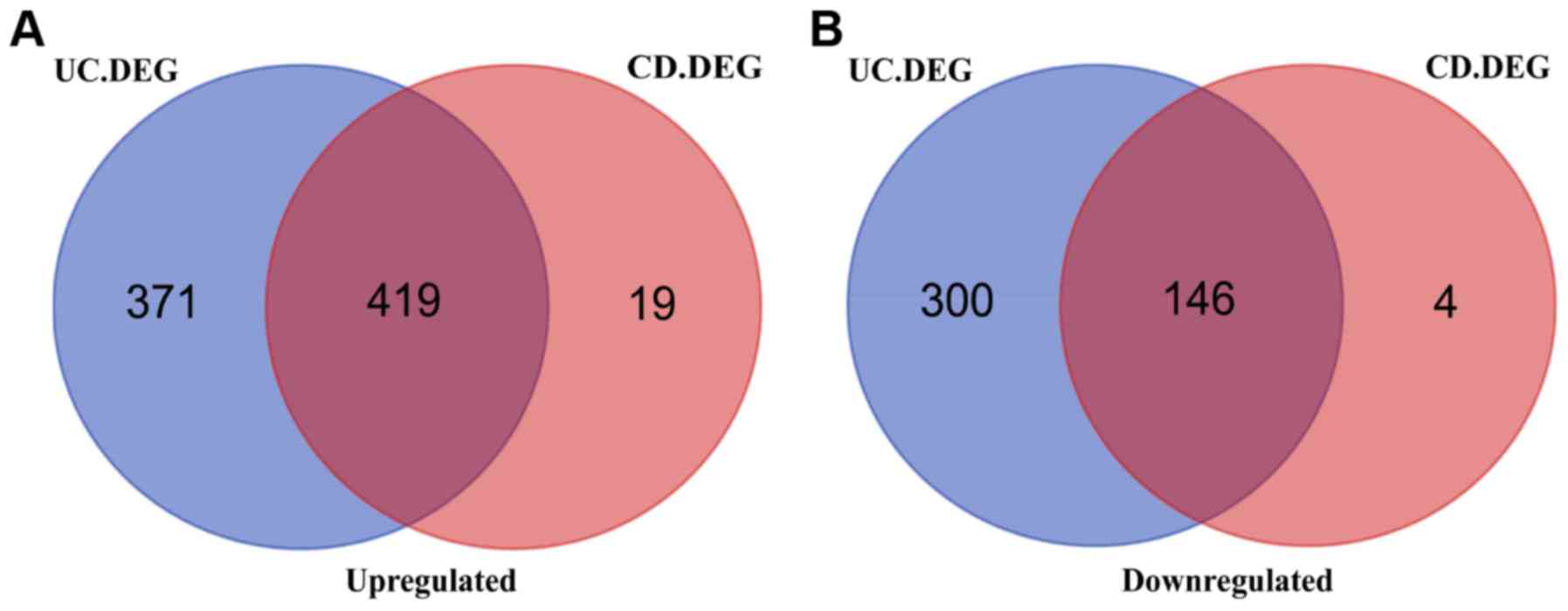
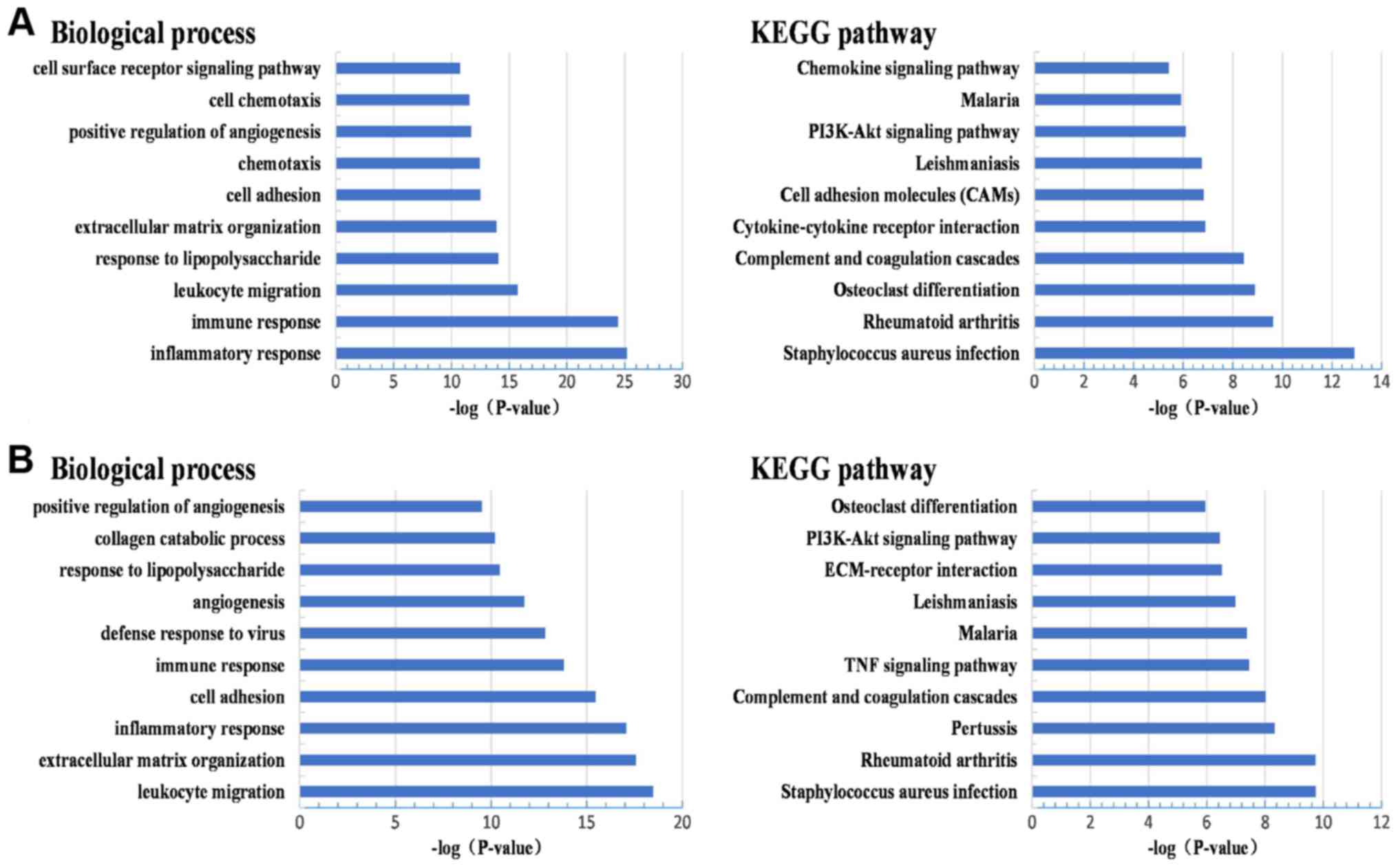
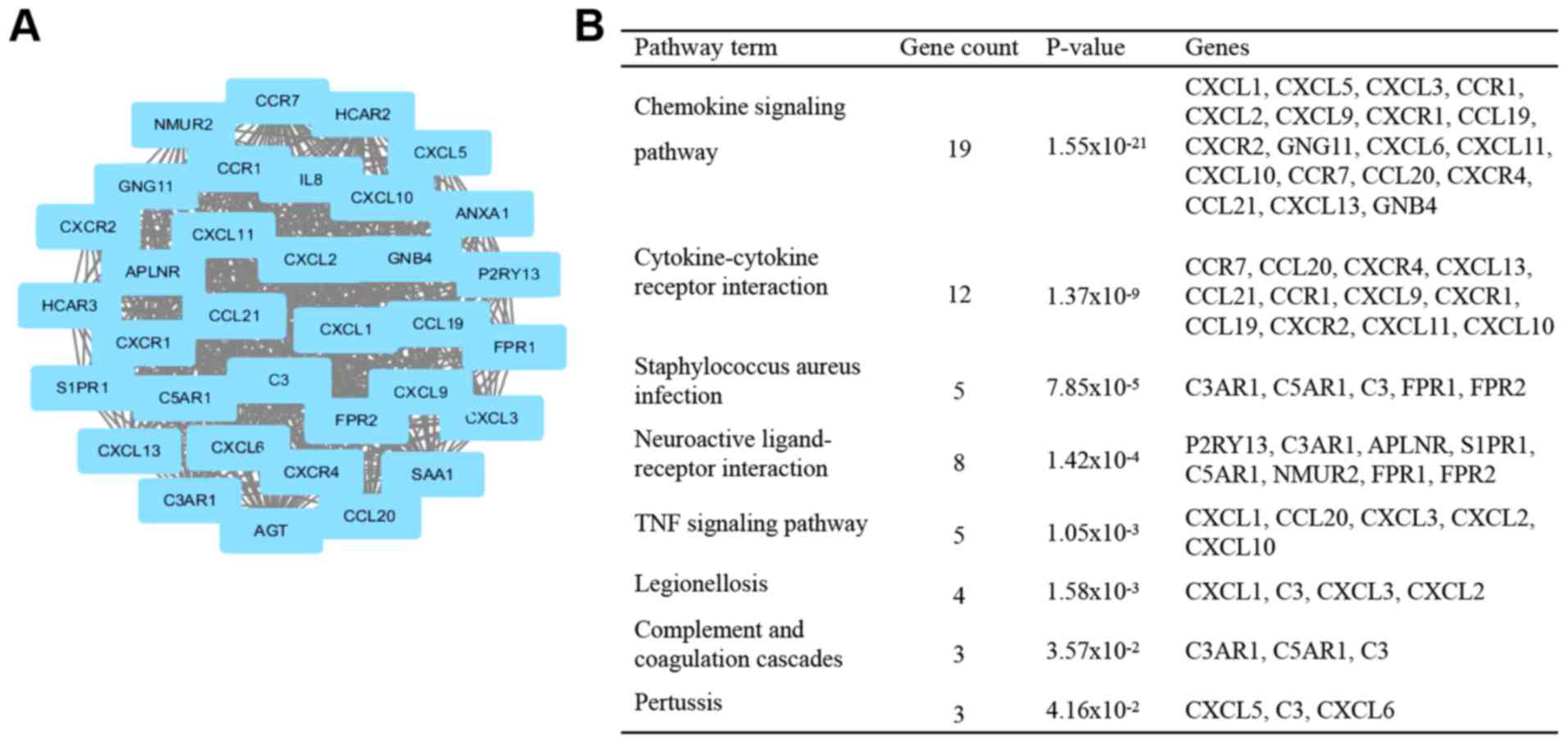
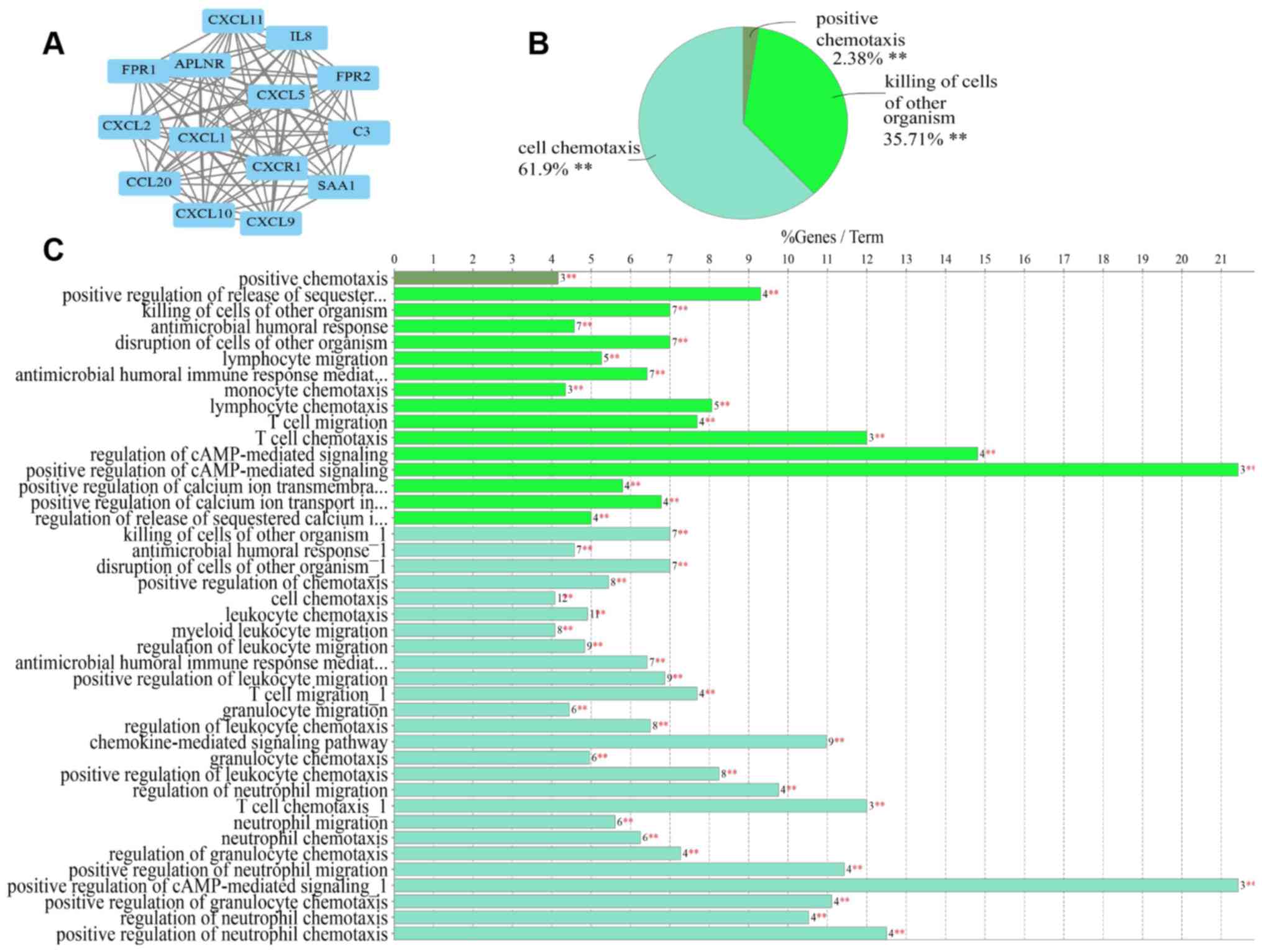
|
1
|
von Stein P, Lofberg R, Kuznetsov NV,
Gielen AW, Persson JO, Sundberg R, Hellstrom K, Eriksson A, Befrits
R, Ost A and von Stein OD: Multigene analysis can discriminate
between ulcerative colitis, Crohn's disease, and irritable bowel
syndrome. Gastroenterology. 134:1869–1881; quiz 2153–2164. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feuerstein JD and Cheifetz AS: Ulcerative
colitis: Epidemiology, diagnosis, and management. Mayo Clin Proc.
89:1553–1563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feuerstein JD and Cheifetz AS: Crohn
disease: Epidemiology, diagnosis, and management. Mayo Clin Proc.
92:1088–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaplan GG and Ng SC: Understanding and
preventing the global increase of inflammatory bowel disease.
Gastroenterology. 152:313–321.e2. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ng SC, Tang W, Ching JY, Wong M, Chow CM,
Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, et al: Incidence and
phenotype of inflammatory bowel disease based on results from the
Asia-pacific Crohn's and colitis epidemiology study.
Gastroenterology. 145:158–165.e2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ng SC, Kaplan G, Banerjee R, Wei SC, Tang
W, Zeng Z, Chen MH, Yang H, Silva JD, Niriella MA, et al: 78
incidence and phenotype of inflammatory bowel disease from 13
countries in Asia-Pacific: Results from the Asia-Pacific Crohn's
and colitis epidemiologic study 2011–2013. Gastroenterology.
150:S212016. View Article : Google Scholar
|
|
7
|
Molodecky NA, Soon IS, Rabi DM, Ghali WA,
Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema
HW and Kaplan GG: Increasing incidence and prevalence of the
inflammatory bowel diseases with time, based on systematic review.
Gastroenterology. 142:46–54.e42; quiz e30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoivik ML, Bernklev T and Moum B: Need for
standardization in population-based quality of life studies: A
review of the current literature. Inflamm Bowel Dis. 16:525–536.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Serna E, Morales JM, Mata M,
Gonzalez-Darder J, San Miguel T, Gil-Benso R, Lopez-Gines C,
Cerda-Nicolas M and Monleon D: Gene expression profiles of
metabolic aggressiveness and tumor recurrence in benign meningioma.
PLoS One. 8:e672912013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dai J, Ma Y, Chu S, Le N, Cao J and Wang
Y: Identification of key genes and pathways in meningioma by
bioinformatics analysis. Oncol Lett. 15:8245–8252. 2018.PubMed/NCBI
|
|
12
|
Li XL, Zhou CY, Sun Y, Su ZY, Wang X, N
Jia E, Zhang Q, Jiang XF, Qi WQ and Xu Y: Bioinformatic analysis of
potential candidates for therapy of inflammatory bowel disease. Eur
Rev Med Pharmacol Sci. 19:4275–4284. 2015.PubMed/NCBI
|
|
13
|
Feng J, Gao Q, Liu Q, Wang F, Lin X, Zhao
Q, Liu J and Li J: Integrated strategy of differentially expressed
genes associated with ulcerative colitis. Mol Med Rep.
16:7479–7489. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujimoto K, Kinoshita M, Tanaka H, Okuzaki
D, Shimada Y, Kayama H, Okumura R, Furuta Y, Narazaki M, Tamura A,
et al: Regulation of intestinal homeostasis by the ulcerative
colitis-associated gene RNF186. Mucosal Immunol. 10:446–459. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van der Goten J, Vanhove W, Lemaire K, Van
Lommel L, Machiels K, Wollants WJ, De Preter V, De Hertogh G,
Ferrante M, Van Assche G, et al: Integrated miRNA and mRNA
expression profiling in inflamed colon of patients with ulcerative
colitis. PLoS One. 9:e1161172014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
UK IBD Genetics Consortium, Barrett JC,
Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E,
Parnell K, Zhang H, et al: Genome-wide association study of
ulcerative colitis identifies three new susceptibility loci,
including the HNF4A region. Nat Genet. 41:1330–1334. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jostins L, Ripke S, Weersma RK, Duerr RH,
McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et
al: Host-microbe interactions have shaped the genetic architecture
of inflammatory bowel disease. Nature. 491:119–124. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vancamelbeke M, Vanuytsel T, Farré R,
Verstockt S, Ferrante M, Van Assche G, Rutgeerts P, Schuit F,
Vermeire S, Arijs I and Cleynen I: Genetic and transcriptomic bases
of intestinal epithelial barrier dysfunction in inflammatory bowel
disease. Inflamm Bowel Dis. 23:1718–1729. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumgart DC and Carding SR: Inflammatory
bowel disease: Cause and immunobiology. Lancet. 369:1627–1640.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Albertson DG and Pinkel D: Genomic
microarrays in human genetic disease and cancer. Hum Mol Genet 12
Spec No. 2:R145–R152. 2003. View Article : Google Scholar
|
|
21
|
Forini F, Nicolini G, Kusmic C, D'Aurizio
R, Rizzo M, Baumgart M, Groth M, Doccini S, Iervasi G and Pitto L:
Integrative analysis of differentially expressed genes and miRNAs
predicts complex T3-mediated protective circuits in a rat model of
cardiac ischemia reperfusion. Sci Rep. 8:138702018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cecchini MJ, Hosein K, Howlett CJ, Joseph
M and Mura M: Comprehensive gene expression profiling identifies
distinct and overlapping transcriptional profiles in non-specific
interstitial pneumonia and idiopathic pulmonary fibrosis. Respir
Res. 19:1532018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tatiya-Aphiradee N, Chatuphonprasert W and
Jarukamjorn K: Immune response and inflammatory pathway of
ulcerative colitis. J Basic Clin Physiol Pharmacol. 30:1–10. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Strober W and Fuss IJ: Proinflammatory
cytokines in the pathogenesis of inflammatory bowel diseases.
Gastroenterology. 140:1756–1767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng X, Szabo S, Khomenko T, Tolstanova G,
Paunovic B, French SW and Sandor Z: Novel pharmacologic approaches
to the prevention and treatment of ulcerative colitis. Curr Pharm
Des. 19:17–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mateescu RB, Bastian AE, Nichita L,
Marinescu M, Rouhani F, Voiosu AM, Benguş A, Tudoraşcu DR and Popp
CG: Vascular endothelial growth factor-key mediator of angiogenesis
and promising therapeutical target in ulcerative colitis. Rom J
Morphol Embryol. 58:1339–1345. 2017.PubMed/NCBI
|
|
27
|
Danese S, Sans M, de la Motte C, Graziani
C, West G, Phillips MH, Pola R, Rutella S, Willis J, Gasbarrini A
and Fiocchi C: Angiogenesis as a novel component of inflammatory
bowel disease pathogenesis. Gastroenterology. 130:2060–2073. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu G and Agarwal P: Human disease-drug
network based on genomic expression profiles. PLoS One.
4:e65362009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bernstein CN, Blanchard JF, Rawsthorne P
and Yu N: The prevalence of extraintestinal diseases in
inflammatory bowel disease: A population-based study. Am J
Gastroenterol. 96:1116–1122. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Veloso FT: Extraintestinal manifestations
of inflammatory bowel disease: Do they influence treatment and
outcome? World J Gastroenterol. 17:2702–2707. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kappelman MD, Galanko JA, Porter CQ and
Sandler RS: Association of paediatric inflammatory bowel disease
with other immune-mediated diseases. Arch Dis Child. 96:1042–1046.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dinca M, Fries W, Luisetto G, Peccolo F,
Bottega F, Leone L, Naccarato R and Martin A: Evolution of
osteopenia in inflammatory bowel disease. Am J Gastroenterol.
94:1292–1297. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schulte C, Dignass AU, Mann K and Goebell
H: Reduced bone mineral density and unbalanced bone metabolism in
patients with inflammatory bowel disease. Inflamm Bowel Dis.
4:268–275. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ciucci T, Ibáñez L, Boucoiran A,
Birgy-Barelli E, Pène J, Abou-Ezzi G, Arab N, Rouleau M, Hébuterne
X, Yssel H, et al: Bone marrow Th17 TNFα cells induce osteoclast
differentiation, and link bone destruction to IBD. Gut.
64:1072–1081. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu XF, Xu R, Ouyang ZJ, Qian C, Shen Y, Wu
XD, Gu YH, Xu Q and Sun Y: Beauvericin ameliorates experimental
colitis by inhibiting activated T cells via downregulation of the
PI3K/Akt signaling pathway. PLoS One. 8:e830132013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Q, Duan X, Fan H, Xu M, Tang Q, Zhang
L, Shou Z, Liu X, Zuo D, Yang J, et al: Oxymatrine protects against
DSS-induced colitis via inhibiting the PI3K/AKT signaling pathway.
Int Immunopharmacol. 53:149–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei J and Feng J: Signaling pathways
associated with inflammatory bowel disease. Recent Pat Inflamm
Allergy Drug Discov. 4:105–117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Latella G, Di Gregorio J, Flati V, Rieder
F and Lawrance IC: Mechanisms of initiation and progression of
intestinal fibrosis in IBD. Scand J Gastroenterol. 50:53–65. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Latella G, Sferra R, Speca S, Vetuschi A
and Gaudio E: Can we prevent, reduce or reverse intestinal fibrosis
in IBD? Eur Rev Med Pharmacol Sci. 17:1283–1304. 2013.PubMed/NCBI
|
|
41
|
Rieder F and Fiocchi C: Intestinal
fibrosis in IBD-a dynamic, multifactorial process. Nat Rev
Gastroenterol Hepatol. 6:228–235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Saddala MS, Lennikov A, Grab DJ, Liu GS,
Tang S and Huang H: Proteomics reveals ablation of PlGF increases
antioxidant and neuroprotective proteins in the diabetic mouse
retina. Sci Rep. 8:167282018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Benjafield AV, Lin RC, Dalziel B, Gosby
AK, Caterson ID and Morris BJ: G-protein beta3 subunit gene splice
variant in obesity and overweight. Int J Obes Relat Metab Disord.
25:777–780. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Principi M, Iannone A, Losurdo G, Mangia
M, Shahini E, Albano F, Rizzi SF, La Fortezza RF, Lovero R,
Contaldo A, et al: Nonalcoholic fatty liver disease in inflammatory
bowel disease: Prevalence and risk factors. Inflamm Bowel Dis.
24:1589–1596. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sartini A, Gitto S, Bianchini M, Verga MC,
Di Girolamo M, Bertani A, Del Buono M, Schepis F, Lei B, De Maria N
and Villa E: Non-alcoholic fatty liver disease phenotypes in
patients with inflammatory bowel disease. Cell Death Dis. 9:872018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bessissow T, Le NH, Rollet K, Afif W,
Bitton A and Sebastiani G: Incidence and predictors of nonalcoholic
fatty liver disease by serum biomarkers in patients with
inflammatory bowel disease. Inflamm Bowel Dis. 22:1937–1944. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Balcueva EA, Wang Q, Hughes H, Kunsch C,
Yu Z and Robishaw JD: Human G protein gamma(11) and gamma(14)
subtypes define a new functional subclass. Exp Cell Res.
257:310–319. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hossain MN, Sakemura R, Fujii M and
Ayusawa D: G-protein gamma subunit GNG11 strongly regulates
cellular senescence. Biochem Biophys Res Commun. 351:645–650. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fang F, Pan J, Xu L, Li G and Wang J:
Identification of potential transcriptomic markers in developing
ankylosing spondylitis: A meta-analysis of gene expression
profiles. Biomed Res Int. 2015:8263162015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Takauji Y, Kudo I, En A, Matsuo R, Hossain
MN, Nakabayashi K, Miki K, Fujii M and Ayusawa D: GNG11 (G-protein
subunit γ 11) suppresses cell growth with induction of reactive
oxygen species and abnormal nuclear morphology in human SUSM-1
cells. Biochem Cell Biol. 95:517–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Farcas A, Gligor F, Bucșa C, Mogoșan C,
Bojiță M and Dumitrașcu D: The current insight on dual
renin-angiotensin system blockade: A data review with a focus on
safety. Farmacia. 63:325–333. 2015.
|
|
52
|
Raizada V, Skipper B, Luo W and Griffith
J: Intracardiac and intrarenal renin-angiotensin systems:
Mechanisms of cardiovascular and renal effects. J Investig Med.
55:341–359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shen L, Zhang T and Lu H: Overexpression
of AGT promotes bronchopulmonary dysplasis via the JAK/STAT signal
pathway. Oncotarget. 8:96079–96088. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dalli J, Norling LV, Renshaw D, Cooper D,
Leung KY and Perretti M: Annexin 1 mediates the rapid
anti-inflammatory effects of neutrophil-derived microparticles.
Blood. 112:2512–2519. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sena AA, Pedrotti LP, Barrios BE, Cejas H,
Balderramo D, Diller A and Correa SG: Lack of TNFRI signaling
enhances annexin A1 biological activity in intestinal inflammation.
Biochem Pharmacol. 98:422–431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Babbin BA, Lee WY, Parkos CA, Winfree LM,
Akyildiz A, Perretti M and Nusrat A: Annexin I regulates SKCO-15
cell invasion by signaling through formyl peptide receptors. J Biol
Chem. 281:19588–19599. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang XD, Corvalan JR, Wang P, Roy CM and
Davis CG: Fully human anti-interleukin-8 monoclonal antibodies:
Potential therapeutics for the treatment of inflammatory disease
states. J Leukoc Biol. 66:401–410. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Skov L, Beurskens FJ, Zachariae CO,
Reitamo S, Teeling J, Satijn D, Knudsen KM, Boot EP, Hudson D,
Baadsgaard O, et al: IL-8 as antibody therapeutic target in
inflammatory diseases: Reduction of clinical activity in
palmoplantar pustulosis. J Immunol. 181:669–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mitsuyama K, Toyonaga A, Sasaki E,
Watanabe K, Tateishi H, Nishiyama T, Saiki T, Ikeda H, Tsuruta O
and Tanikawa K: IL-8 as an important chemoattractant for
neutrophils in ulcerative colitis and Crohn's disease. Clin Exp
Immunol. 96:432–436. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Luther J, Dave M, Higgins PD and Kao JY:
Association between Helicobacter pylori infection and
inflammatory bowel disease: A meta-analysis and systematic review
of the literature. Inflamm Bowel Dis. 16:1077–1084. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rokkas T, Gisbert JP, Niv Y and O'Morain
C: The association between Helicobacter pylori infection and
inflammatory bowel disease based on meta-analysis. United European
Gastroenterol J. 3:539–550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu XW, Ji HZ, Yang MF, Wu L and Wang FY:
Helicobacter pylori infection and inflammatory bowel disease
in Asians: A meta-analysis. World J Gastroenterol. 21:4750–4756.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Varga MG, Piazuelo MB, Romero-Gallo J,
Delgado AG, Suarez G, Whitaker ME, Krishna US, Patel RV, Skaar EP,
Wilson KT, et al: TLR9 activation suppresses inflammation in
response to Helicobacter pylori infection. Am J Physiol
Gastrointest Liver Physiol. 311:G852–G858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Owyang SY, Luther J, Owyang CC, Zhang M
and Kao JY: Helicobacter pylori DNA's anti-inflammatory
effect on experimental colitis. Gut microbes. 3:168–171. 2012.
View Article : Google Scholar : PubMed/NCBI
|