Introduction
Inflammatory bowel diseases (IBDs) are chronic
idiopathic inflammatory disorders of the gastrointestinal tract
with two distinct phenotypes: Ulcerative colitis (UC) and Crohn's
disease (CD) (1,2). UC is characterized by relapsing and
remitting mucosal inflammation extending from the rectum to
proximal segments of the colon (2).
By contrast, CD may affect the entire digestive tract and cause
transmural inflammation (3). The
prevalence of IBDs is increasing steadily in Western countries, and
developing countries have had a rapidly increasing incidence in
recent years (4,5). For instance, the incidence of IBDs in
China has reached 3.3 per 100,000 and exhibits a further increasing
trend (6). Furthermore, IBD
frequently progresses into a long-term health condition with a
variety of complications and reduces patients' health-associated
quality of life (7,8). Therefore, it is essential to expand the
current understanding of the pathogenesis of IBDs to develop
effective drugs and therapies for its treatment.
Despite great advances toward the understanding of
the pathophysiology of IBDs, early diagnosis, therapeutic
intervention and the underlying molecular mechanisms of IBD remain
a challenge. Furthermore, in spite of certain differences, UC and
CD share overlapping genetic and clinical features, making it
challenging to diagnose and distinguish between them. Therefore,
elucidation of the unique characteristics of UC or CD is also vital
for the development of more precise diagnostic and effective
therapeutic strategies to improve patient outcome.
Recently, with the continuous development of
bioinformatics and molecular biology, microarray technology has
been widely used for exploring the molecular mechanisms of various
diseases (9–11). In previous decades, analysis of the
expression profiles of gene microarrays was used to identify
several key genes and diagnostic biomarkers of IBDs, including
several differentially expressed genes (DEGs) involved in different
pathways, biological processes or molecular functions (12,13).
Furthermore, several key genes and candidate biomarkers of IBDs,
including Ring finger protein 186, Cadherin 11, Intercellular
adhesion molecule 1 (ICAM1) and Hepatocyte nuclear factor 4 alpha,
have been identified by bioinformatics analyses (14–17).
However, previous studies have mainly focused on identifying
candidate genes for UC or CD, while the potential intrinsic links
among DEGs have not been extensively investigated.
In the present study, a gene expression dataset
(GSE75214) was downloaded from the Gene Expression Omnibus (GEO)
database. The DEGs were identified in UC patients and CD patients
compared with controls, respectively. Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analyses performed in the
DAVID database were applied to determine the functional enrichment
and significant pathways associated with the DEGs. In addition, a
protein-protein interaction (PPI) network of common DEGs was
constructed to identify the critical genes and significant modules.
By means of analyzing of the biological functions and pathways
shared between and unique to UC and CD, the present study provided
further insight into the pathogenesis of UC and CD at the molecular
level, which may facilitate the diagnosis and provide potential
molecular targets for UC and CD.
Materials and methods
Microarray data
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) stores original
submitter-supplied records as well as curated datasets. The gene
expression dataset GSE75214 was obtained from the GEO database.
This dataset had been generated using the platform GPL6244
{(HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array [transcript
(gene) version]} and was deposited by Vancamelbeke et al
(18) in the GEO database. A total
of 194 samples are included in this dataset, comprising colon
tissues from 97 patients with UC (74 with active disease and 23
with inactive disease), 34 patients with CD (8 with active colonic
disease and 26 with inactive disease), and 11 controls, as well as
the (neo-) terminal ileum of 67 CD patients and 11 controls. The
baseline characteristic data of the subjects was published
previously by Vancamelbeke et al (18). The present study focused on colonic
mucosal biopsies (74 active UC, 8 active CD patients and 11
controls) and excluded the ileal biopsies. Thus, a total of 93
chips were available for the subsequent analysis.
Identification of DEGs
Data pre-processing was performed using GEO2R
(https://www.ncbi.nlm.nih.gov/geo/geo2r/) and was
applied to screen DEGs between the following groups: UC vs.
Controls and CD vs. Controls. GEO2R is an interactive web tool that
compares two groups of samples under the same experimental
conditions and is able to analyze almost any GEO series (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The adjusted
P<0.01 and |log2 fold change (FC)| >1 (i.e., FC
>2) were selected as the threshold for each group.
GO and KEGG pathway enrichment
analyses
The Database for Annotation, Visualization and
Integrated Discovery (DAVID 6.8, http://david.ncifcrf.gov/summary.jsp) is an online
program that may be used to systematically extract biological
meaning from large lists of genes or proteins. In order to analyze
the screened DEGs at the functional level, GO enrichment and KEGG
pathway analysis was performed using the DAVID online tool.
P<0.05 was set as the cut-off criterion.
PPI network analysis
To further evaluate the functional interactions
among DEGs, a PPI network was used. The DEGs were mapped using the
Search Tool for the Retrieval of Interacting Genes (STRING; version
11.0; http://string-db.org/cgi/input.pl) and only the
interaction pairs with a PPI combined score of >0.7 were
selected as significant. Subsequently, the PPI network was
constructed using Cytoscape software (https://cytoscape.org/; version 3.6.1). The top 10
essential nodes ranked in the network of respective DEGs were
calculated using the Cytoscape plugin CytoHubba.
Module analysis
The plug-in Molecular Complex Detection (MCODE) was
used to identify the densely connected regions of the PPI network
in Cytoscape. MCODE was then applied to screen modules of the PPI
network with the following parameters: Degree cutoff, 2; node score
cutoff, 0.2; k-core, 2; and maximum depth, 100. In addition, the
functions and pathways of the DEGs in each module were assessed
using ClueGo, with P<0.05 being considered to indicate
significance.
Results
Identification of DEGs
The gene expression dataset GSE75214 was downloaded
from the GEO database. DEGs between the disease samples and the
controls were determined using the GEO2R tool. As presented in
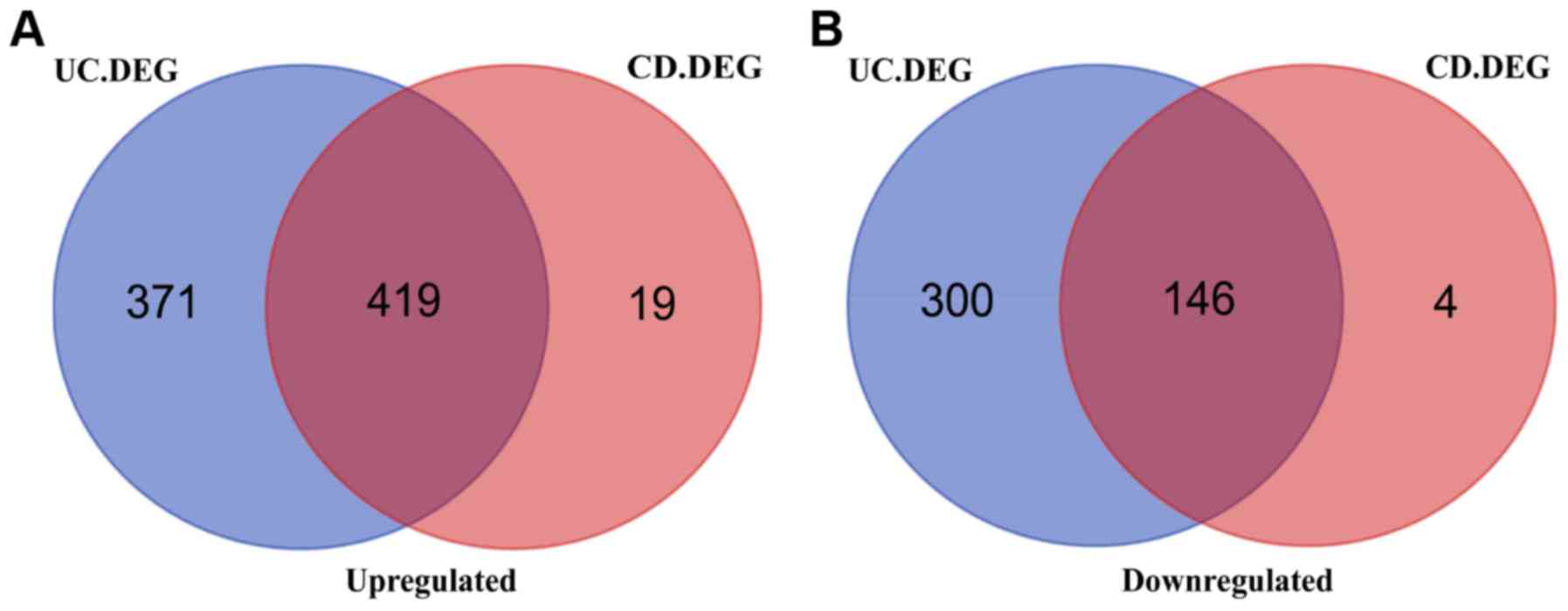
Fig. 1, a total of 1,236 DEGs were
identified in the UC group using the threshold of P<0.05 and
|log2FC| >1, including 790 upregulated genes and 446
downregulated genes in the UC vs. control samples. The top 10 up-
and downregulated genes for UC vs. the control are listed in
Table I.
 | Table I.Top 10 up- and downregulated
differentially expressed genes in ulcerative colitis vs.
control. |
Table I.
Top 10 up- and downregulated
differentially expressed genes in ulcerative colitis vs.
control.
| Gene symbol | Log
fold-change | Adjusted
P-value |
|---|
| Upregulated |
|
|
|
SLC6A14 | 5.72 |
3.96×10−23 |
|
MMP3 | 4.80 |
1.09×10−12 |
|
REG1A | 4.63 |
1.61×10−10 |
|
DUOX2 | 4.34 |
1.31×10−15 |
|
TINP3 | 4.32 |
2.15×10−16 |
|
DUOXA2 | 4.32 |
2.84×10−18 |
|
IDO1 | 4.23 |
2.77×10−16 |
|
S100A8 | 4.15 |
1.01×10−11 |
|
MMP1 | 4.14 |
5.77×10−11 |
|
REG1B | 4.08 |
4.31×10−7 |
| Downregulated |
|
|
|
AQP8 | −5.52 |
8.44×10−24 |
|
HMGCS2 | −4.76 |
5.65×10−17 |
|
SLC26A2 | −4.52 |
1.45×10−19 |
|
CLDN8 | −3.85 |
2.95×10−15 |
|
ABCG2 | −3.80 |
3.77×10−19 |
|
PCK1 | −3.71 |
6.10×10−15 |
|
TMIGD1 | −3.71 |
7.83×10−11 |
|
OTOP2 | −3.37 |
2.22×10−16 |
|
GUCA2A | −3.32 |
1.02×10−9 |
|
UGT2A3 | −3.24 |
2.04×10−26 |
Similarly, comparison of the CD group with the
healthy control samples identified 588 DEGs, including 438
upregulated and 150 downregulated genes. The top 10 DEGs for CD vs.
control samples are listed in Table
II. Of note, most of the DEGs identified in UC were also
differentially expressed in CD. A total of 565 overlapping DEGs
were identified between UC and CD vs. the controls (Fig. 1). Thus, a total of 671 and 23
specific DEGs remained for UC and CD, respectively (Fig. 1).
 | Table II.Top 10 up- and downregulated
differentially expressed genes in Crohn's disease vs. control. |
Table II.
Top 10 up- and downregulated
differentially expressed genes in Crohn's disease vs. control.
| Gene symbol | Log
fold-change | Adjusted
P-value |
|---|
| Upregulated |
|
|
|
SLC6A14 | 4.77 |
2.34×10−7 |
|
REG1A | 4.68 |
1.12×10−3 |
|
MMP3 | 4.64 |
1.59×10−4 |
|
REG1B | 4.29 |
1.28×10−3 |
|
MMP1 | 4.28 |
3.91×10−4 |
|
S100A8 | 4.24 |
1.29×10−5 |
|
IDO1 | 4.13 |
1.17×10−6 |
|
DUOX2 | 3.95 |
1.25×10−4 |
|
CHI3L1 | 3.73 |
5.93×10−6 |
|
REG3A | 3.63 |
3.53×10−3 |
| Downregulated |
|
|
|
AQP8 | −3.47 |
6.29×10−4 |
|
ABCG2 | −3.05 |
4.30×10−5 |
|
CYP2B7P | −2.86 |
2.66×10−5 |
|
HMGCS2 | −2.69 |
1.36Ex10−3 |
|
SLC30A10 | −2.51 |
2.34×10−4 |
|
PRKG2 | −2.48 |
2.52×10−4 |
|
CYP2B6 | −2.44 |
1.33×10−3 |
|
SLC16A9 | −2.40 |
1.03×10−4 |
|
ABCB1 | −2.28 |
5.02×10−5 |
|
APOBEC3B | −2.26 |
5.99×10−6 |
GO and pathway enrichment analysis of
DEGs
To further perform a systematic characterization and
explore the biological functions of the identified DEGs in UC and
CD vs. control samples, functional annotation and pathway analyses,
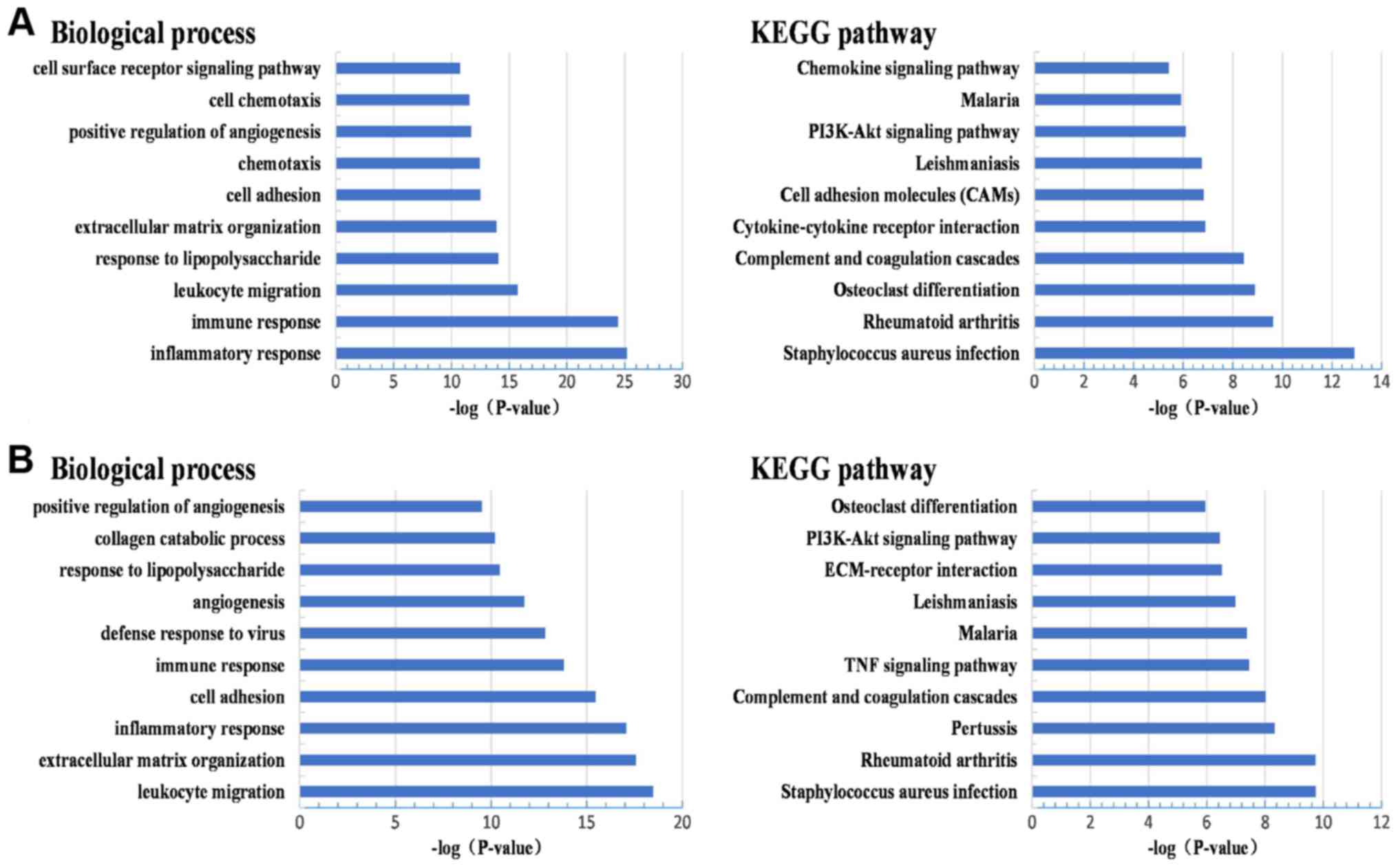
including GO and KEGG analyses, were performed using DAVID. The top
10 GO terms in the category biological process are presented in
Fig. 2. The results of the GO
analysis indicated that most of the DEGs in the UC and CD groups
were simultaneously enriched in inflammatory response, immune
response, leukocyte migration, response to lipopolysaccharides,
extracellular matrix organization, cell adhesion and positive
regulation of angiogenesis (Fig.
2A). In addition, several DEGs in UC were enriched in
chemotaxis, cell chemotaxis and cell surface receptor signaling
pathways (Fig. 2A). Similarly,
certain DEGs in the CD group were also closely associated with
defense response to viruses and collagen catabolic process
(Fig. 2B).
Subsequently, KEGG pathway analysis indicated that
the DEGs in UC and CD were primarily enriched in Staphylococcus
aureus infection, rheumatoid arthritis, complement and
coagulation cascades, malaria, leishmaniasis, the PI3K/Akt
signaling pathway and osteoclast differentiation (Fig. 2A and B). In addition, certain KEGG
pathways, including cytokine-cytokine receptor interaction, cell
adhesion molecules and chemokine signaling pathway, were commonly
involved in the development of UC, whereas certain other pathways
[extracellular matrix (ECM)-receptor interaction, tumor necrosis
factor (TNF) signaling pathway and pertussis] were mainly involved
in the pathogenesis of CD (Fig. 2A and
B). Therefore, several GO terms in the category biological
process and KEGG pathways exhibited significant differences between
UC and CD, indicating that UC and CD have different pathological
characteristics.
Construction of PPI network and
identification of hub genes
To further explore the association between DEGs at
the protein level, the PPI networks were constructed based on the
interactions of DEGs. DEGs for UC and CD, as well as their
overlapping DEGs, were mapped to PPI networks and visualized by
Cytoscape software, respectively. With the pre-defined criterion of
a combined score of >0.7, the PPI network of the overlapping
DEGs consisted of 307 nodes and 1,205 edges. The top 10 hub nodes
with a high degree of interaction in this network are presented in
Table III. These hub genes were
interleukin Interleukin 6 (IL6), cluster of differentiation 44,
IL8, IL1B, ICAM1, Signal transducer and activator of transcription
1 (STAT1), Jun proto-oncogene, Transforming growth factor beta 1,
Janus kinase 2 (JAK2) and Matrix metallopeptidase 2 (MMP2), which
were identified as the candidate genes. Similarly, the PPI networks
for UC and CD individually were constructed, including 631 and 312
nodes, and 3,231 and 1,218 edges, respectively. Among these nodes,
the top 10 hub genes with the highest degree of interaction are
provided in Table III. Of note, G
protein subunit gamma 11 (GNG11), G protein subunit beta 4 (GNB4),
Angiotensinogen (AGT), Phosphoinositide-3-kinase regulatory subunit
3 (PIK3R3) and C-C motif chemokine receptor 7 (CCR7) were highly
expressed in UC but not in CD. These specific hub genes may be
regarded as candidate marker genes for UC. However, the hub genes
for CD overlapped with the DEGs for UC, and there were no specific
hub genes for CD. Most of the hub genes in CD were mainly
associated with the chemokine signaling pathway, leukocyte
migration and intestinal fibrosis-associated pathways. Several of
the candidate genes identified may be used as biomarkers for the
differential diagnosis of CD and UC. The hub genes, GNG11 and GNB4,
that were identified in UC tissues may therefore serve to
distinguish UC from CD. However, certain candidate genes, including
JAK2 and MMP2, that were identified in CD tissues may not
differentiate UC from CD.
 | Table III.Top ten genes with the highest degree
of interaction in the protein-protein interaction network using
respective DEGs for UC, CD and UC + CD vs. controls. |
Table III.
Top ten genes with the highest degree
of interaction in the protein-protein interaction network using
respective DEGs for UC, CD and UC + CD vs. controls.
| DEG | Degree of
interaction |
|---|
| UC |
|
|
IL6 | 88 |
|
IL8 | 67 |
|
GNB4 | 62 |
|
GNG11 | 62 |
|
AGT | 59 |
|
CXCR4 | 54 |
|
ANXA1 | 54 |
|
PIK3R3 | 53 |
|
SAA1 | 52 |
|
CCR7 | 51 |
| CD |
|
|
IL6 | 63 |
|
CD44 | 39 |
|
IL8 | 37 |
|
IL1B | 36 |
|
ICAM1 | 34 |
|
STAT1 | 33 |
|
TGFB1 | 30 |
|
JAK2 | 29 |
|
VCAM1 | 28 |
| C3 | 27 |
| UC and CD |
|
| IL6 | 63 |
|
CD44 | 39 |
|
IL8 | 38 |
|
IL1B | 36 |
|
ICAM1 | 34 |
|
STAT1 | 32 |
|
JUN | 31 |
|
TGFB1 | 31 |
|
JAK2 | 29 |
|
MMP2 | 28 |
Module analysis of the PPI
network
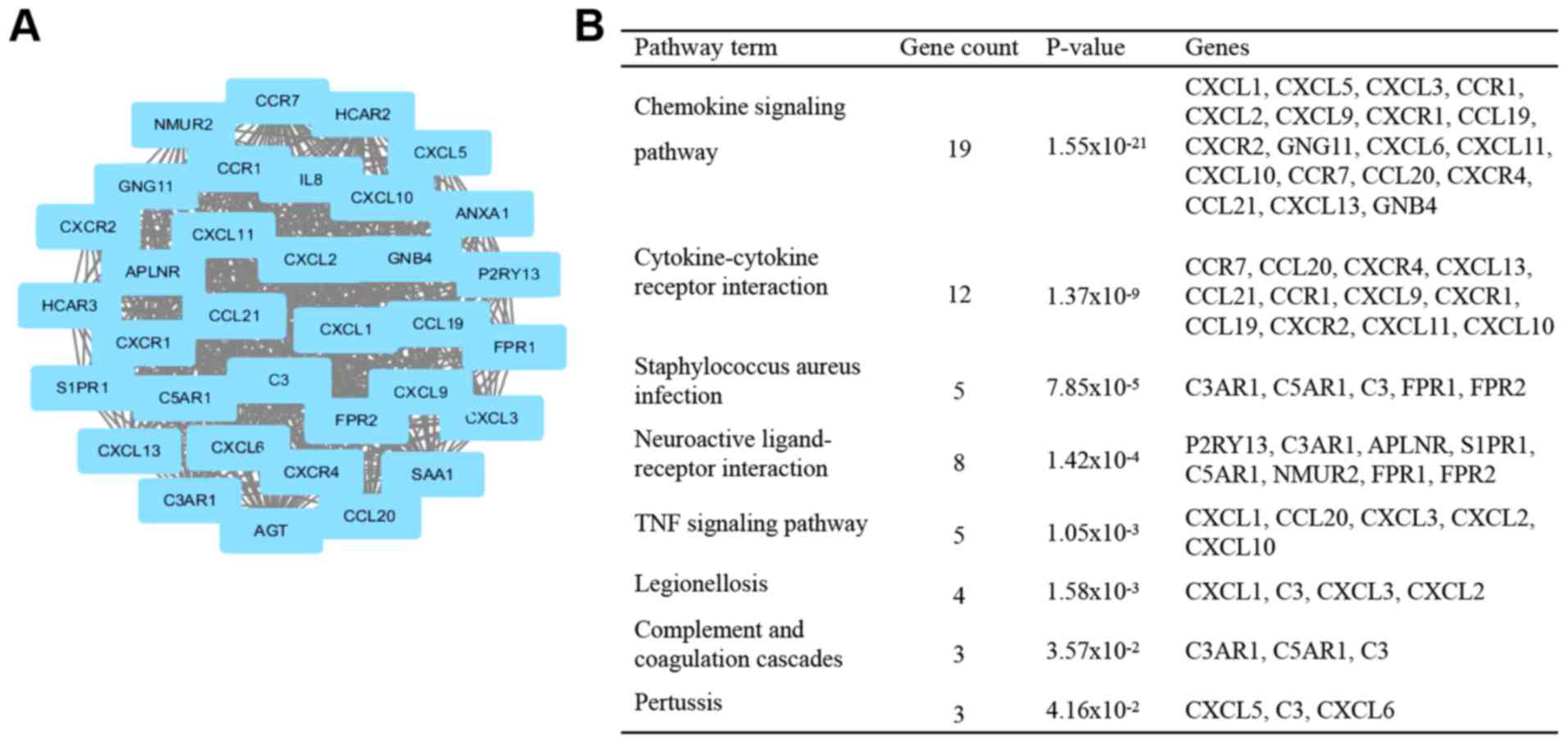
With the application of the MCODE plugin of
Cytoscape, the top modules of the PPI networks for UC and CD were
selected, and a functional enrichment analysis of DEGs in the
respective functional modules was (Figs.
3–5). KEGG pathway enrichment
analysis revealed that the DEGs in UC and CD in the modules were
primarily associated with the chemokine signaling pathway,
cytokine-cytokine receptor interaction, Staphylococcus
aureus infection, TNF signaling pathway and legionellosis
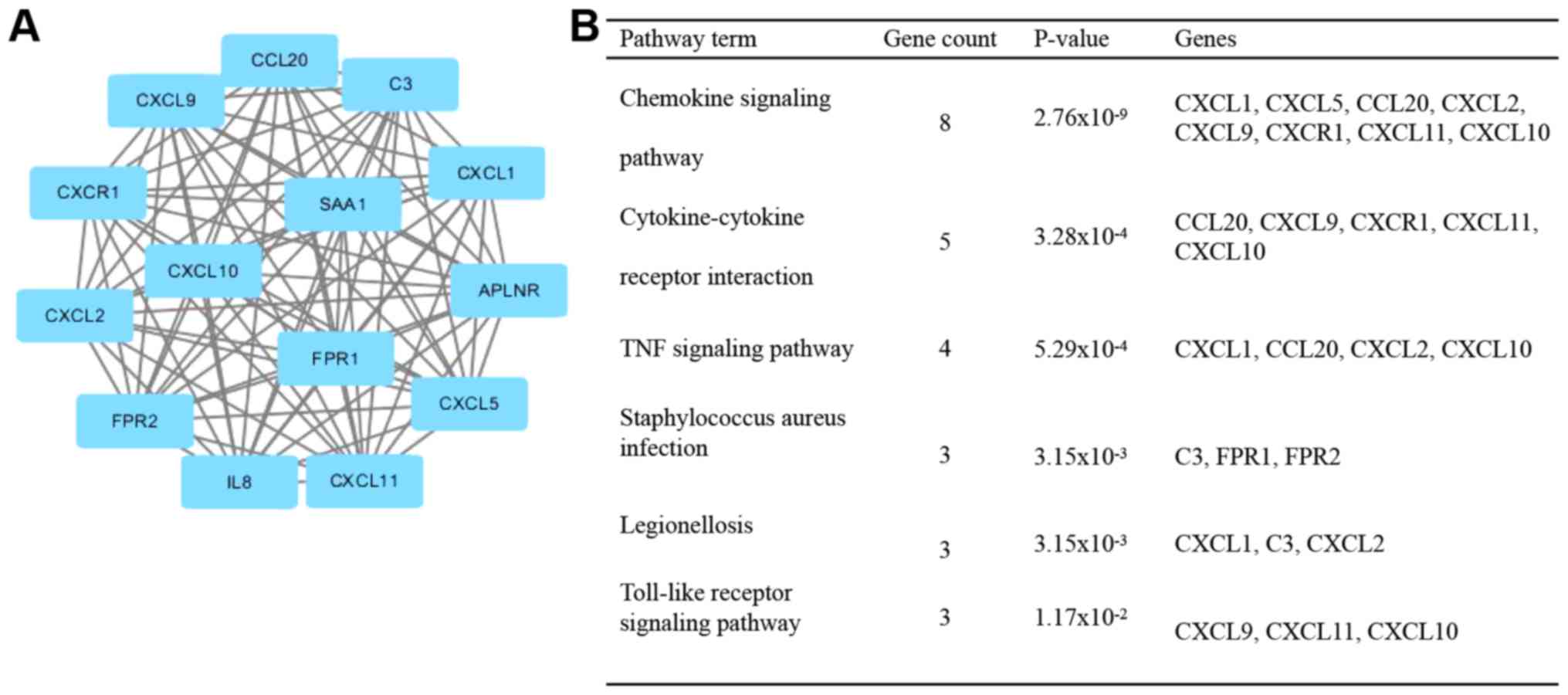
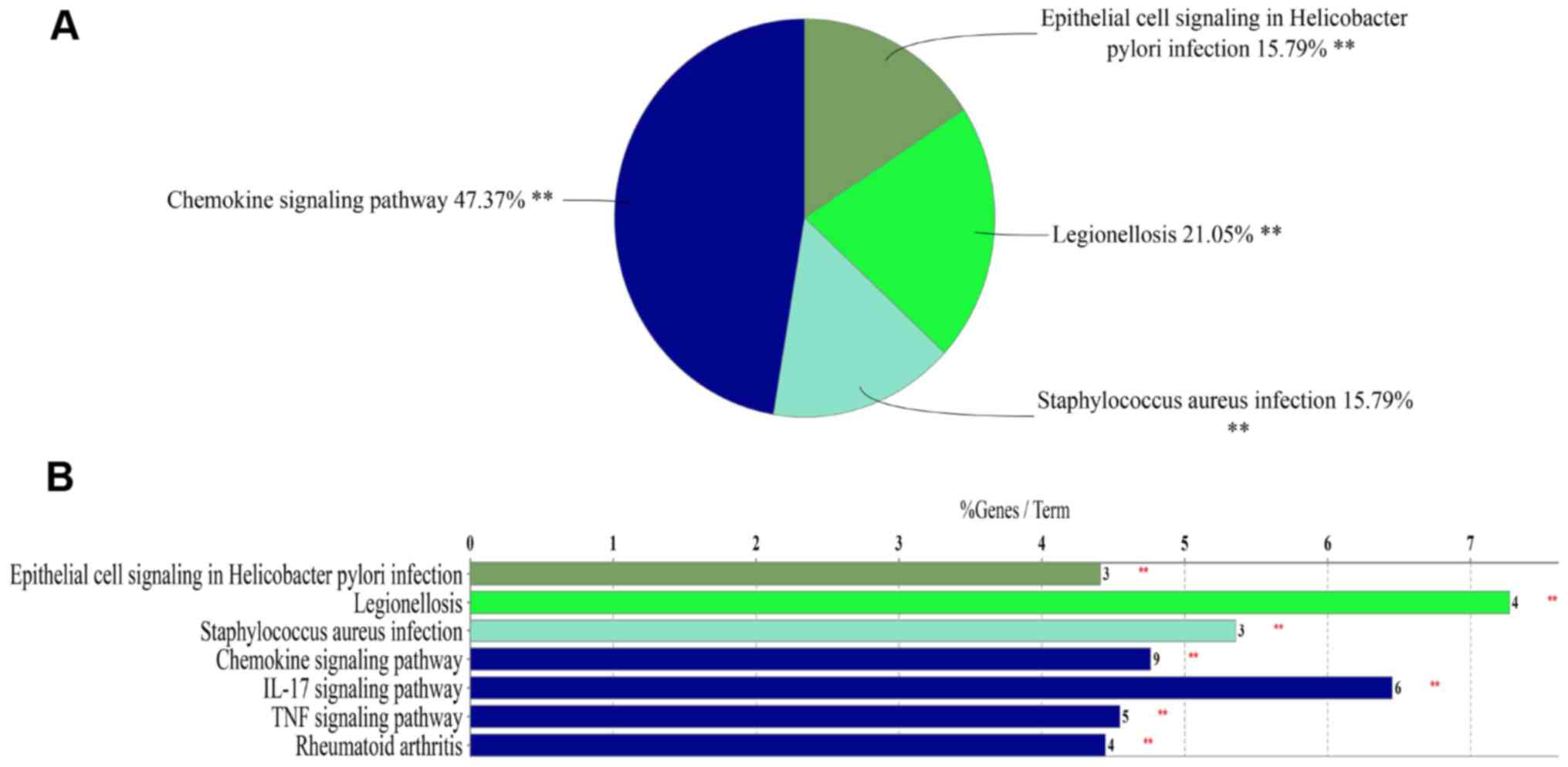
(Figs. 3 and 4).
While UC and CD are different diseases, they have
various pathological characteristics in common; therefore, the
biological functions and pathways shared between UC and CD were
then analyzed. A total of 565 overlapped DEGs were identified for
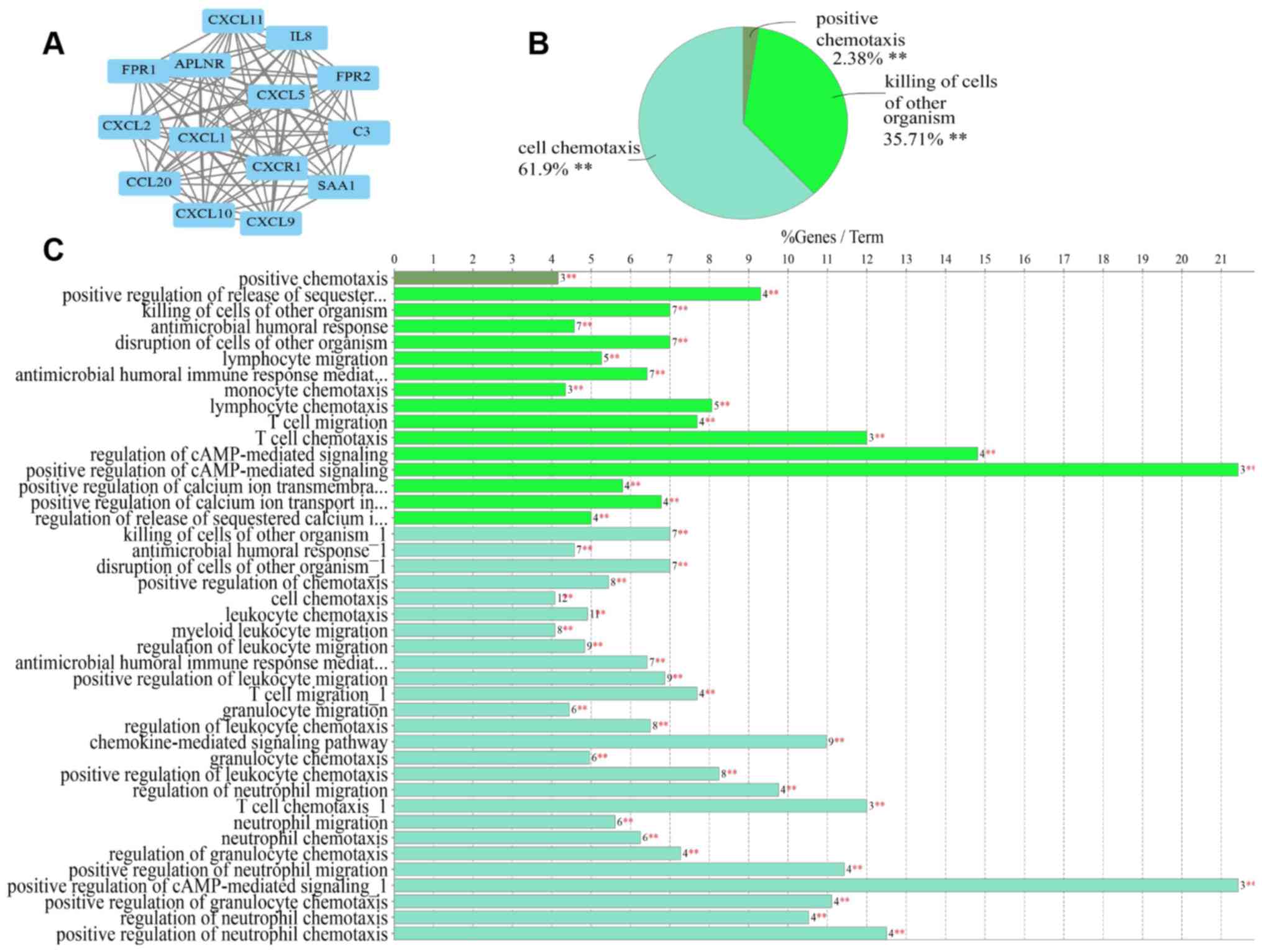
the UC and CD groups. Subsequently, one significant module was
obtained from the PPI network of the overlapped DEGs (Fig. 5). The results of the GO analysis
indicated that genes in the top module were primarily associated
with cell chemotaxis, killing of cells of other organisms and
positive chemotaxis (Fig. 5). KEGG
pathway enrichment analysis of the DEGs included in the top module
revealed that these genes were mainly associated with chemokine
signaling pathway, Staphylococcus aureus infection,
legionellosis and epithelial cell signaling in Helicobacter
pylori infection (Fig. 6).
Discussion
UC and CD are the two major types of inflammatory
bowel disease, whose etiology involves a complex interaction
between genetics, environmental factors, infectious microbes and
dysregulated immune responses (19).
Although research on IBDs has achieved great progress in the past
decade, the pathogenesis of IBDs has yet to be fully elucidated due
to the complex and poorly understood interactions among these
factors. Therefore, advancement of the understanding of the
molecular mechanisms of IBDs based on microarray technology, which
has developed rapidly and has been widely used to examine the
pathogenic processes of various diseases (20–22), may
provide potential diagnostic and therapeutic targets for UC and
CD.
In the present study, 1,236 DEGs were identified in
the UC group, including 790 upregulated and 446 downregulated
genes, and in the CD group, 588 DEGs, including 438 upregulated and
150 downregulated genes, were identified. The UC and CD groups had
565 DEGs in common, indicating that the two disorders have an
important and overlapping genetic component. GO analysis indicated
that the identified DEGs for UC and CD are accumulated in similar
functional terms in the category biological process, including
inflammatory response, immune response, leukocyte migration,
response to lipopolysaccharide, ECM organization and cell adhesion.
Previous studies have demonstrated that the aforementioned GO terms
are potentially important events in the pathogenesis of IBDs. For
instance, immuno-inflammatory responses, leukocyte migration, cell
chemotaxis and cell adhesion have roles in the pathogenesis of IBDs
(19,23,24). In
addition, accumulating evidence has demonstrated that angiogenesis
has a pathogenic role in IBDs (25–27).
Growth of new blood vessels is an important component of the
inflammatory process, and exacerbation of inflammation in a vicious
circle that aggravates mucosal damage and remodeling (26). Therefore, several significant DEGs of
the angiogenesis pathway may be used as therapeutic targets for
adjuvant therapy.
Furthermore, KEGG pathway analysis revealed that UC
and CD also shared the top enriched pathways, including
Staphylococcus aureus infection, rheumatoid arthritis,
complement and coagulation cascades, PI3K/Akt signaling pathway and
osteoclast differentiation. Previous studies have demonstrated that
infectious microbes have role in the development of IBDs. The
present study indicated that UC as well as CD were associated with
Staphylococcus aureus infection, malaria and leishmaniasis.
Hu and Agarwal (28) reported that
malaria drugs may be used to treat CD, indicating a potential
association between malaria and IBDs. In addition, a number of
extra-intestinal diseases have been associated with IBD. For
instance, patients with IBD were reported to have an increased
prevalence of rheumatoid arthritis (29–31).
Previous studies have also demonstrated that patients with IBDs
have an increased risk of developing osteopenia and osteoporosis,
which is a common complication of IBDs and associated with an
impaired quality of life (32,33).
Ciucci et al (34) reported
that CD4+ T-cell subsets contributed to the
differentiation of osteoclasts and maintained a vicious circle
linking inflammation and bone destruction. The PI3K/Akt pathway is
associated with cell cycle progression, cell death and cell growth
(35). Recent studies have reported
that the PI3K/Akt pathway is involved in the pathogenesis of IBDs
by exerting pro-inflammatory effects and activating T cells
(36–38). In the present study, several GO terms
in the category biological process and KEGG pathways exhibited
significant differences between UC and CD, including chemotaxis,
collagen catabolic process, cytokine-cytokine receptor interaction
and ECM-receptor interaction. Previous studies have demonstrated
that intestinal fibrosis with stricture formation is a common
outcome of IBDs and may occur in UC as well as in CD, but is more
prevalent in CD (39,40). Intestinal fibrosis is characterized
by abnormal deposition of ECM proteins and involves collagen
catabolic processes, ECM organization and ECM-receptor interaction
(41), which is consistent with the
results of the present study. Therefore, further investigating
these biological processes and pathways may aid in gaining a better
understanding of the molecular mechanisms of UC and CD.
The gene expression profile of GSE75214 analyzed in
the present study was submitted by Vancamelbeke et al
(18). However, the significant DEGs
in the colon of patients with UC and patients with CD vs. controls
and the enrichment analysis of DEGs in the present study were
inconsistent with the results of the original study. There may be
two reasons for these differences. One reason is that the original
study had a different aim, namely to take an in-depth view on the
genetic and transcriptomic bases of intestinal epithelial barrier
defects in IBD, so its major focus was to evaluate the intestinal
epithelial barrier, and the results were filtered from the
genome-wide comparative analyses for the gene probe sets
representing the selected barrier genes (18). By contrast, the aim of the present
study was to identify genetic signatures in patients with IBDs and
elucidate the potential associated molecular mechanisms underlying
IBD subtypes, so the microarray dataset was used to obtain general
genetic alterations in UC and CD patients vs. controls. The other
reason is that the research objectives were different. The original
study investigated the expression levels of barrier genes using
colonic or ileal mucosal tissue from patients with UC and CD
(active and inactive status) (18),
whereas the present study only evaluated genetic alterations in
colonic mucosa of patients with active IBD from the data of the
original study. The abovementioned points are likely to be
accountable for this difference.
Clinical discrimination between UC and CD has long
been a problem due to a substantial overlap not only in
pathological characteristics but also in the clinical symptoms.
Therefore, it is necessary to identify discriminative key genes
that may be used for diagnostics and screening of IBD subtypes.
Subsequently, the respective PPI networks for IBD subtypes were
constructed and the potential key genes involved in the
pathogenesis of UC or CD were identified. The top 10 hub genes of
the UC group included IL6, IL8, GNB4, GNG11, AGT, C-X-C motif
chemokine receptor 4 (CXCR4), annexin A1 (ANXA1),
phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3), serum
amyloid A1 and CCR7, which were mainly associated with inflammatory
response. Among these genes, GNG11, GNB4, AGT, PIK3R3 and CCR7 were
highly expressed in UC but not in CD. These hub genes were
disease-specific, and may be used as biomarkers for differential
diagnosis of UC and CD. Furthermore, the identified hub genes GNB4,
GNG11 and AGT have not been previously reported to be associated
with IBD. GNB4 is classified as a G protein that has a crucial role
in the insulin signaling pathway (42). Incessant activation of G proteins
(polymorphism) results in insulin resistance and ultimately
increases hepatic glucose output (43). An increasing number of studies have
indicated that non-alcoholic fatty liver disease (NAFLD) is common
in IBDs (44,45). A recent study reported that NAFLD may
be detected in up to 33.6% of IBD patients, frequently in the
absence of metabolic risk factors (46). Therefore, it may be speculated that
GNB4 has a vital role in IBD with NAFLD. GNG11 belongs to the G
protein gamma family and has a role in the transmembrane signaling
system and cellular senescence (47,48). A
previous study reported that GNG11 was upregulated in ankylosing
spondylitis, which was involved in immune system processes
(49). Another study indicated that
expression of GNG11 induced the generation of reactive oxygen
species (50). Therefore, GNG11 may
have a role in UC via the immune response and oxidative stress.
Angiotensinogen (AGT) is involved in the production of angiotensin
II, which is the major mediator of the action of the
rennin-angiotensin system (RAS) (51), whereas the RAS mediates the
regulation of sodium homeostasis, blood pressure and inflammation
(52). Overexpression of AGT
activates the Jak2/STAT3 signaling pathway in cells, leading to
inflammation (53). Combined with
the present results, the above information allows for the
speculation that AGT has a role in the pathogenesis of IBD via this
signaling pathway.
In addition, the hub genes of the CD group
overlapped with the DEGs in the UC group, in which there were no
specific hub genes. Most of the hub genes in CD were mainly
associated with the chemokine signaling pathway, leukocyte
migration and intestinal fibrosis-associated pathways. Based on
these results, UC and CD exhibited significant differences
regarding several hub genes and biological pathways. Several other
hub genes may be potential therapeutic targets for IBDs. For
instance, ANXA1 is a phospholipid-binding protein with potent
anti-inflammatory activities (54).
A previous study reported that the antagonist of ANXA1 receptors
exacerbated the colitis outcome in TNF receptor 1−/−
mice (55). Another study revealed
that ANXA1 has a critical role in the regulation of mucosal
regeneration and healing (56).
These results suggest that ANXA1 may serve a pivotal role in early
recovery from colitis. IL-8 is one of the major mediators of the
inflammatory response in IBD. IL-8 functions as an important
chemoattractant for neutrophils and is also a potent angiogenic
factor (57). IL-8 is a key mediator
in colonic inflammation, and is closely associated with disease
activity of IBDs (58). Therefore,
the increased expression of IL-8 may initiate or perpetuate IBDs.
Recent study demonstrated that antibody against IL-8 effectively
neutralizes IL-8-dependent neutrophil activation and migration in
inflammatory diseases (59).
Collectively, these hub genes may serve as promising candidates for
targeted biological therapy.
Although UC and CD are different diseases, they
share similar pathological characteristics. It may be useful to
elucidate the shared genetic characteristics and biological
pathways belonging to UC and CD. The hub genes of the overlapping
DEGs in UC and CD were also identified. These hub genes were mainly
enriched in pathways associated with inflammation and ECM
organization. Therefore, it may be speculated that these hub genes
have important functions in IBDs and have implications in the
progression of these diseases. Overall, the hub genes identified
constitute potential molecular targets and diagnostic biomarkers of
UC or CD.
Furthermore, modules analysis of the PPI network
revealed that the development of UC was associated with neuroactive
ligand-receptor interaction and the TNF signaling pathway, whereas
the pathogenesis of CD was also associated with the Toll-like
receptor (TLR) signaling pathway. In addition, the top module
analysis of the PPI network revealed that the overlapping DEGs in
UC and CD were predominantly enriched in immune-associated
biological processes, including chemotaxis and killing of cells of
other organisms, which was consistent with the aforementioned
results of the GO analysis of the present study. Furthermore,
functional pathway enrichment analysis revealed that the
pathogenesis of IBDs was mainly associated with chemokine signaling
pathway and infectious microbes, including Staphylococcus
aureus infection, legionellosis and epithelial cell signaling
in Helicobacter pylori infection. Previous studies have
reported a protective benefit of H. pylori infection against
the development of IBDs (60–62).
H. pylori may induce immune tolerance and limit inflammatory
responses by TLR9 activation (63,64).
These results indicate the importance of H. pylori-induced
activation of TLR9-mediated mechanisms in the pathogenesis of
IBDs.
Of note, the present study has several limitations
that should be emphasized. One limitation is the insufficient
sample size in the dataset used for analysis. Hence, in order to
achieve a more conclusive results, further analysis using a larger
sample size is required. The results of comprehensive analysis
combined with microarrays chips will be more reliable. Analysis of
multiple transcriptomic datasets has the likelihood of discovering
robust candidates for diagnosis and treatment. Therefore, it is
recommended to use meta-analysis to evaluate the comprehensive
effect. However, further factors also require to be taken into
account. Different studies may be heterogeneous due to the
variation of microarray platforms, laboratories and technicians.
Furthermore, it should be ensured that the data format and class
labels are consistent across datasets. In addition, due to the
differences in study design, measurement errors and insufficient
information [disease type (CD and UC), biopsy location (colon and
ileum) and disease activity status (inflamed and uninflamed)], the
integrated analysis of multi-chip data may be a major challenge. A
second limitation is the lack of experimental verification. In the
present study, the results of a chip expression profile were
analyzed using a bioinformatics method analysis and were not
validated by reverse-transcription-quantitative (RT-q) PCR.
Furthermore, in the original study, only the differential
expression of the epithelial barrier genes MUC1, MUC4 and TFF1,
CLDN1, CLDN8 and OCLN, DSG3 and MAGI1 were confirmed by RT-qPCR
(18). Therefore, future studies
with a larger amount of clinical samples and experimental
validation are required. Despite these limitations, the present
study provided novel insight into the mechanisms of IBDs.
In conclusion, the present study used an integrated
analysis method to identify DEGs, as well as biological functions
and pathways shared between and unique to UC and CD, thereby
enhancing the current understanding of the pathogenesis of IBDs and
the molecular mechanisms of UC and CD. In addition, these results
may provide potential biomarkers for the differential diagnosis of
UC and CD, as well as potential therapeutic targets for the
development of novel IBD treatments. However, the present study
only included a bioinformatics analysis and no experiments were
performed to further confirm these biomarkers. Therefore, further
experiments and analyses of larger datasets are required to confirm
the ability of the above candidate genes in differentiating UC from
CD.
Acknowledgements
The authors would like to thank Miss Juan Hua
(Department of Cardiology, The Sixth Hospital of Wuhan, Affiliated
Hospital of Jianghan University) for her support and language
editing of this article.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81570486
and 81330014).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LZ and XH designed the study. CC and JT analyzed the
microarray datasets and interpreted the results. JH and WQ selected
and downloaded the gene expression profile from the GEO. CC wrote
and edited the manuscript. All authors read the content and
approved the final version.
Ethics approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
von Stein P, Lofberg R, Kuznetsov NV,
Gielen AW, Persson JO, Sundberg R, Hellstrom K, Eriksson A, Befrits
R, Ost A and von Stein OD: Multigene analysis can discriminate
between ulcerative colitis, Crohn's disease, and irritable bowel
syndrome. Gastroenterology. 134:1869–1881; quiz 2153–2164. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feuerstein JD and Cheifetz AS: Ulcerative
colitis: Epidemiology, diagnosis, and management. Mayo Clin Proc.
89:1553–1563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feuerstein JD and Cheifetz AS: Crohn
disease: Epidemiology, diagnosis, and management. Mayo Clin Proc.
92:1088–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaplan GG and Ng SC: Understanding and
preventing the global increase of inflammatory bowel disease.
Gastroenterology. 152:313–321.e2. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ng SC, Tang W, Ching JY, Wong M, Chow CM,
Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, et al: Incidence and
phenotype of inflammatory bowel disease based on results from the
Asia-pacific Crohn's and colitis epidemiology study.
Gastroenterology. 145:158–165.e2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ng SC, Kaplan G, Banerjee R, Wei SC, Tang
W, Zeng Z, Chen MH, Yang H, Silva JD, Niriella MA, et al: 78
incidence and phenotype of inflammatory bowel disease from 13
countries in Asia-Pacific: Results from the Asia-Pacific Crohn's
and colitis epidemiologic study 2011–2013. Gastroenterology.
150:S212016. View Article : Google Scholar
|
|
7
|
Molodecky NA, Soon IS, Rabi DM, Ghali WA,
Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema
HW and Kaplan GG: Increasing incidence and prevalence of the
inflammatory bowel diseases with time, based on systematic review.
Gastroenterology. 142:46–54.e42; quiz e30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoivik ML, Bernklev T and Moum B: Need for
standardization in population-based quality of life studies: A
review of the current literature. Inflamm Bowel Dis. 16:525–536.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Serna E, Morales JM, Mata M,
Gonzalez-Darder J, San Miguel T, Gil-Benso R, Lopez-Gines C,
Cerda-Nicolas M and Monleon D: Gene expression profiles of
metabolic aggressiveness and tumor recurrence in benign meningioma.
PLoS One. 8:e672912013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dai J, Ma Y, Chu S, Le N, Cao J and Wang
Y: Identification of key genes and pathways in meningioma by
bioinformatics analysis. Oncol Lett. 15:8245–8252. 2018.PubMed/NCBI
|
|
12
|
Li XL, Zhou CY, Sun Y, Su ZY, Wang X, N
Jia E, Zhang Q, Jiang XF, Qi WQ and Xu Y: Bioinformatic analysis of
potential candidates for therapy of inflammatory bowel disease. Eur
Rev Med Pharmacol Sci. 19:4275–4284. 2015.PubMed/NCBI
|
|
13
|
Feng J, Gao Q, Liu Q, Wang F, Lin X, Zhao
Q, Liu J and Li J: Integrated strategy of differentially expressed
genes associated with ulcerative colitis. Mol Med Rep.
16:7479–7489. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujimoto K, Kinoshita M, Tanaka H, Okuzaki
D, Shimada Y, Kayama H, Okumura R, Furuta Y, Narazaki M, Tamura A,
et al: Regulation of intestinal homeostasis by the ulcerative
colitis-associated gene RNF186. Mucosal Immunol. 10:446–459. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van der Goten J, Vanhove W, Lemaire K, Van
Lommel L, Machiels K, Wollants WJ, De Preter V, De Hertogh G,
Ferrante M, Van Assche G, et al: Integrated miRNA and mRNA
expression profiling in inflamed colon of patients with ulcerative
colitis. PLoS One. 9:e1161172014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
UK IBD Genetics Consortium, Barrett JC,
Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E,
Parnell K, Zhang H, et al: Genome-wide association study of
ulcerative colitis identifies three new susceptibility loci,
including the HNF4A region. Nat Genet. 41:1330–1334. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jostins L, Ripke S, Weersma RK, Duerr RH,
McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et
al: Host-microbe interactions have shaped the genetic architecture
of inflammatory bowel disease. Nature. 491:119–124. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vancamelbeke M, Vanuytsel T, Farré R,
Verstockt S, Ferrante M, Van Assche G, Rutgeerts P, Schuit F,
Vermeire S, Arijs I and Cleynen I: Genetic and transcriptomic bases
of intestinal epithelial barrier dysfunction in inflammatory bowel
disease. Inflamm Bowel Dis. 23:1718–1729. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumgart DC and Carding SR: Inflammatory
bowel disease: Cause and immunobiology. Lancet. 369:1627–1640.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Albertson DG and Pinkel D: Genomic
microarrays in human genetic disease and cancer. Hum Mol Genet 12
Spec No. 2:R145–R152. 2003. View Article : Google Scholar
|
|
21
|
Forini F, Nicolini G, Kusmic C, D'Aurizio
R, Rizzo M, Baumgart M, Groth M, Doccini S, Iervasi G and Pitto L:
Integrative analysis of differentially expressed genes and miRNAs
predicts complex T3-mediated protective circuits in a rat model of
cardiac ischemia reperfusion. Sci Rep. 8:138702018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cecchini MJ, Hosein K, Howlett CJ, Joseph
M and Mura M: Comprehensive gene expression profiling identifies
distinct and overlapping transcriptional profiles in non-specific
interstitial pneumonia and idiopathic pulmonary fibrosis. Respir
Res. 19:1532018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tatiya-Aphiradee N, Chatuphonprasert W and
Jarukamjorn K: Immune response and inflammatory pathway of
ulcerative colitis. J Basic Clin Physiol Pharmacol. 30:1–10. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Strober W and Fuss IJ: Proinflammatory
cytokines in the pathogenesis of inflammatory bowel diseases.
Gastroenterology. 140:1756–1767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng X, Szabo S, Khomenko T, Tolstanova G,
Paunovic B, French SW and Sandor Z: Novel pharmacologic approaches
to the prevention and treatment of ulcerative colitis. Curr Pharm
Des. 19:17–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mateescu RB, Bastian AE, Nichita L,
Marinescu M, Rouhani F, Voiosu AM, Benguş A, Tudoraşcu DR and Popp
CG: Vascular endothelial growth factor-key mediator of angiogenesis
and promising therapeutical target in ulcerative colitis. Rom J
Morphol Embryol. 58:1339–1345. 2017.PubMed/NCBI
|
|
27
|
Danese S, Sans M, de la Motte C, Graziani
C, West G, Phillips MH, Pola R, Rutella S, Willis J, Gasbarrini A
and Fiocchi C: Angiogenesis as a novel component of inflammatory
bowel disease pathogenesis. Gastroenterology. 130:2060–2073. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu G and Agarwal P: Human disease-drug
network based on genomic expression profiles. PLoS One.
4:e65362009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bernstein CN, Blanchard JF, Rawsthorne P
and Yu N: The prevalence of extraintestinal diseases in
inflammatory bowel disease: A population-based study. Am J
Gastroenterol. 96:1116–1122. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Veloso FT: Extraintestinal manifestations
of inflammatory bowel disease: Do they influence treatment and
outcome? World J Gastroenterol. 17:2702–2707. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kappelman MD, Galanko JA, Porter CQ and
Sandler RS: Association of paediatric inflammatory bowel disease
with other immune-mediated diseases. Arch Dis Child. 96:1042–1046.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dinca M, Fries W, Luisetto G, Peccolo F,
Bottega F, Leone L, Naccarato R and Martin A: Evolution of
osteopenia in inflammatory bowel disease. Am J Gastroenterol.
94:1292–1297. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schulte C, Dignass AU, Mann K and Goebell
H: Reduced bone mineral density and unbalanced bone metabolism in
patients with inflammatory bowel disease. Inflamm Bowel Dis.
4:268–275. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ciucci T, Ibáñez L, Boucoiran A,
Birgy-Barelli E, Pène J, Abou-Ezzi G, Arab N, Rouleau M, Hébuterne
X, Yssel H, et al: Bone marrow Th17 TNFα cells induce osteoclast
differentiation, and link bone destruction to IBD. Gut.
64:1072–1081. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu XF, Xu R, Ouyang ZJ, Qian C, Shen Y, Wu
XD, Gu YH, Xu Q and Sun Y: Beauvericin ameliorates experimental
colitis by inhibiting activated T cells via downregulation of the
PI3K/Akt signaling pathway. PLoS One. 8:e830132013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Q, Duan X, Fan H, Xu M, Tang Q, Zhang
L, Shou Z, Liu X, Zuo D, Yang J, et al: Oxymatrine protects against
DSS-induced colitis via inhibiting the PI3K/AKT signaling pathway.
Int Immunopharmacol. 53:149–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei J and Feng J: Signaling pathways
associated with inflammatory bowel disease. Recent Pat Inflamm
Allergy Drug Discov. 4:105–117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Latella G, Di Gregorio J, Flati V, Rieder
F and Lawrance IC: Mechanisms of initiation and progression of
intestinal fibrosis in IBD. Scand J Gastroenterol. 50:53–65. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Latella G, Sferra R, Speca S, Vetuschi A
and Gaudio E: Can we prevent, reduce or reverse intestinal fibrosis
in IBD? Eur Rev Med Pharmacol Sci. 17:1283–1304. 2013.PubMed/NCBI
|
|
41
|
Rieder F and Fiocchi C: Intestinal
fibrosis in IBD-a dynamic, multifactorial process. Nat Rev
Gastroenterol Hepatol. 6:228–235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Saddala MS, Lennikov A, Grab DJ, Liu GS,
Tang S and Huang H: Proteomics reveals ablation of PlGF increases
antioxidant and neuroprotective proteins in the diabetic mouse
retina. Sci Rep. 8:167282018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Benjafield AV, Lin RC, Dalziel B, Gosby
AK, Caterson ID and Morris BJ: G-protein beta3 subunit gene splice
variant in obesity and overweight. Int J Obes Relat Metab Disord.
25:777–780. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Principi M, Iannone A, Losurdo G, Mangia
M, Shahini E, Albano F, Rizzi SF, La Fortezza RF, Lovero R,
Contaldo A, et al: Nonalcoholic fatty liver disease in inflammatory
bowel disease: Prevalence and risk factors. Inflamm Bowel Dis.
24:1589–1596. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sartini A, Gitto S, Bianchini M, Verga MC,
Di Girolamo M, Bertani A, Del Buono M, Schepis F, Lei B, De Maria N
and Villa E: Non-alcoholic fatty liver disease phenotypes in
patients with inflammatory bowel disease. Cell Death Dis. 9:872018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bessissow T, Le NH, Rollet K, Afif W,
Bitton A and Sebastiani G: Incidence and predictors of nonalcoholic
fatty liver disease by serum biomarkers in patients with
inflammatory bowel disease. Inflamm Bowel Dis. 22:1937–1944. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Balcueva EA, Wang Q, Hughes H, Kunsch C,
Yu Z and Robishaw JD: Human G protein gamma(11) and gamma(14)
subtypes define a new functional subclass. Exp Cell Res.
257:310–319. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hossain MN, Sakemura R, Fujii M and
Ayusawa D: G-protein gamma subunit GNG11 strongly regulates
cellular senescence. Biochem Biophys Res Commun. 351:645–650. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fang F, Pan J, Xu L, Li G and Wang J:
Identification of potential transcriptomic markers in developing
ankylosing spondylitis: A meta-analysis of gene expression
profiles. Biomed Res Int. 2015:8263162015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Takauji Y, Kudo I, En A, Matsuo R, Hossain
MN, Nakabayashi K, Miki K, Fujii M and Ayusawa D: GNG11 (G-protein
subunit γ 11) suppresses cell growth with induction of reactive
oxygen species and abnormal nuclear morphology in human SUSM-1
cells. Biochem Cell Biol. 95:517–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Farcas A, Gligor F, Bucșa C, Mogoșan C,
Bojiță M and Dumitrașcu D: The current insight on dual
renin-angiotensin system blockade: A data review with a focus on
safety. Farmacia. 63:325–333. 2015.
|
|
52
|
Raizada V, Skipper B, Luo W and Griffith
J: Intracardiac and intrarenal renin-angiotensin systems:
Mechanisms of cardiovascular and renal effects. J Investig Med.
55:341–359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shen L, Zhang T and Lu H: Overexpression
of AGT promotes bronchopulmonary dysplasis via the JAK/STAT signal
pathway. Oncotarget. 8:96079–96088. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dalli J, Norling LV, Renshaw D, Cooper D,
Leung KY and Perretti M: Annexin 1 mediates the rapid
anti-inflammatory effects of neutrophil-derived microparticles.
Blood. 112:2512–2519. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sena AA, Pedrotti LP, Barrios BE, Cejas H,
Balderramo D, Diller A and Correa SG: Lack of TNFRI signaling
enhances annexin A1 biological activity in intestinal inflammation.
Biochem Pharmacol. 98:422–431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Babbin BA, Lee WY, Parkos CA, Winfree LM,
Akyildiz A, Perretti M and Nusrat A: Annexin I regulates SKCO-15
cell invasion by signaling through formyl peptide receptors. J Biol
Chem. 281:19588–19599. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang XD, Corvalan JR, Wang P, Roy CM and
Davis CG: Fully human anti-interleukin-8 monoclonal antibodies:
Potential therapeutics for the treatment of inflammatory disease
states. J Leukoc Biol. 66:401–410. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Skov L, Beurskens FJ, Zachariae CO,
Reitamo S, Teeling J, Satijn D, Knudsen KM, Boot EP, Hudson D,
Baadsgaard O, et al: IL-8 as antibody therapeutic target in
inflammatory diseases: Reduction of clinical activity in
palmoplantar pustulosis. J Immunol. 181:669–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mitsuyama K, Toyonaga A, Sasaki E,
Watanabe K, Tateishi H, Nishiyama T, Saiki T, Ikeda H, Tsuruta O
and Tanikawa K: IL-8 as an important chemoattractant for
neutrophils in ulcerative colitis and Crohn's disease. Clin Exp
Immunol. 96:432–436. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Luther J, Dave M, Higgins PD and Kao JY:
Association between Helicobacter pylori infection and
inflammatory bowel disease: A meta-analysis and systematic review
of the literature. Inflamm Bowel Dis. 16:1077–1084. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rokkas T, Gisbert JP, Niv Y and O'Morain
C: The association between Helicobacter pylori infection and
inflammatory bowel disease based on meta-analysis. United European
Gastroenterol J. 3:539–550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu XW, Ji HZ, Yang MF, Wu L and Wang FY:
Helicobacter pylori infection and inflammatory bowel disease
in Asians: A meta-analysis. World J Gastroenterol. 21:4750–4756.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Varga MG, Piazuelo MB, Romero-Gallo J,
Delgado AG, Suarez G, Whitaker ME, Krishna US, Patel RV, Skaar EP,
Wilson KT, et al: TLR9 activation suppresses inflammation in
response to Helicobacter pylori infection. Am J Physiol
Gastrointest Liver Physiol. 311:G852–G858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Owyang SY, Luther J, Owyang CC, Zhang M
and Kao JY: Helicobacter pylori DNA's anti-inflammatory
effect on experimental colitis. Gut microbes. 3:168–171. 2012.
View Article : Google Scholar : PubMed/NCBI
|