Introduction
Tumor metastasis is the main cause of mortality
among patients with different types of cancers (1). Glioma as one of the main type of
central nervous system tumor accounts for more than 80% cases of
all malignant brain tumors (2).
Glioma causes a series of clinical symptoms, including vomiting,
cranial nerve disorders, seizures, headaches and loss of vision
(3,4). However, these clinical symptoms can
easily be ignored or misdiagnosed. Therefore, the existence of
distant tumor metastasis is common by the time of diagnosis
(5). Investigating how to prevent
and treat glioma metastasis is a major task for clinical
researchers as the efficacy of treatment is usually poor due to the
unknown pathogenesis of glioma (3–5).
As a double edged sword in cancer biology,
transforming growth factor (TGF)-β signaling inhibits tumor growth
at the early stage of tumor development, but promotes tumor
metastasis in late stages (6). TGF-β
signaling also has been demonstrated to enhance the invasion and
metastasis of glioma, and the inhibition of TGF-β signaling has
been considered to be a therapeutic target for glioma (7). Successful TGF-β signaling transduction
requires the involvement of different long non-coding (lnc)RNAs
(8,9), which are a subgroup of non-coding RNAs
composed of more than 200 nucleotides and have critical functions
in cancer biology (10). NEF is a
novel lncRNA and has been determined to have a tumor suppressive
role in hepatocellular carcinoma (11). In the current study, the authors
hypothesized that lncRNA NEF may inhibit glioma cell migration and
invasion by downregulating TGF-β1.
Materials and methods
Patients and specimens
A total of 53 patients with glioma were selected in
Yichang Second People's Hospital (Yichang, China) between March
2015 and January 2018. Inclusion criteria: i) Patients
pathologically diagnosed with glioma and treated for the first
time; ii) patients willing to participate. Exclusion criteria: i)
Patients complicated with other severe diseases; ii) patients who
received treatment prior to admission. These patients included 30
males and 23 females; the age ranged was from 29 to 69 years, with
a mean age of 49.1±5.3 years. At the same time, a total of 56
healthy controls were also included to serve as control group. The
control group included 31 males and 25 females; the age ranged was
from 28 to 68 years, with a mean age of 48.8±6.1 years. Biopsies of
tumor tissues (100–200 mg) and adjacent heathy tissues (100–200 mg)
were obtained from each patient with glioma. Blood samples (5 ml)
were collected from patients with glioma as well as participants in
the control group on the day of admission. No significant
differences in gender, age and other basic clinical data between
the patient group and the control group were identified (Table I). The ethics committee of Yichang
Second People's Hospital approved the current study. All
participants signed informed consent forms.
 | Table I.Associations between the expression of
NEF in the blood and clinicopathological data of patients with
glioma. |
Table I.
Associations between the expression of
NEF in the blood and clinicopathological data of patients with
glioma.
|
|
|
| Number of
patients |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Characteristic | Groups | Cases | High-expression | Low-expression | χ2 | P-value |
|---|
| Age, years | >50 | 28 | 16 | 12 | 0.91 | 0.34 |
|
| ≤50 | 25 | 11 | 14 |
|
|
| Sex | Male | 30 | 14 | 16 | 0.51 | 0.48 |
|
| Female | 23 | 13 | 10 |
|
|
| Primary tumor
diameter, cm | >2 | 26 | 15 | 11 | 0.93 | 0.33 |
|
| ≤2 | 27 | 12 | 15 |
|
|
| Tumor distant
metastasis | Yes | 31 | 12 | 19 | 4.47 | 0.03 |
|
| No | 22 | 15 | 7 |
|
|
Cell culture
Hs 683 (ATCC® HTB-138™) and CCD-25Lu
(ATCC® CCL-215™) human glioma cell lines were bought
from American Type Culture Collection (Manassas, VA, USA). Cells
were culture in Eagle's Minimum Essential Medium (cat. no.
30-2003™; American Type Culture Collection) supplemented with 10%
fetal bovine serum (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany
and maintained at 37°C in a 5% CO2-humidified
incubator.
In cases of TGF-β1 treatment, cells were pre-treated
with 5, 10, 20 and 30 ng/ml TGF-β1 in culture medium for 24 h prior
to subsequent experimentation.
Cell transfection
Full-length cDNA for NEF was PCR amplified
introducing EcoRI restriction sites and cloned into the
pIRSE2 vector (Clontech Laboratories, Inc., Mountainview, CA, USA)
by Sangon Biotech Co., Ltd. (Shanghai, China). pIRSE2-NEF vectors
(10 nM) were mixed with Lipofectamine® 2000 reagent
(cat. no. 11668-019; Invitrogen, Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) to form vector-reagent complexes and used to
transfect cells. Cells (1×105 cells in each well of a
six-well plate) were subsequently transfected with vector-reagent
complexes at 37°C for 5 h. Cells without transfection were used as
control cells. Cells transfected with empty vectors were used as
negative control cells. Transfection efficiency >200% compared
with control cells was achieved in each experiment.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and blood samples
using TRIzol® reagent (Invitrogen, Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA was reverse transcribed into cDNA using SuperScript III Reverse
Transcriptase (Thermo Fisher Scientific, Inc.) using the following
thermocycling conditions: 25°C for 5 min, 50°C for 20 min and 75°C
for 5 min. qPCR was subsequently performed using the
SYBR® Green Real-Time PCR Master mix (Thermo Fisher
Scientific, Inc.). The following primer pairs were used for the
qPCR: lncRNA-NEF forward, 5′-CTGCCGTCTTAAACCAACCC-3′ and reverse,
5′-GCCCAAACAGCTCCTCAATT-3′; β-actin forward,
5′-GACCTCTATGCCAACACAGT-3′ and reverse, 5′-AGTACTTGCGCTCAGGAGGA-3′.
The following thermocycling conditions were used: Initial
denaturation at 95°C for 42 sec; 40 cycles of 95°C for 22 sec and
56.5°C for 38 sec. lncRNA-NEF levels were quantified using the
2−ΔΔCq method and normalized to β-actin as the internal
control (12).
ELISA
An ELISA kit was used to measure blood levels of
TGF-β1 (cat. no. RAB0460-1KT; Sigma-Aldrich; Merck KGaA) according
to the manufacturer's protocol.
In vitro cell migration and invasion
assays
Transwell migration and invasion assays were
performed to examine in vitro cell migration and invasion.
Following transfection, Hs 683 and CCD-25Lu cell suspensions with a
density of 5×104 cells/ml were made. In the migration
assay, 5×103 cells in 0.1 ml serum-free Eagle's Minimum
Essential Medium were seeded into the upper chamber, while the
lower chamber was filled with RPMI-1640 medium (Thermo Fisher
Scientific, Inc.) supplemented with 20% fetal calf serum
(Sigma-Aldrich; Merck KGaA). Membranes were collected after cell
culture was performed for 24 h in an incubator at 37°C with 5%
CO2. Membranes were stained with 0.5% crystal violet
(Sigma-Aldrich; Merck KGaA) for 30 min at 25°C. Cells were counted
under an optical microscope (magnification, 40×; CX33, Olympus
Corporation, Tokyo, Japan). The cell invasion assay was performed
using the same protocol, however the upper chambers were precoated
with Matrigel (cat. no. 356234; EMD Millipore, Billerica, MA,
USA).
Western blot analysis
Following transfection, total protein was extracted
from Hs 683 and CCD-25Lu cells using radioimmunoprecipitation assay
buffer (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Total protein was quantified using a
bicinchoninic acid assay and 30 µg protein/well was separated using
SDS-PAGE on a 12% gel. The separated proteins were transferred onto
polyvinylidene difluoride membranes and blocked for 2 h with 5%
skimmed milk. The membranes were incubated with primary antibodies
against TGF-β1 (cat. no. ab92486) and GAPDH (cat. no. ab9485; both
1:1,000; Abcam) overnight at 4°C. Following primary incubation,
membranes were further incubated with anti-rabbit immunoglobulin G
horseradish peroxidase-conjugated secondary antibodies (1:1,000;
cat. no. MBS435036; MyBioSource) for 2 h at room temperature.
Protein bands were visualized using Pierce™ ECL Western Blotting
Substrate (Thermo Fisher Scientific, Inc.) and signals were
detected using MYECL™ Imager (Thermo Fisher Scientific, Inc.).
Protein expression was quantified using ImageJ software (version
1.6; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Graphpad Prism 6 software was used for all data
analysis. Gene expression, and cell migration and invasion data
were recorded as mean ± standard deviation. Data were compared by
t-test (between two groups) or one-way analysis of variance
followed by a Fisher's Least Significant Difference test (among
multiple groups). Receiver operating characteristic (ROC) curve
analysis was performed to evaluate the diagnostic value of blood
NEF for glioma. Pearson's correlation coefficient was used to
analyze the correlations between blood NEF and TGF-β1 expression
levels. Chi-square test was performed to analyze the associations
between blood NEF expression and clinicopathological data of
patients. P<0.05 was considered to be statistically
significant.
Results
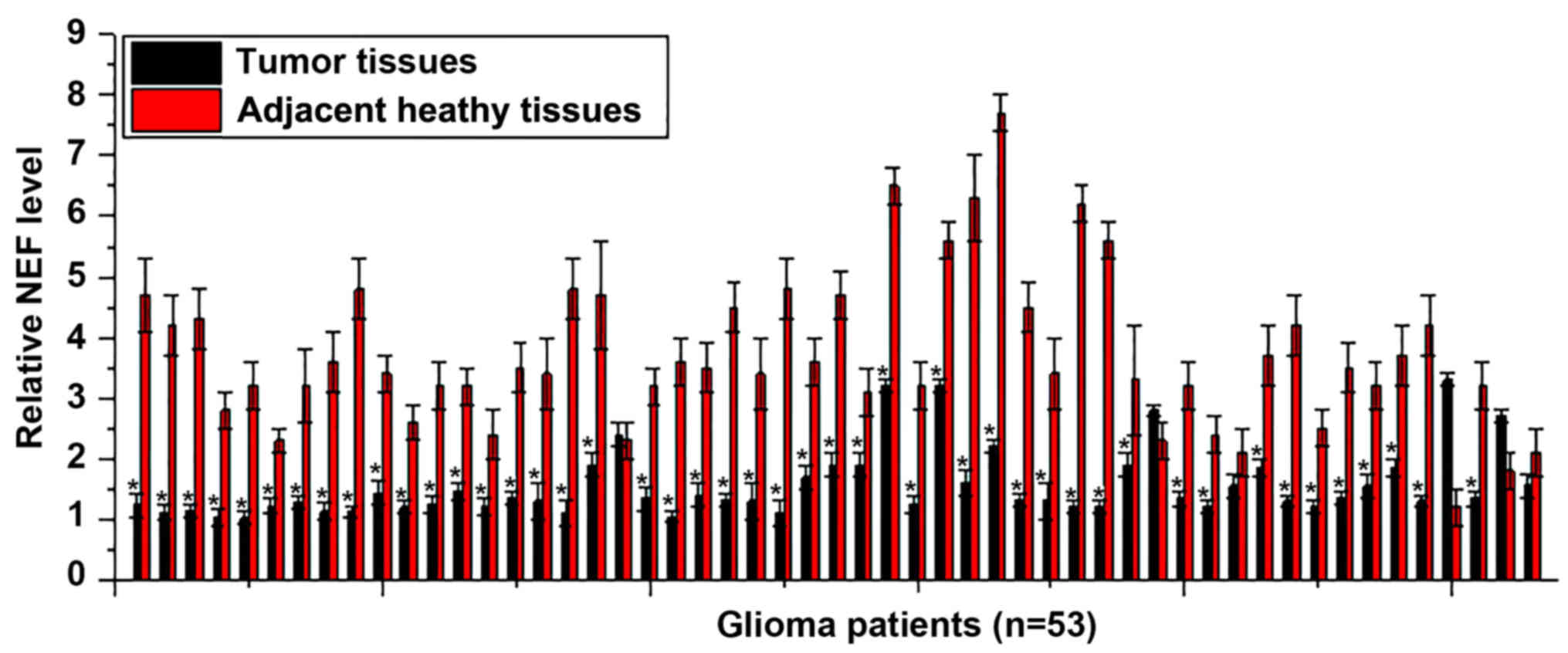
NEF expression is lower in tumor
tissues compared with adjacent healthy tissues in the majority of
patients with glioma
Differential expression of a gene in tumor tissues
and adjacent healthy tissues indicated its involvement in cancers.
Therefore, the expression of NEF in tumor tissues and adjacent
healthy tissues of 53 patients with glioma was detected. As
presented in Fig. 1, significantly
lower expression levels of NEF in tumor tissues was identified in
47 out of 53 patients when compared with adjacent healthy tissues,
accounting for 88.7% of the patients. Therefore, the downregulation
of lncRNA NEF is likely involved in the pathogenesis of glioma.
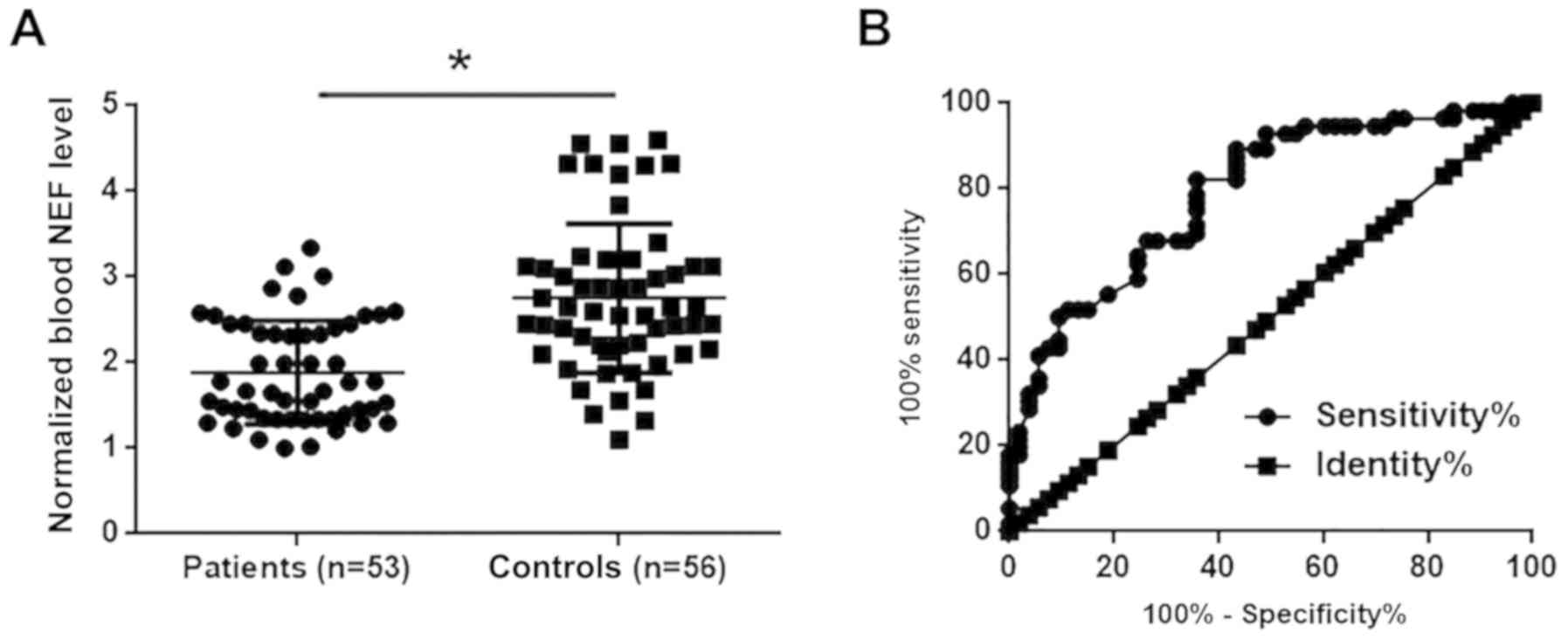
Low blood levels of NEF in glioma
patients effectively distinguishes patients from healthy
controls
Blood levels of NEF in glioma patients and healthy
controls were measured by RT-qPCR. As presented in Fig. 2A, blood levels of NEF were
significantly lower in patients with glioma compared with healthy
controls (P<0.05). ROC curve analysis was performed to evaluate
the diagnostic value of blood NEF for glioma. As presented in
Fig. 2B, area under the curve was
0.7908, with standard error of 0.04271 and 95% confident interval
of 0.7070–0.8745.
Blood levels of NEF significantly
associate with distant tumor metastasis
Patients were divided into high (n=27) and low
(n=26) expression groups based on NEF expression cut-off score of
2.03. A chi-square test was performed to analyze the association
between the blood NEF levels and clinicopathological data of
patients with glioma. No significant associations were identified
between NEF blood levels, and patients' age, sex and tumor size
(Table I). By contrast, a
significant association between blood levels of NEF and the
existence of distant tumor metastasis was observed.
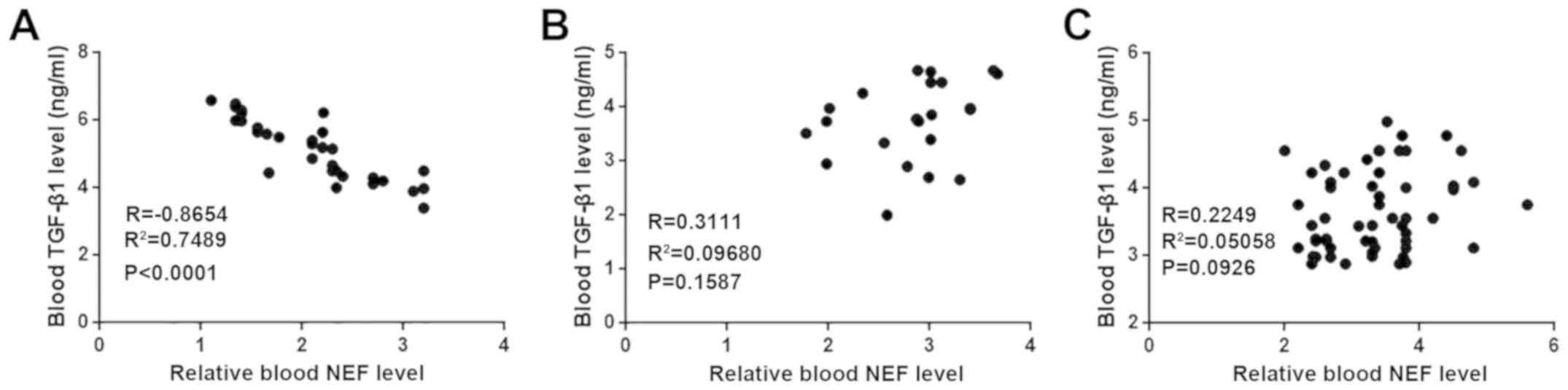
Blood levels of NEF are significantly
correlated with TGF-β1 in patients with metastatic glioma
Pearson's correlation coefficient was used to
analyze the correlations between blood NEF and TGF-β1 levels. Blood
levels of NEF were significantly correlated with TGF-β1 in patients
with metastatic glioma (Fig. 3A). In
contrast, no significant correlations between blood NEF and TGF-β1
were identified in patients with non-metastatic glioma (Fig. 3B) and healthy controls (Fig. 3C).
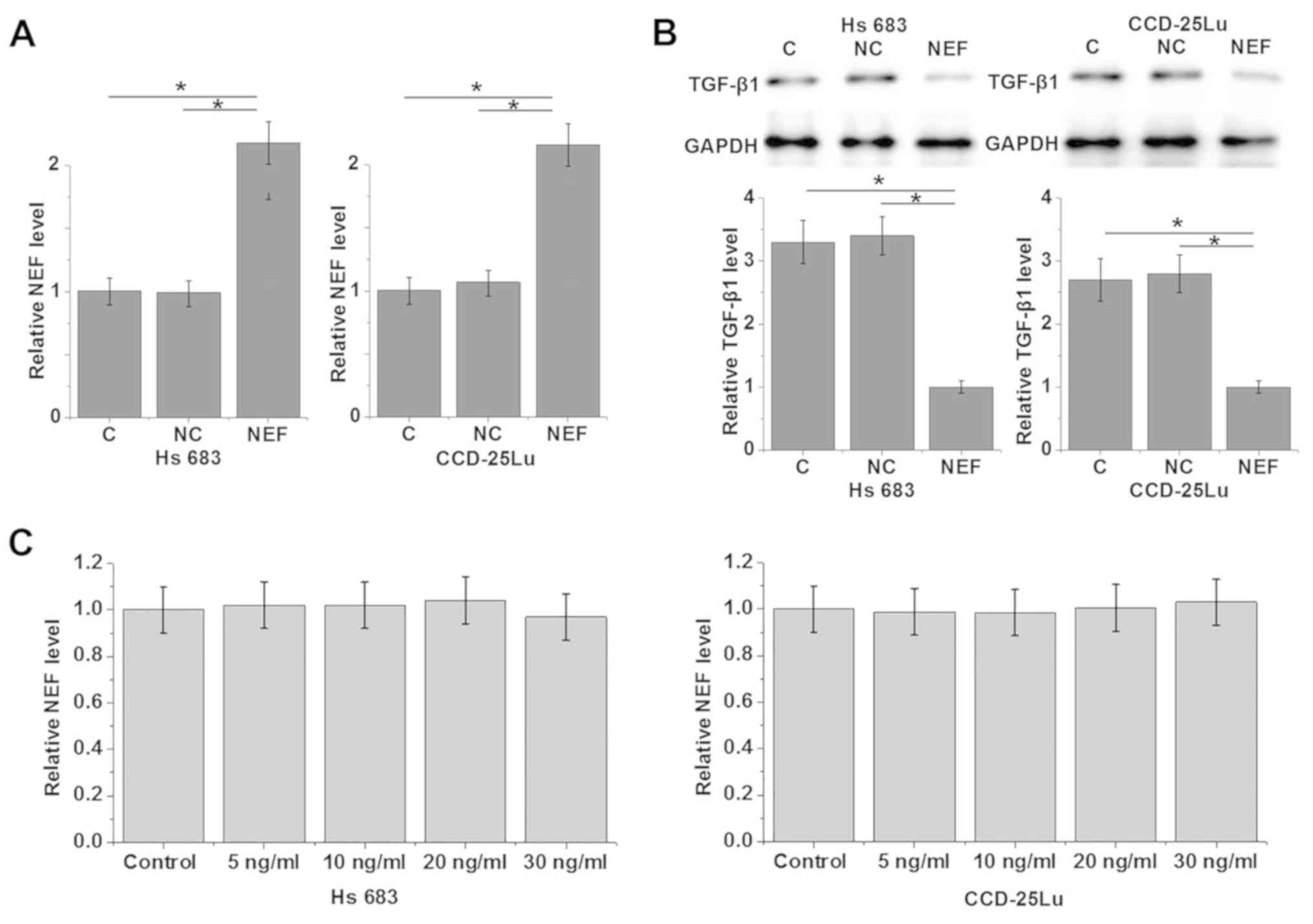
NEF is likely an upstream inhibitor of
TGF-β1 in glioma cells
To further investigate the interactions between NEF
and TGF-β1, NEF expression vectors were transfected into Hs 683 and
CCD-25Lu cells. RT-qPCR revealed that the expression of NEF was
significantly upregulated in both cell lines following transfection
compared with control and negative control groups (Fig. 4A; P<0.05). Compared with control
and negative control cells, cells with NEF overexpression
demonstrated significantly downregulated TGF-β1 protein expression
(all P<0.05; Fig. 4B). By
contrast, treatment with exogenous TGF-β1 at concentrations of 5,
10, 20 and 30 ng/ml demonstrated no significant effects on NEF
expression (Fig. 4C). Therefore NEF
is likely an upstream inhibitor of TGF-β1 in glioma cells.
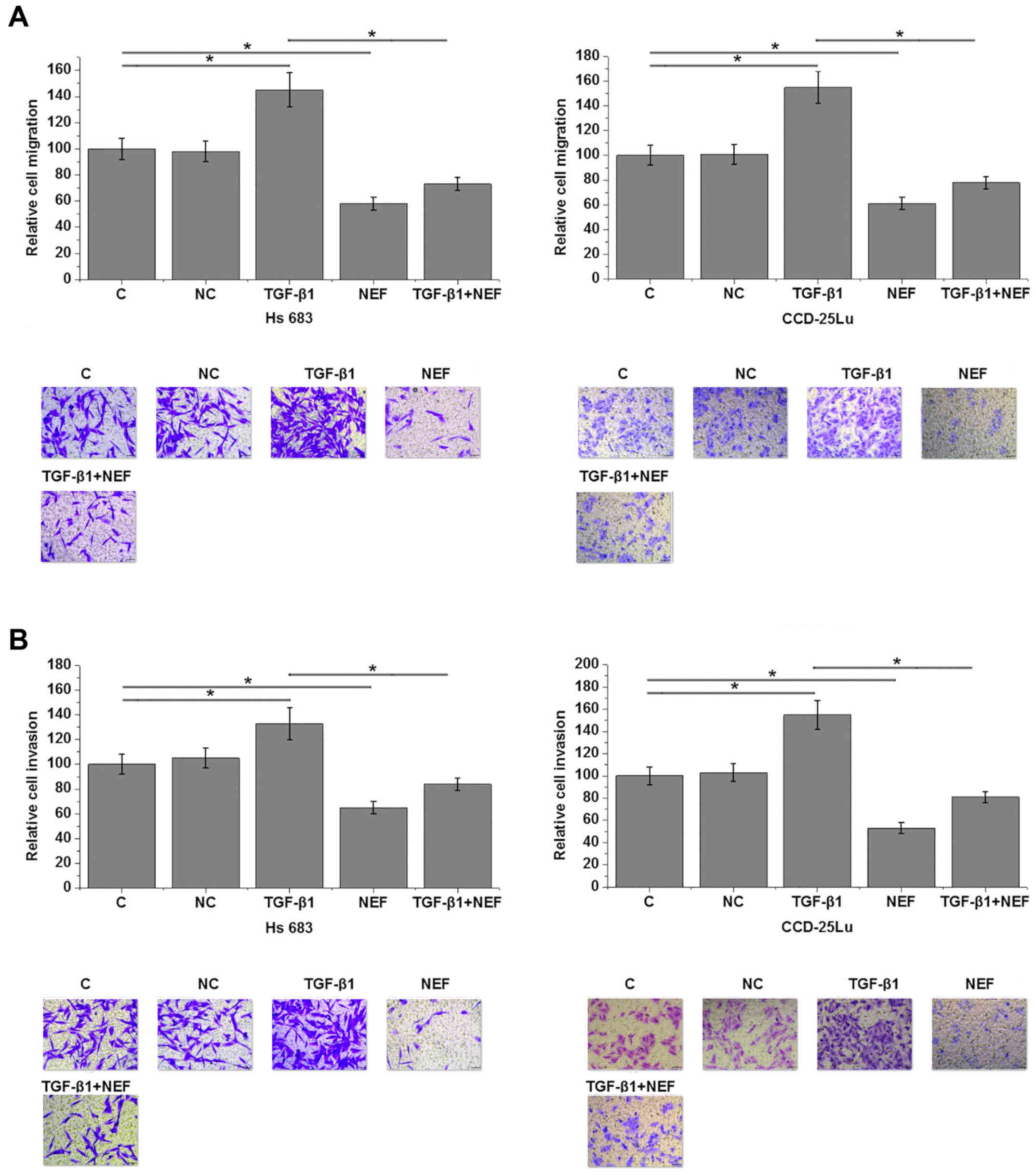
NEF overexpression promotes the
migration and invasion of glioma cells
Cell migration and invasion data demonstrated that,
compared with control, cells with NEF overexpression significantly
inhibited cell migration (all P<0.05; Fig. 5A) and invasion (all P<0.05;
Fig. 5B). In addition, treatment
with exogenous TGF-β1 at a dosage of 10 ng/ml significantly
promoted cell migration (all P<0.05; Fig. 5A) and invasion (all P<0.05;
Fig. 5B) compared with control
cells. Exogenous TGF-β1 treatment also significantly reduced the
inhibitory effects of NEF overexpression on cell migration (all
P<0.05; Fig. 5A) and invasion
(all P<0.05; Fig. 5B) compared
with cells treated with exogenous TGF-β1 alone.
Discussion
lncRNA NEF suppresses epithelial to mesenchymal
transition (EMT) in hepatocellular carcinoma, which in turn
inhibits tumor invasion and metastasis (11). The current study suggests that lncRNA
NEF also serves a tumor suppression role in glioma by inhibiting
glioma cell migration and invasion. The actions of lncRNA NEF may
be mediated by the activation of the TGF-β signaling pathway, which
is critical for EMT in a variety of types of cancers (12).
The development of glioma altered the expression of
a large set of lncRNAs and the expression pattern of differentially
expressed lncRNAs has been demonstrated to determine the clinical
phenotype of glioma (13).
Upregulation of lncRNA MALAT1 was observed in patients with glioma
and the overexpression of this lncRNA at least partially
contributed to the malignant nature of this disease (14). By contrast, lncRNA TUG1 is
downregulated in human glioma and the overexpression of TUG1
promoted cancer cell apoptosis, indicating the potential
application of lncRNA TUG1 as a therapeutic target for glioma
(15). In the current study, the
authors examined the expression of lncRNA NEF in the tumor tissues
and adjacent heathy tissues of glioma patients, and in the blood of
patients and healthy controls due to difficulties in collecting
brain biopsies from healthy controls. It was observed that NEF was
downregulated in tumor tissues compared with adjacent heathy
tissues and that blood levels of NEF in patients with glioma were
lower compared with healthy controls, indicating the potential role
of lncRNA NEF in glioma.
Early diagnosis is critical for the survival of
patients with glioma as a considerable proportion of patients are
diagnosed following distant tumor metastasis (5). With the advantages of less invasive
techniques, circulating biomarkers have been increasing used in the
clinical diagnosis of human diseases, such as glioma (16). In contrast to other cancers, glioma
cells migrate through cerebrospinal fluid (17). In the current study, blood was used
instead of cerebrospinal fluid to detect circulating NEF due to
difficulties in enrolling enough volunteers willing to donate
cerebrospinal fluid. However, blood biomarkers have also been
revealed to have value in diagnosing glioma (18). The current study indicated that low
blood levels of lncRNA NEF can be used to effectively distinguish
glioma patients from healthy controls. Therefore, NEF may be used
to improve the diagnosis of glioma.
The current study observed a significant association
between blood levels of NEF and the existence of distant tumor
metastasis in patients with glioma, indicating the potential
involvement of NEF in glioma. TGF-β signaling is a key regulator in
the invasion and metastasis of glioma cells (7). The authors of the current study
observed a significant negative correlation between blood NEF and
TGF-β1 in patients with metastatic glioma, but not in patients with
non-metastatic glioma and healthy controls. Also, TGF-β1 treatment
did not affect NEF expression, suggesting that NEF is upstream of
TGF-β1 in the metastasis of glioma. TGF-β signaling has been
demonstrated to be activated in the growth and metastasis of glioma
(7). In vitro cell experiment
data also supported this hypothesis through the following
observations: i) NEF overexpression promoted TGF-β1 expression; ii)
exogenous TGF-β1 did not have an effect on NEF expression; iii) NEF
overexpression inhibited and exogenous TGF-β1 promoted the
migration and invasion of glioma cells; iv) exogenous TGF-β1
treatment reduced the inhibitory effects of NEF overexpression on
cell migration and invasion. Therefore, NEF may inhibit glioma
metastasis by inactivating TGF-β signaling.
Due to the low incidence rate of glioma, the sample
size in the current study is relative low. Future studies will try
to enroll more patients to further confirm the conclusions of the
current study. In conclusion, lncRNA NEF was downregulated in
glioma and NEF overexpression may inhibit the metastasis of glioma
by serving as an upstream inhibitor of the TGF-β signaling
pathway.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
QH and HC designed the experiments. QH, HC, BZ and
CC performed the experiments. WY and YY analyzed the data. HC
prepared the manuscript. All other authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Yichang Second People's Hospital (Yichang, China). All participants
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mehlen P and Puisieux A: Metastasis: A
question of life or death. Nat Rev Cancer. 6:449–458. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mishra MV, Andrews DW, Glass J, Evans JJ,
Dicker AP, Shen X and Lawrence YR: Characterization and outcomes of
optic nerve gliomas: A population-based analysis. J Neurooncol.
107:591–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stark AM, van de Bergh J, Hedderich J,
Mehdorn HM and Nabavi A: Glioblastoma: Clinical characteristics,
prognostic factors and survival in 492 patients. Clin Neurol
Neurosurg. 114:840–845. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer-a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han J, Alvarez-Breckenridge CA, Wang QE
and Yu J: TGF-β signaling and its targeting for glioma treatment.
Am J Cancer Res. 5:945–955. 2015.PubMed/NCBI
|
|
8
|
Yuan J, Yang F, Wang F, Ma JZ, Guo YJ, Tao
QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang WC, Ren JL, Wong CW, Chan SO, Waye
MM, Fu WM and Zhang JF: LncRNA-NEF antagonized epithelial to
mesenchymal transition and cancer metastasis via cis-regulating
FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene.
37:1445–1456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Tian XJ and Xing J: Signal
transduction pathways of EMT induced by TGF-β, SHH, and WNT and
their crosstalks. J Clin Med. 5:E412016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P
and Wu JW: Long noncoding RNA MALAT1 associates with the malignant
status and poor prognosis in glioma. Tumor Biol. 36:3355–3359.
2015. View Article : Google Scholar
|
|
15
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kros JM, Mustafa DM, Dekker LJ, Sillevis
Smitt PA, Luider TM and Zheng PP: Circulating glioma biomarkers.
Neuro Oncol. 17:343–360. 2015.PubMed/NCBI
|
|
17
|
Pan W, Gu W, Nagpal S, Gephart MH and
Quake SR: Brain tumor mutations detected in cerebral spinal fluid.
Clin Chem. 61:514–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Westphal M and Lamszus K: Circulating
biomarkers for gliomas. Nat Rev Neurol. 11:556–566. 2015.
View Article : Google Scholar : PubMed/NCBI
|