recurring seizures | (42) |
| FOS | Regulation of CA3
neuronal excitability and survival | (43,44) |
Discussion
Pharmaco-resistant epilepsy remains a major clinical
issue with elusive underlying mechanisms. In the present study, a
total of 1,153 DEGs between VPA responders and non-responders
(P<0.05) were initially identified. Of these genes, 123
upregulated and 60 downregulated genes fulfilled the criterion of
P<0.001. This number was higher than that in the study of Tang
et al (45), which may be
accounted for by the different methods used. In the latter study,
oligonucleotide microarrays were employed, containing probe sets
for more than 12,000 genes and ESTs. However, previous studies have
concluded that microarray platforms suffer from technical issues
including cross-hybridization, nonspecific hybridization and
limited range of detection of individual probes (46,47). As
a result, genes with expression below or near the background level
may exhibit increased variability and as such, calculated
fold-changes for these genes may be difficult to detect with
statistical significance. RNA-seq avoids such technical issues and
exhibits a broader dynamic range. In the analysis of the current
study, specific DEGs (dual specificity phosphatase 1, ribosomal
protein S6 kinase A1 and aldehyde dehydrogenase 2 family member)
reported by Tang et al (45)
were also identified.
In the present study, most of the DEGs were
implicated in the immune and inflammatory response, and associated
with cytokine-cytokine receptor interactions, as also identified in
epileptic patients by Floriano-Sánchez et al (32). Inflammation is increasingly
recognized as an important pathogenetic factor in epilepsy.
Evidence suggests the presence of all of the hallmarks of a chronic
inflammatory state, i.e., infiltration of leukocytes, reactive
gliosis, as well as overexpression of cytokines and their target
proteins, in the brain of pharmaco-resistant epileptic patients and
animal models (48). CCL3, CCL4 and
CXCL9 are the chemokines that guide directional migration of
leukocytes and have an important role in the inflammation of the
central nervous system. Several studies have demonstrated increased
mRNA and protein expression of CCL3, CCL4 and CXCL9 in the cortex
and hippocampus of epileptic rats and drug-refractory patients
(49–51). The present study revealed that the
expression of CCL3, CCL4 and CXCL9 was higher in the blood of VPA
non-responsive vs. responsive pediatric patients, which was
consistent with the results obtained by Srivastava et al
(52). The reason for the
overexpression of CCL3, CCL4 and CXCL9 in the brain and blood of
drug-refractory patients remains elusive. It has been reported that
TNF-α and IL-1β induce the expression of CCL3 and CCL4 through
NF-κB and activate inflammation via the mTOR signaling pathway
(41,53). Of note, VPA was demonstrated to
reduce the amount of leukocytes and inhibit the expression of TNF-α
(54,55). The present study indicated that the
mRNA levels of TNF-α, IL-1β, NF-κB and IL-1 receptor-associated
kinase 2 (a regulator of TNF-α), were significantly higher in VPA
non-responders than in responders, which suggested that CCL3 and
CCL4 overexpression was associated with the high expression of
TNF-α and IL-1β, and indicated that the transcriptional states of
CCL3, CCL4, CXCL9, TNF-α and IL-1β are potential markers for
monitoring the patients' resistance to VPA.
Previous reports have revealed that increased TNF-α
may result in the inhibition of cytochrome P450 family 2 subfamily
D member 6 (CYP2D6) and CYP2C19 expression, and may results in the
enhanced expression of CYP3A4 and CYP2C9 (the enzymes responsible
for VPA metabolism in the brain) (56). Furthermore, TNF-α has been reported
to induce the overexpression of transporter (P-gp), which is
associated with AED efficacy (57).
However, Feng et al (58)
indicated that there was no difference in the plasma VPA
concentration between responders and non-responders, suggesting
that the role of TNF-α in the efficacy of VPA may be independent of
its effects on drug metabolism and P-gp.
CCL3, CCL4, IL-1β and TNF-α are able to increase the
permeability of the blood-brain barrier (59,60),
possibly resulting in their passage into the brain and
cerebrospinal fluid (61).
Furthermore, overexpression of CCL3 and TNF-α was identified to
induce the influx of Ca2+ and to enhance the expression
of N-methyl-D-aspartate receptor (NMDAR), leading to increased
excitatory neurotransmission and contributing to the development of
epileptic seizures and excitotoxicity (62,63).
Previous studies have demonstrated an association between NMDAR and
the efficacy of AEDs. Zellinger et al (64) indicated that blocking the
glycine-binding site of the NR2B subunit of NMDAR may increase the
sensitivity to AEDs. Hung et al (65) confirmed that a specific mutation
(−200 T>G) of NR2B is associated with s sustained dosage of VPA.
Therefore, it may be speculated that NMDAR functionally links CCL3,
CCL4, IL-1β and TNF-α with VPA efficacy. However, further study is
required for verification.
The protein expression of FOS, an immediate early
gene and recognized biomarker of neuronal activity, is activated
during spontaneous seizure (66–68).
Previous studies have indicated that the expression of FOS rapidly
increases 1.5 h after seizure stimulation by pentylenetetrazol and
in amygdala-kindled seizures (69).
However, the expression profile of FOS during seizures is complex
and varies depending on the seizure type and status. FOS expression
is increased in generalized seizure but not change in focal
seizures. With respect to seizure status, high FOS mRNA expression
was detected in rats at 1 h after the injection of kainic acid,
while it tended to be low after 6 h (70). Furthermore, Madsen et al
(71) identified a large increase in
FOS expression at 2 h after a kindling stimulus, while after 18 h,
the expression was lower than that observed upon a sham
stimulation, and reached the control levels after 3 weeks. Of note,
all plasma samples in the present study were collected during a
non-seizure period which may account for the slightly decreased FOS
expression.
In conclusion, the chemokines CCL3, CCL4, CXCL9,
TNF-α and IL-1β may participate in processes associated with VPA
resistance and serve as potential biomarkers for monitoring the
efficacy of VPA. The study also revealed numerous critical pathways
and sub-modules of potential pathogenetic relevance, deserving
further investigation.
Acknowledgements
Not applicable.
Funding
The current study was supported by Important
Discipline of Shanghai (grant no. 20162B0305) and the National
Natural Science Foundation of China (grant nos. 81370776 and
81874325).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contribution
YW analyzed the data and drafted the manuscript. ZL
designed the study and revised the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the Children's Hospital of Fudan University (Shanghai,
China) in 2016 (no. 136). Written informed consent was obtained
from the guardians of patients prior to enrolment.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Singh A and Trevick S: The epidemiology of
global epilepsy. Neurol Clin. 34:837–847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lv RJ, Shao XQ, Cui T and Wang Q:
Significance of MDR1 gene C3435T polymorphism in predicting
childhood refractory epilepsy. Epilepsy Res. 132:21–28. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Voll A, Hernández-Ronquillo L, Buckley S
and Téllez-Zenteno JF: Predicting drug resistance in adult patients
with generalized epilepsy: A case-control study. Epilepsy Behav.
53:126–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rakitin A, Kõks S and Haldre S: Valproate
modulates glucose metabolism in patients with epilepsy after first
exposure. Epilepsia. 56:e172–e175. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fernando-Dongas MC, Radtke RA,
Vanlandingham KE and Husain AM: Characteristics of valproic acid
resistant juvenile myoclonic epilepsy. Seizure. 9:385–388. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nevitt ST, Sudell M, Weston J, Tudur Smith
C and Marson AG: Antiepileptic drug monotherapy for epilepsy: A
network meta-analysis of individual participant data. Cochrane
Database Syst Rev. 6:CD01141212017.
|
|
7
|
Brigo F, Igwe SC and Lattanzi S:
Ethosuximide, sodium valproate or lamotrigine for absence seizure
in children and adolescents. Cochrane Database Syst Rev.
2:CD0030322019.PubMed/NCBI
|
|
8
|
Yasiry Z and Shorvon SD: The relative
effectiveness of five antiepileptic drugs in treatment of
benzodiazepine-resistant convulsive status epilepticus: A
meta-analysis of published studies. Seizure. 23:167–174. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trinka E, Höfler J, Zerbs A and Brigo F:
Efficacy and safety of intravenous valproate for status
epilepticus: A systematic review. CNS Drugs. 28:623–639. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gesche J, Khanevski M, Solberg C and Beier
CP: Resistance to valproic acid as predictor of treatment
resistance in genetic generalized epilepsies. Epilepsia.
58:E64–E69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luna-Tortós C, Fedrowitz M and Löscher W:
Evaluation of transport of common antiepileptic drugs by human
multidrug resistance-associated proteins (MRP1, 2 and 5) that are
overexpressed in pharmacoresistant epilepsy. Neuropharmacology.
58:1019–1032. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baltes S, Fedrowitz M, Tortos CL, Potschka
H and Löscher W: Valproic acid is not a substrate for
P-glycoprotein or multidrug resistance proteins 1 and 2 in a number
of in vitro and in vivo transport assays. J Pharmacol Exp Ther.
320:331–343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moerman L, Wyffels L, Slaets D, Raedt R,
Boon P and De Vos F: Antiepileptic drugs modulate P-glycoproteins
in the brain: A mice study with (11)C-desmethylloperamide. Epilepsy
Res. 94:18–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Römermann K, Helmer R and Löscher W: The
antiepileptic drug lamotrigine is a substrate of mouse and human
breast cancer resistance protein (ABCG2). Neuropharmacology.
93:7–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou L, Cao Y, Long H, Long L, Xu L, Liu
Z, Zhang Y and Xiao B: ABCB1, ABCC2, SCN1A, SCN2A, GABRA1 gene
polymorphisms and drug resistant epilepsy in the Chinese Han
population. Pharmazie. 70:416–420. 2015.PubMed/NCBI
|
|
16
|
Balan S, Sathyan S, Radha SK, Joseph V,
Radhakrishnan K and Banerjee M: GABRG2, rs211037 is associated with
epilepsy susceptibility, but not with antiepileptic drug resistance
and febrile seizures. Pharmacogenet Genomics. 23:605–610. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh E, Pillai KK and Mehndiratta M:
Characterization of a lamotrigine-resistant kindled model of
epilepsy in mice: Evaluation of drug resistance mechanisms. Basic
Clin Pharmacol Toxicol. 115:373–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Glauser TA, Holland K, O'Brien VP,
Keddache M, Martin L, Clark PO, Cnaan A, Dlugos D, Hirtz DG6
Shinnar S, et al: Pharmacogenetics of antiepileptic drug efficacy
in childhood absence epilepsy. Ann Neurol. 81:444–453. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv N, Qu J, Long H, Zhou L, Cao Y, Long L,
Liu Z and Xiao B: Association study between polymorphisms in the
CACNA1A, CACNA1C, and CACNA1H genes and drug-resistant epilepsy in
the Chinese Han population. Seizure. 30:64–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mou P, Chen Z, Jiang L, Cheng J and Wei R:
PTX3: A potential biomarker in thyroid associated ophthalmopathy.
Biomed Res Int. 2018:59619742018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waters WR, Maggioli MF, Palmer MV, Thacker
TC, McGill JL, Vordermeier HM, Berney-Meyer L, Jacobs WR Jr and
Larsen MH: Interleukin-17A as a Biomarker for bovine tuberculosis.
Clin Vaccine Immunol. 23:168–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Satoh J, Yamamoto Y, Asahina N, Kitano S
and Kino Y: RNA-Seq data mining: Downregulation of NeuroD6 serves
as a possible biomarker for alzheimer's disease brains. Dis
Markers. 2014:1231652014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pong WW, Walker J, Wylie T, Magrini V, Luo
J, Emnett RJ, Choi J, Cooper ML, Griffith M, Griffith OL, et al:
F11R is a novel monocyte prognostic biomarker for malignant glioma.
PLoS One. 8:e775712013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dixit AB, Banerjee J, Srivastava A,
Tripathi M, Sarkar C, Kakkar A, Jain M and Chandra PS: RNA-seq
analysis of hippocampal tissues reveals novel candidate genes for
drug refractory epilepsy in patients with MTLE-HS. Genomics.
107:178–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Griffin NG, Wang Y, Hulette CM, Halvorsen
M, Cronin KD, Walley NM, Haglund MM, Radtke RA, Skene JH, Sinha SR
and Heinzen EL: Differential gene expression in dentate granule
cells in mesial temporal lobe epilepsy with and without hippocampal
sclerosis. Epilepsia. 57:376–385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liew CC, Ma J, Tang HC, Zheng R and
Dempsey AA: The peripheral blood transcriptome dynamically reflects
system wide biology: A potential diagnostic tool. J Lab Clin Med.
147:126–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Borovecki F, Lovrecic L, Zhou J, Jeong H,
Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV and
Krainc D: Genome-wide expression profiling of human blood reveals
biomarkers for Huntington's disease. Proc Natl Acad Sci USA.
102:11023–11028. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jergil M, Forsberg M, Salter H, Stockling
K, Gustafson AL, Dencker L and Stigson M: Short-time gene
expression response to valproic acid and valproic acid analogs in
mouse embryonic stem cells. Toxicol Sci. 121:328–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dozawa M, Kono H, Sato Y, Ito Y, Tanaka H
and Ohshima T: Valproic acid, a histone deacetylase inhibitor,
regulates cell proliferation in the adult zebrafish optic tectum.
Dev Dyn. 243:1401–1415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Van Lint C, Emiliani S and Verdin E: The
expression of a small fraction of cellular genes is changed in
response to histone hyperacetylation. Gene Expr. 5:245–253.
1996.PubMed/NCBI
|
|
31
|
Rakitin A, Kõks S, Reimann E, Prans E and
Haldre S: Changes in the peripheral blood gene expression profile
induced by 3 months of valproate treatment in patients with newly
diagnosed epilepsy. Front Neurol. 6:1882015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Floriano-Sánchez E, Brindis F,
Ortega-Cuellar D, Ignacio-Mejía I, Moreno-Arriola E, Romero-Morelos
P, Ceballos-Vasquez E, Córdova-Espinoza MG, Arregoitia-Sarabia CK,
Sandoval-Pacheco R, et al: Differential gene expression profile
induced by valproic acid (VPA) in pediatric epileptic patients.
Genes (Basel). 9(pii): E3282018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwan P, Arzimanoglou A, Berg AT, Brodie
MJ, Allen Hauser W, Mathern G, Moshé SL, Perucca E, Wiebe S and
French J: Definition of drug resistant epilepsy: Consensus proposal
by the ad hoc task force of the ILAE commission on therapeutic
Strategies. Epilepsia. 51:1069–1077. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berg AT, Berkovic SF, Brodie MJ,
Buchhalter J, Cross JH, van Emde Boas W, Engel J, French J, Glauser
TA, Mathern GW, et al: Revised terminology and concepts for
organization of seizures and epilepsies: Report of the ILAE
commission on classification and terminology, 2005–2009. Epilepsia.
51:676–685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brahma R, Gurumayum S, Naorem LD,
Muthaiyan M, Gopal J and Venkatesan A: Identification of hub genes
and pathways in Zika Virus infection using RNA-Seq Data: A
network-based computational approach. Viral Immunol. 31:321–332.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kan AA, de Jager W, de Wit M, Heijnen C,
van Zuiden M, Ferrier C, van Rijen P, Gosselaar P, Hessel E, van
Nieuwenhuizen O and de Graan PN: Protein expression profiling of
inflammatory mediators in human temporal lobe epilepsy reveals
co-activation of multiple chemokines and cytokines. J
Neuroinflammation. 9:2072012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Louboutin JP, Chekmasova A, Marusich E,
Agrawal L and Strayer DS: Role of CCR5 and its ligands in the
control of vascular inflammation and leukocyte recruitment required
for acute excitotoxic seizure induction and neural damage. FASEB J.
25:737–753. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pocock JM and Liddle AC: Microglial
signalling cascades in neurodegenerative disease. Prog Brain Res.
132:555–565. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chui R and Dorovini-Zis K: Regulation of
CCL2 and CCL3 expression in human brain endothelial cells by
cytokines and lipopolysaccharide. J Neuroinflammation. 7:12010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
De Simoni MG, Perego C, Ravizza T, Moneta
D, Conti M, Marchesi F, De Luigi A, Garattini S and Vezzani A:
Inflammatory cytokines and related genes are induced in the rat
hippocampus by limbic status epilepticus. Eur J Neurosci.
12:2623–2633. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin W, Zhang J, Lou D, Chavkin C and Xu M:
C-fos-deficient mouse hippocampal CA3 pyramidal neurons exhibit
both enhanced basal and kainic acid-induced excitability. Neurosci
Lett. 331:151–154. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Zhang D, McQuade JS, Behbehani M,
Tsien JZ and Xu M: c-fos regulates neuronal excitability and
survival. Nat Genet. 30:416–420. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang Y, Glauser TA, Gilbert DL, Hershey
AD, Privitera MD, Ficker DM, Szaflarski JP and Sharp FR: Valproic
acid blood genomic expression patterns in children with epilepsy-a
pilot study. Acta Neurol Scand. 109:159–168. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Minnier J, Pennock ND, Guo Q, Schedin P
and abd Harrington CA: RNA-Seq and expression arrays: Selection
guidelines for genome-wide expression profiling. Methods Mol Bio.
1783:7–33. 2018. View Article : Google Scholar
|
|
47
|
Zhao S, Fung-Leung WP, Bittner A, Ngo K
and Liu X: Comparison of RNA-Seq and microarray in transcriptome
profiling of activated T cells. PLoS One. 9:e786442014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walker L and Sills GJ: Inflammation and
epilepsy: The foundations for a new therapeutic approach in
epilepsy? Epilepsy Curr. 12:8–12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guzik-Kornacka A, Sliwa A, Plucinska G and
Lukasiuk K: Status epilepticus evokes prolonged increase in the
expression of CCL3 and CCL4 mRNA and protein in the rat brain. Acta
Neurobiol Exp (Wars). 71:193–207. 2011.PubMed/NCBI
|
|
50
|
Owens GC, Huynh MN, Chang JW, McArthur DL,
Hickey MJ, Vinters HV, Mathern GW and Kruse CA: Differential
expression of interferon-γ and chemokine genes distinguishes
Rasmussen encephalitis from cortical dysplasia and provides
evidence for an early Th1 immune response. J Neuroinflammation.
10:562013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Arisi GM, Foresti ML, Katki K and Shapiro
LA: Increased CCL2, CCL3, CCL5, and IL-1β cytokine concentration in
piriform cortex, hippocampus, and neocortex after
pilocarpine-induced seizures. J Neuroinflammation. 12:1292015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Srivastava A, Dixit AB, Paul D, Tripathi
M, Sarkar C, Chandra PS and Banerjee J: Comparative analysis of
cytokine/chemokine regulatory networks in patients with hippocampal
sclerosis (HS) and focal cortical dysplasia (FCD). Sci Rep.
7:159042017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Saber S, Mahmoud AAA, Helal NS, El-Ahwany
E and Abdelghany RH: Renin-angiotensin system inhibition
ameliorates CCl4-induced liver fibrosis in mice through
the inactivation of nuclear transcription factor kappa B. Can J
Physiol Pharmacol. 96:569–576. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Guenther S, Bauer S, Hagge M, Knake S,
Olmes DG, Tackenberg B, Rosenow F and Hamer HM: Chronic valproate
or levetiracetam treatment does not influence cytokine levels in
humans. Seizure. 23:666–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ichiyama T, Okada K, Lipton JM, Matsubara
T, Hayashi T and Furukawa S: Sodium valproate inhibits production
of TNF-alpha and IL-6 and activation of NF-kappaB. Brain Res.
857:246–251. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ghosh C, Hossain M, Solanki J, Najm IM,
Marchi N and Janigro D: Overexpression of pregnane X and
glucocorticoid receptors and the regulation of cytochrome P450 in
human epileptic brain endothelial cells. Epilepsia. 58:576–585.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lee NY, Riechmann P and Kang YS: The
changes of P-glycoprotein activity by interferon-γ and tumor
necrosis factor-α in primary and immortalized human brain
microvascular endothelial cells. Blomol Ther (Seoul). 20:293–298.
2012. View Article : Google Scholar
|
|
58
|
Feng W, Mei S, Zhu L, Yu Y, Yang W, Gao B,
Wu X, Zhao Z and Feng F: Effects of UGT2B7, SCN1A and CYP3A4 on the
therapeutic response of sodium valproate treatment in children with
generalized seizures. Seizure. 58:96–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mantle JL and Lee KH: A differentiating
neural stem cell-derived astrocytic population mitigates the
inflammatory effects of TNF-α and IL-6 in an iPSC-based blood-brain
barrier model. Neurobiol Dis. 119:113–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Alluri H, Grimsley M, Anasooya Shaji C,
Varghese KP, Zhang SL, Peddaboina C, Robinson B, Beeram MR, Huang
JH and Tharakan B: Attenuation of blood-brain barrier breakdown and
hyperpermeability by calpain inhibition. J Biol Chem.
291:26958–26969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sakuma H, Tanuma N, Kuki I, Takahashi Y,
Shiomi M and Hayashi M: Intrathecal overproduction of
proinflammatory cytokines and chemokines in febrile
infection-related refractory status epilepticus. J Neurol Neurosurg
Psychiatry. 86:820–822. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kuijpers M, van Gassen KL, de Graan PN and
Gruol D: Chronic exposure to the chemokine CCL3 enhances neuronal
network activity in rat hippocampal cultures. J Neuroimmunol.
229:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Anaparti V, Pascoe CD, Jha A, Mahood TH,
Ilarraza 3, Unruh H, Moqbel R and Halayko AJ: Tumor necrosis factor
regulates NMDA receptor-mediated airway smooth muscle contractile
function and airway responsiveness. Am J Physiol Lung Cell Mol
Physiol. 311:L467–480. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zellinger C, Salvamoser JD, Soerensen J,
van Vliet EA, Aronica E, Gorter J and Potschka H: Pre-treatment
with the NMDA receptor glycine-binding site antagonist L-701,324
improves pharmacosensitivity in a mouse kindling model. Epilepsy
Res. 108:634–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hung CC, Ho JL, Chang WL, Tai JJ, Hsieh
TJ, Hsieh YW and Liou HH: Association of genetic variants in six
candidate genes with valproic acid therapy optimization.
Pharmacogenomics. 12:1107–1117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gautier NM and Glasscock E: Spontaneous
seizures in Kcna1-null mice lacking voltage-gated Kv1.1 channels
activate Fos expression in select limbic circuits. J Neurochem.
135:157–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Szyndler J, Maciejak P, Turzyńska D,
Sobolewska A, Taracha E, Skórzewska A, Lehner M, Bidziński A, Hamed
A, Wisłowska-Stanek A, et al: Mapping of c-Fos expression in the
rat brain during the evolution of pentylenetetrazol-kindled
seizures. Epilepsy Behav. 16:216–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cunningham JT, Mifflin SW, Gould GG and
Frazer A: Induction of c-Fos and DeltaFosB immunoreactivity in rat
brain by Vagal nerve stimulation. Neuropsychopharmacology.
33:1884–1895. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Deransart C Lê BT, Marescaux C and
Depaulis A: Role of the subthalamo-nigral input in the control of
amygdala-kindled seizures in the rat. Brain Res. 807:78–83. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Bozzi Y, Vallone D and Borrelli E:
Neuroprotective role of dopamine against hippocampal cell death. J
Neurosci. 20:8643–8649. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Madsen TM, Bolwig TG and Mikkelsen JD:
Differential regulation of c-Fos and FosB in the rat brain after
amygdala kindling. Cell Mol Neurobiol. 26:87–100. 2006. View Article : Google Scholar : PubMed/NCBI
|