Introduction
Hepatocellular carcinoma (HCC), which often develops
as a result of hepatitis and liver cirrhosis, is one of the most
common types of cancer worldwide. The majority of patients are
diagnosed at an advanced stage, which results in poor survival
rates (1). The mortality rate for
patients with HCC is second only to that of lung cancer (2–4).
Forkhead box P3 (FOXP3) is a member of the
transcription factor fork head protein family, and it serves a
vital role in the generation of regulatory T cells (Tregs), which
function largely in immunosuppression (5,6).
Aberrant FOXP3 expression leads to autoimmune disease and benign or
malignant tumor formation. Previous studies regarding FOXP3
expression and function have been performed primarily in Tregs
(7,8). However, FOXP3 expression levels are
also elevated in a number of tumor cell types, including HCC
(9,10). Additionally, the expression levels
and cellular distribution of FOXP3 vary widely within the tumor and
its microenvironment (11–13). By inhibiting the secretion of certain
cytokines, such as transforming growth factor (TGF)-β1, TGF-β2, and
interleukin-10, FOXP3 controls the growth of melanoma tumors
(14). FOXP3+ Tregs
affect the development and progression of hepatocellular carcinoma
(15,16). FOXP3 may also act as a tumor
suppressor in HCC by regulating the TGF-β/Smad2/3 signaling pathway
(17).
Chemokines are a superfamily of small-molecules that
bind to G protein-coupled receptors on the cell membrane and
promote cell migration through the guanine nucleotide binding
protein-mediated signaling cascade (18). Chemokines are grouped into C-X-C
motif chemokine (CXC), CX3C, CC and C families based on their amino
acid sequences. CXC ligand (CXCL) 12 is a CXC chemotactic factor
that was originally identified in bone marrow stroma and termed
stromal cell-derived factor-1 (SDF-1) (19). CXC receptor (CXCR) 4 belongs to the G
protein-coupled receptor family and selectively binds CXCL12
(18,19). The CXCL12/CXCR4 signaling pathway
mediates proliferation, migration and migration in a variety of
tumor cells, including lung (20)
and gastric cancer (21), oral
squamous cell carcinoma (22), and
HCC (23,24). CXCR7 was originally cloned from the
cDNA library of the dog thyroid, and was considered to be an orphan
receptor (25); CXCL12 and CXCL11
were later identified as ligands of CXCR7 (26). Of note, the CXCL12/CXCR7 axis also
serves as a therapeutic target in the control of cell survival,
cell adhesion and tumor development (27).
In a study of the molecular mechanisms of HCC
progression, MHCC-97H cells are frequently used, as they share
pathological and genetic characteristics of malignant liver tumors
(28). In the present study,
MHCC-97H cells were transfected with FOXP3-short hairpin (sh)RNAs
to detect changes in cell proliferation, apoptosis and migration.
In addition, the effect of FOXP3 silencing on chemokine/chemokine
receptor expression was investigated.
Materials and methods
Cell culture
MHCC97-H cells were obtained from the Cell Bank of
The Chinese Academy of Sciences and cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; cat. no. 04-007-1A;
Biological Industries) and 100 U/ml penicillin and 100 µg/ml
streptomycin (Beijing Solarbio Science & Technology Co., Ltd.)
in 5% CO2 at 37°C. Cells were used for experiments at
60% confluence.
shRNA construction
The mRNA sequence of FOXP3 was obtained from the
NCBI database (https://www.ncbi.nlm.nih.gov/gene/20371), and three
shRNA sequences were designed accordingly (Table I). The sense strands included a
BamHI restriction site and the antisense strands included an
EcoRI restriction site. Double chains were formed and
inserted into pGreenPuro vectors (cat. no. SI505A-1-SBI; System
Biosciences). A total of three recombinant vectors were
constructed: Sh-FOXP3-1-pGreenPuro, sh-FOXP3-2-pGreenPuro and
sh-FOXP3-3. Single colonies were selected from plates harboring
MHCC97-H cells with either sh-FOXP3-1-pGreenPuro,
sh-FOXP3-2-pGreenPuro or sh-FOXP3-3-pGreenPuro. The vectors were
prepared with a Plasmid Minipreparation kit (cat. no. KL060;
Shanghai Kang Lang Biological technology Co., Ltd.) and detected
using 2% agarose gel electrophoresis with ethidium bromide
staining.
 | Table I.Sequences of FOXP3 shRNAs. |
Table I.
Sequences of FOXP3 shRNAs.
| Primer | Sequence
(5′→3′) |
|---|
| sh-FOXP3-F1 |
GATCCGCCACATTTCATGCACCAGCTCTCGAGAGCTGGTGCATGAAATGTGGCTTTTTG |
| sh-FOXP3-R1 |
AATTCGCCACATTTCATGCACCAGCTCTCGAGAGCTGGTGCATGAAATGTGGCG |
| sh-FOXP3-F2 |
GATCCGCACTGACCAAGGCTTCATCTCTCGAGAGATGAAGCCTTGGTCAGTGCTTTTTG |
| sh-FOXP3-R2 |
AATTCGCACTGACCAAGGCTTCATCTCTCGAGAGATGAAGCCTTGGTCAGTGCG |
| sh-FOXP3-F3 |
GATCCGCATGTTTGCCTTCTTCAGAACTCGAGTTCTGAAGAAGGCAAACATGCTTTTTG |
| sh-FOXP3-R3 |
AATTCGCATGTTTGCCTTCTTCAGAACTCGAGTTCTGAAGAAGGCAAACATGCG |
| sh-NC-F |
GATCCTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG |
| sh-NC-R |
AATTCAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAAG |
The experiments were performed in three groups: A
control group, a scrambled negative control (NC) group, and a
sh-FOXP3-1-pGreenPuro group. Using the Lipofectamine®
3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
DNA-liposome complexes were prepared at 4°C to a final volume of 1
µg/µl and added to MHCC97-H cells (1 µg/ml). Transfection was
performed for 1 h at room temperature. Further experiments were
performed 12, 24, 48 or 72 h post-transfection.
Reverse transcription quantitative-PCR
(RT-qPCR)
Total RNA was extracted using an Ultrapure RNA
extraction kit (CoWin Biosciences Co., Ltd.), and RNA purity was
assessed by measuring optical density (OD) at 280/260 nm. Total RNA
(1 µg) was reverse transcribed into cDNA using an Avian
Myeloblastosis Virus Reverse-Transcriptase kit (cat. no. KL041;
Shanghai Kang Lang Biological technology Co., Ltd.). qPCR was
performed in 25 µl sample volumes including, 9.5 µl RNase-Free
dH2O, 1 µl cDNA/DNA, 2 µl primers and 12.5 µl UltraSYBR
Mixture (cat. no. 00081405; CWBIO Corporation) with the following
thermocycling conditions: 35 cycles of denaturation at 94°C for 45
sec, annealing at 59°C for 45 sec and extension at 72°C for 60 sec.
The 2−ΔΔcq method was used for quantification (29). The primers used are presented in
Table II.
 | Table II.Primer sequences used for reverse
transcription--quantitative PCR. |
Table II.
Primer sequences used for reverse
transcription--quantitative PCR.
| Gene | Sequence
(5′→3′) |
|---|
| FOXP3 | F:
GTGGCATCATCCGACAAGG |
|
| R:
AGCGTGGCGTAGGTGAAAG |
| CXCR4 | F:
TAAAATCTTCCTGCCCACC |
|
| R:
CGCCAACATAGACCACCTT |
| CXCR7 | F:
CTCTTCGGCAGCATTTTCT |
|
| R:
CGTGACGGTCTTCAGGTAGTA |
| CXCL11 | F:
ATGTTCAAAAGAGGACGCTG |
|
| R:
GTTACTTGGGTACATTATGGAGG |
| CXCL12 | F:
ACTCCAAACTGTGCCCTTCA |
|
| R:
CCACTTTAGCTTCGGGTCAAT |
| GAPDH | F:
GAAGGTCGGAGTCAACGGAT |
|
| R:
CCTGGAAGATGGTGATGGG |
Western blotting
Following transfection, MHCC97-H cells
(5×103/ml) were collected for western blot analysis.
Protein was isolated using a protein isolation kit (ReadyPrep; GE
Healthcare Life Sciences) and concentrations were quantified using
a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol and 20 µg protein/lane was
separated using SDS-PAGE on 10% gel. The proteins were transferred
to a nitrocellulose membrane. Non-specific protein binding was
blocked with 5% non-fat milk at room temperature for 2 h. The
membranes were incubated with anti-GAPDH (1:1,000; cat. no.
TA802519; OriGene Technologies, Inc.), anti-CXCR4 (1:500; cat. no.
PA1237; Boster Biological Technology), anti-CXCR7 (1:500; cat. no.
ab138509; Abcam), anti-CXCL11 (1:500; cat. no. abs139437; Absin
Bioscience, Inc.) or anti-CXCL12 (1:500; cat. no. ab155090; Abcam)
antibodies at 4°C overnight. The membranes were rinsed with 0.1%
PBS + 0.1% Tween-20 and incubated with a horseradish
peroxidase-labeled goat anti-rabbit immunoglobulin G (H+L)
secondary antibody (1:100; ZB-2301; OriGene Technologies, Inc.) at
room temperature for 2 h. The signal was detected using an Enhanced
Chemiluminescence Detection kit (Thermo Fisher Scientific, Inc.)
and ChemiDoc™ XRS system (Bio-Rad Laboratories, Inc.). Densitometry
was performed using Quantity One software (version 1.4.6; Bio-Rad
Laboratories, Inc.).
Cell Counting Kit (CCK)-8 assay
Following transfection for 12, 24, 48 and 72 h, 10
µl of media containing CCK-8 (Gibco; Thermo Fisher Scientific,
Inc.) were added to each well (3×103/ml). An additional
4 h of incubation at 37°C was performed following each transfection
at 12, 24, 48 and 72 h. OD was detected using a microplate reader
(Thermo Fisher Scientific, Inc.) at 490 nm, and cell viability was
determined based on the OD values.
Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL)
MHCC97-H cells were fixed in 4% paraformaldehyde for
30 min at room temperature and incubated with PBS + 0.1% Tween-20
containing 0.3% Triton X-100 for 5 min at room temperature. TUNEL
solution was added to each well and the cells were incubated at
37°C for 60 min. Nuclei were stained with DAPI (5 mg/ml) for 3 min
at room temperature, following which slides were covered with
mounting medium (cat. no. P0126; Beyotime Institute of
Biotechnology). Images of the cells were captured using
fluorescence microscopy at 5 fields of view.
Transwell assay
Following transfection, 3×103 MHCC97-H
cells were seeded in the upper chamber of Transwell plates
(Hyclone; GE Healthcare Life Sciences) with serum-free DMEM. The
lower chamber contained DMEM with 10% FBS. At 48 h, the cells in
the lower chamber were fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 1% crystal violet (Beijing
Solarbio Science & Technology Co., Ltd.) for 5 min at room
temperature. Images were captured using a light microscope
(Magnification, ×200). At least five random fields of view in each
image were counted.
Statistical analysis
Data are presented as the mean ± standard deviation
with six independent replicates. Statistical analyses were
performed using SPSS software (version 17; SPSS, Inc.) and data
were analyzed by one-way analysis of variance followed by a
Bonferroni post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characterization of FOXP3 shRNAs
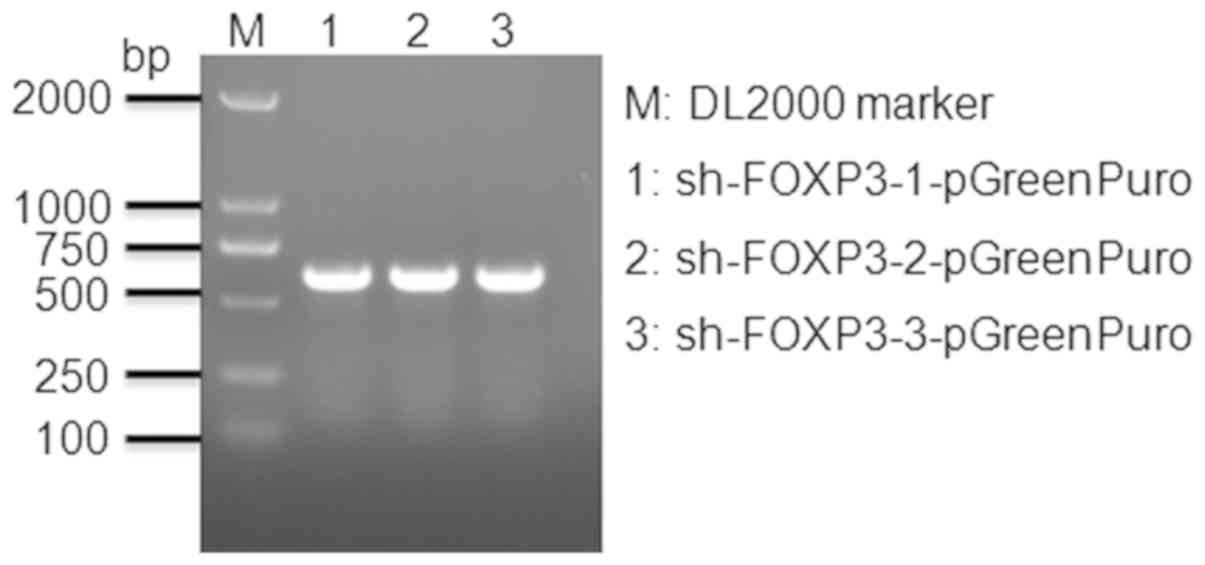
Vectors were detected by 2% agarose gel
electrophoresis (Fig. 1). The band
sizes reflected the predicted sizes of the constructs.
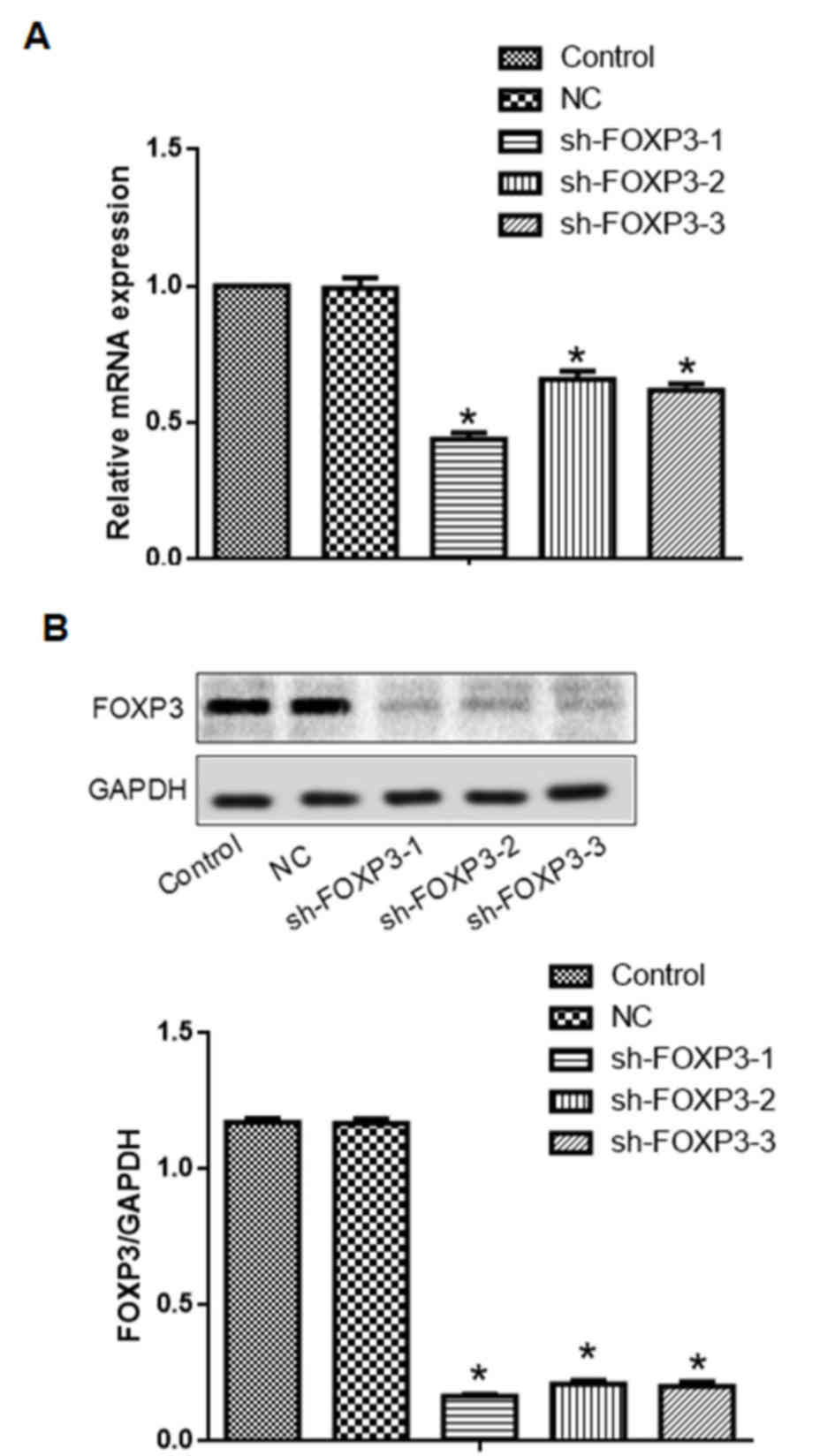
Transfection with either one of the three sh-FOXP3
sequences reduced FOXP3 mRNA and protein expression levels in
MHCC97-H cells (Fig. 2). The
sh-FOXP3-1 shRNA exhibited the strongest reduction of FOXP3 mRNA
and protein expression levels. Therefore, sh-FOXP3-1 was selected
for further experiments.
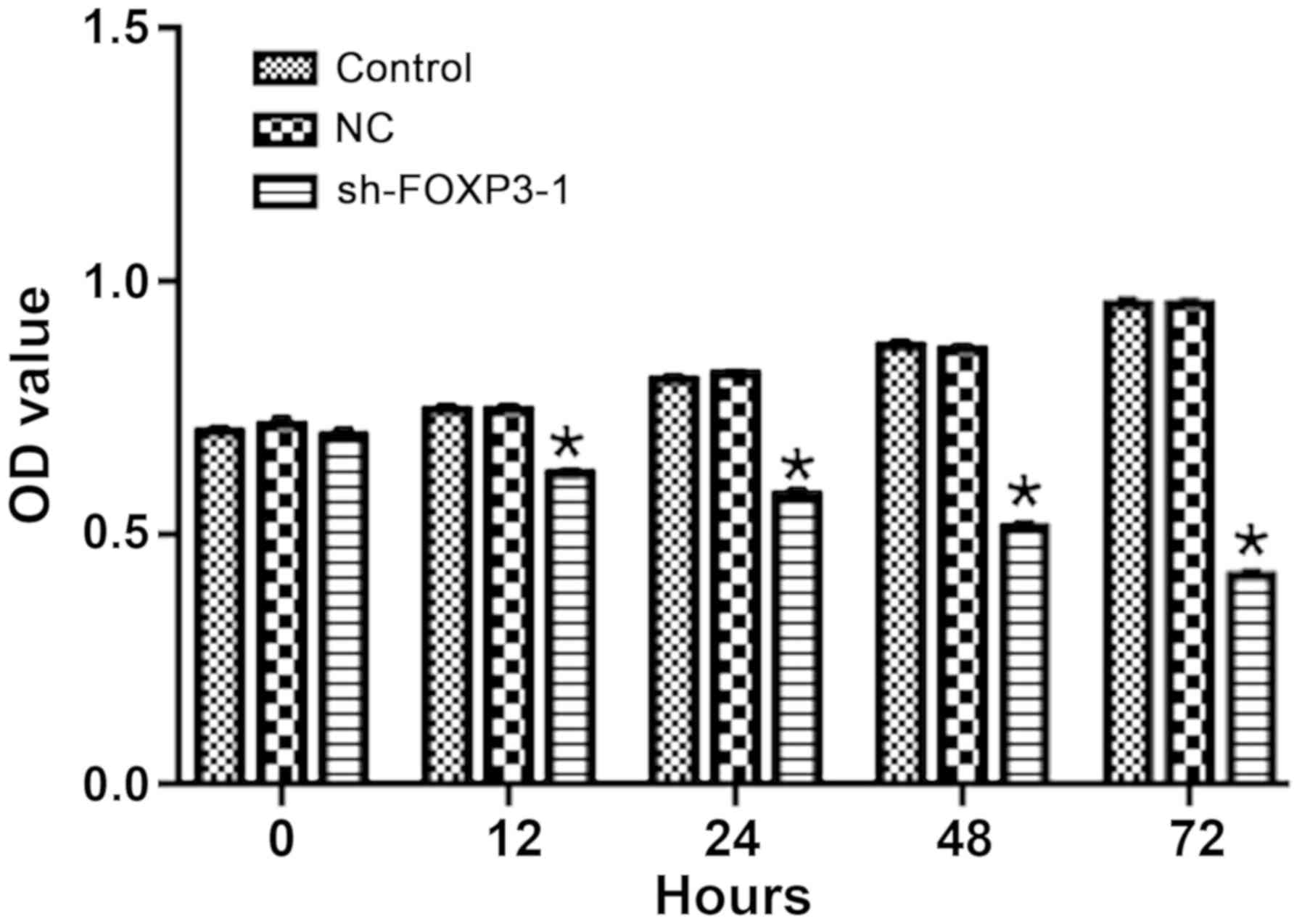
FOXP3 silencing inhibits cell
proliferation in MHCC97-H cells
The OD of MHCC-97H cells treated with sh-FOXP3-1 was
significantly lower compared with the negative control group
following transfection for 12, 24, 48 and 72 h (Fig. 3), which suggested that FOXP3
silencing may inhibit cell proliferation in MHCC97-H cells. The OD
values at 72 h were: Control, 0.96; NC, 0.96; sh-FOXP3-1, 0.42
[F(2, 15)=20.5; overall comparison between the 3 groups,
P<0.0001].
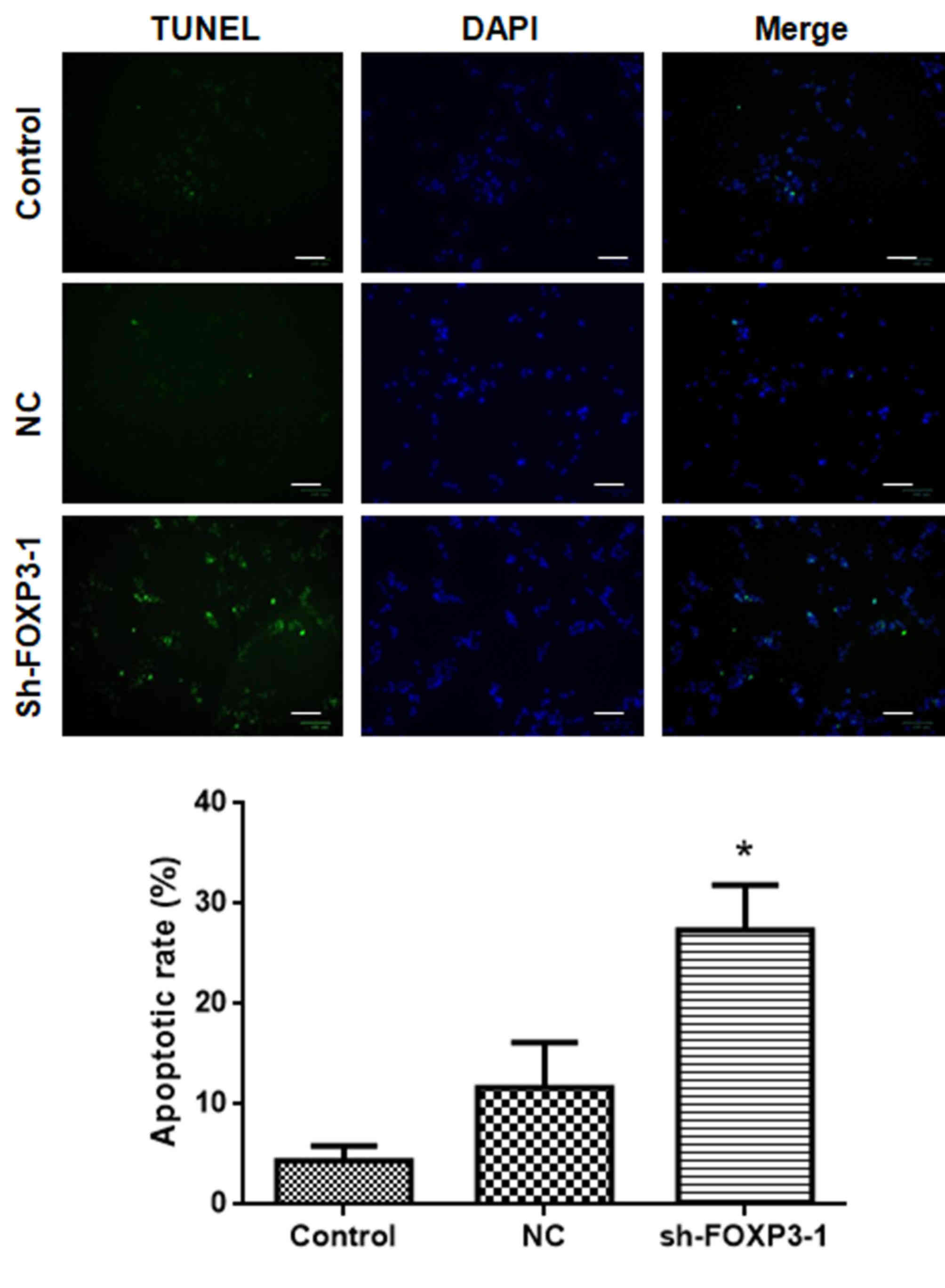
FOXP3 silencing promotes apoptosis in
MHCC97-H cells
The results of the TUNEL assay revealed that the
number of apoptotic cells following treatment with sh-FOXP3-1 was
significantly higher compared with that in the negative control
group (Fig. 4). The percentages of
apoptotic cells in each group were: Control, 4.0%; NC, 11.0%;
sh-FOXP3-1, 26.6% [F(2, 15)=20.5; P<0.05). These data suggested
that FOXP3 silencing may promote apoptosis in MHCC97-H cells.
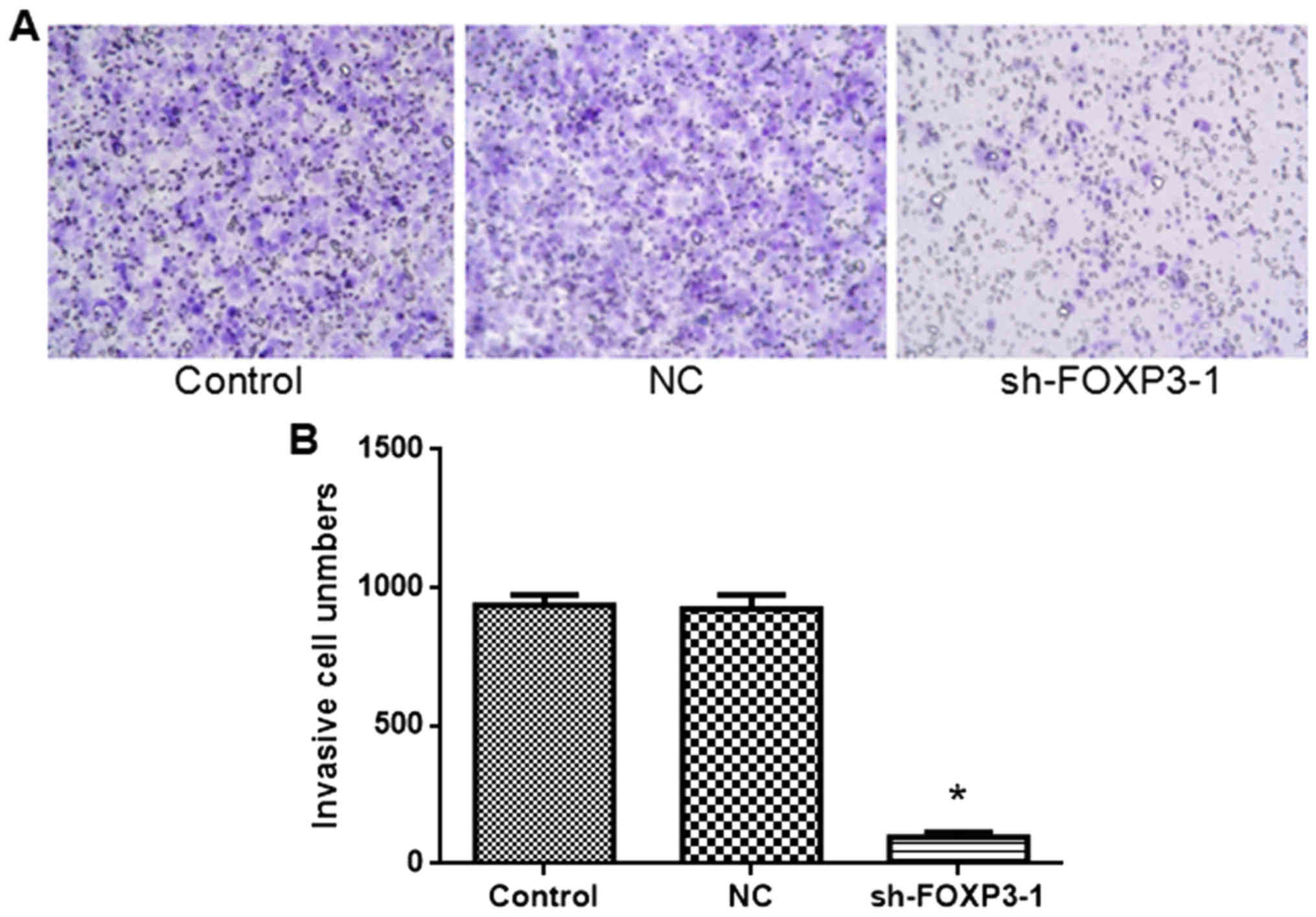
FOXP3 silencing inhibits the migration
of MHCC97-H cells
The rate of migration of cells transfected with
sh-FOXP3-1 was significantly lower compared with the negative
control group (Fig. 5). The number
of invasive cells was: Control, 936; NC, 923; sh-FOXP3-1, 96 [F(2,
15)=473, P<0.05). These data suggest that FOXP3 silencing may
inhibit migration in MHCC97-H cells.
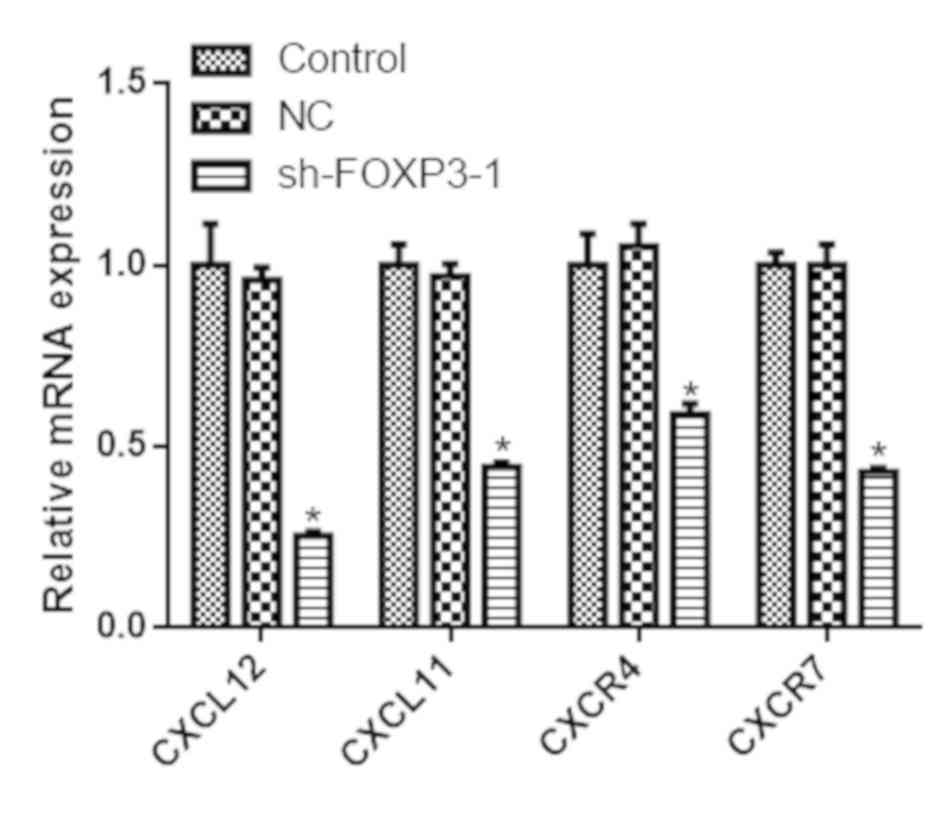
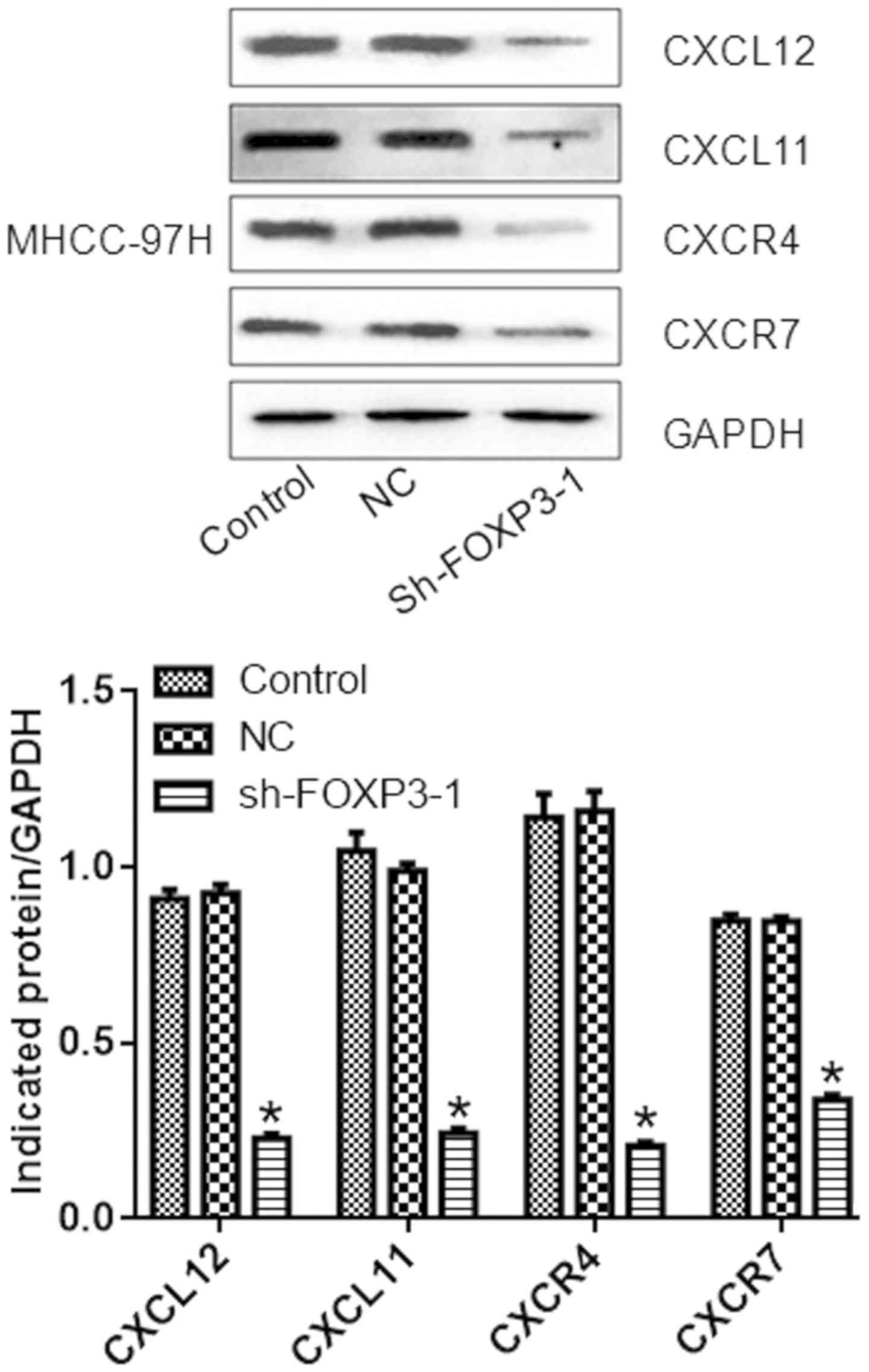
FOXP3 silencing reduces the expression
of chemokines and their receptors in MHCC97-H cells
The mRNA expression levels of CXCL12, CXCL11, CXCR4
and CXCR7 were reduced in cells transfected with sh-FOXP3-1
compared with the negative control group (P<0.05; Fig. 6). Similarly, the protein levels of
CXCL12, CXCL11, CXCR4, and CXCR7 were reduced in cells treated with
sh-FOXP3-1 compared with the NC group (P<0.05; Fig. 7).
Discussion
HCC is primarily treated with radiotherapy and liver
transplantation; however, the rates of recurrence are ~54% at a
median time of 22 months from primary resection (30,31).
FOXP3, a nuclear transcription factor linked to the X chromosome
that belongs to the forkhead/winged helix transcription factor
family, was originally identified by cloning the Scurfin gene. The
head domain of FOXP3 binds to DNA-specific loci to regulate the
activation and expression of its target genes (5,6). FOXP3
consists of 431 amino acids and its functional structure includes a
forkhead DNA binding zone at the C terminal, a C2H2 zinc finger
structure at the N terminal, and a leucine zipper sequence
(32). In humans, FOXP3 is primarily
responsible for the immunosuppressive function of Tregs, and is
frequently used as a Treg-specific marker. It serves an important
role in regulating the growth and function of CD4+
CD25+ Tregs, as well as maintaining immune tolerance and
the stability of the immune responses by inducing the development
of CD4+ CD25+ Tregs, controlling the
production of CD4+ Tregs and determining Treg inhibitory
function (33).
FOXP3 is expressed in various types of tumor cells
(34). The expression pattern of
FOXP3, including expression levels, subcellular localization and
subtypes, in tumor cells is different compared with that in normal
cells or Tregs (34). Cunha et
al (35) highlighted the
complexity of FOXP3 expression in tumor cells, as well as the
diverse roles it serves in tumor progression. Wang et al
(36) previously characterized 10
HCC cell lines with high expression levels of FOXP3. Niu et
al (37) demonstrated that
melanoma cells underwent immune escape by expressing FOXP3.
Silencing of FOXP3 by FOXP3-siRNA has been demonstrated to mitigate
the expression of B7-H1 and TGF-β, whereas FOXP3 overexpression
increased the expression of these molecules (38). Co-culturing FOXP3+
melanoma cells with CD4+ CD25− Tregs strongly
inhibited the proliferation of T cells, which was partially
reversed by the specific silencing of FOXP3 and effectively
enhanced by FOXP3 overexpression (39). Therefore, FOXP3 not only serves an
important role in Treg function, but may also modulate tumor cell
behavior directly.
In the present study, three shRNAs were constructed
to silence FOXP3 expression in HCC cells; sh-FOXP3-1 exhibited the
strongest silencing effects in an HCC cell line. The results
indicated that silencing of FOXP3 expression may inhibit the
proliferation of HCC cells. Apoptosis was also promoted in response
to sh-FOXP3-1, whereas migration was inhibited. Collectively, these
data demonstrate that the silencing of FOXP3 in tumor cells may
regulate HCC proliferation, migration and apoptosis.
Compared with CXCL12/CXCR4, the CXCL12/CXCR7
signaling has a more pronounced effect on HCC progression (40). Downregulation of CXCR7 results in the
reduction of HCC cell proliferation, as well as the inhibition of
lung metastasis (41). Sutton et
al (42) demonstrated that
CXCL12 stimulates the proliferation, migration and migration of HCC
cells, and that the combination of CXCL12 and CXCR4 induces the
aggregation and redistribution of cytoskeletal proteins, which
enhances the rates of migration and migration. In addition, CXCR7
binds CXCL11 and CXCL12 with high affinity, thus serving important
roles in the progression of HCC (43). In the present study, the mRNA and
protein levels of CXCL12, CXCL11, CXCR4 and CXCR7 were
significantly decreased in MHCC97-H cells treated with sh-FOXP3-1
compared with the control groups. These results suggested that
FOXP3 may serve an important role in the growth and metastasis of
HCC, potentially through the chemokine/chemokine receptor axis.
This may be one of the primary mechanisms underlying the FOXP3
silencing-mediated regulation of tumor growth, inhibition of
oncogene transcription and induction of tumor suppressor gene
expression.
FOXP3 binds to the transcription sites of the CXCR4
and CXCR7 genes (44); CXCR4 and
CXCR7 are associated with tumor migration and metastasis (24). Overbeck-Zubrzycka et al
(45) have demonstrated that FOXP3
affects tumor migration and metastasis by modulating the
CXCL12/CXCR4 pathway. In the present study, FOXP3 silencing
prevented tumor migration potentially through inhibition of CXCR7.
Pre-clinical studies have also demonstrated that CXCL12/CXCR4 and
CXCL12/CXCR7 inhibitors display potent anti-tumor properties
(46), which suggests that
chemokines and their receptors represent new potential targets for
cancer therapy.
In the present study, FOXP3 silencing inhibited the
expression of chemokines and chemokine receptors associated with
cell proliferation, migration and apoptosis in an HCC cell line.
However, a direct link between tumor progression and the activities
of chemokines and chemokine receptors in response to FOXP3
silencing has not been established. Future studies will focus on
detecting the downstream molecules of chemokines and their
receptors that are regulated by FOXP3.
In conclusion, FOXP3 silencing may inhibit the
expression of chemokines and chemokine receptors, including
CXCL12/CXCR4, CXCL12/CXCR7 and CXCL11/CXCR7, and may be associated
with the inhibition of cell proliferation and migration, as well as
the induction of apoptosis. The inhibition of chemokine and
chemokine receptor expression may represent an important mechanism
underlying the effects of FOXP3 silencing in HCC cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Development Fund Project of Shenzhen (grant nos.
JCYJ20150403091443302 and JCYJ20160428164539088), the Sanming
Project of Medicine in Shenzhen (grant no. SZSM201612021), the
Science and Technology Developing Project of Guangdong Province
(grant no. 2017B090904010) and Research Foundation of Peking
University Shenzhen Hospital (grant no. JCYJ2018002).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XO, XL and JL conceived and designed the study. XO,
GZ, PT, JC, ZL, YX, AW and XL performed the experiments. XO, XL and
JL wrote the paper. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singh AK, Kumar R and Pandey AK:
Hepatocellular carcinoma: Causes, mechanism of progression and
biomarkers. Curr Chem Genom Transl Med. 12:9–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Glantzounis GK, Kyrochristos ID, Ziogas
DE, Lykoudis EG and Roukos DH: Novel translational therapeutic
strategy by sequencing primary liver cancer genomes. Future Oncol.
13:1049–1052. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang B, Petrick JL, Kelly SP, Graubard BI,
Freedman ND and McGlynn KA: Adiposity across the adult life course
and incidence of primary liver cancer: The NIH-AARP cohort. Int J
Cancer. 141:271–278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lan L, Zhao F, Cai Y, Wu RX and Meng Q:
Epidemiological analysis on mortality of cancer in China, 2015.
Zhonghua Liu Xing Bing Xue Za Zhi. 39:32–34. 2018.(In Chinese).
PubMed/NCBI
|
|
5
|
Sakaguchi S, Yamaguchi T, Nomura T and Ono
M: Regulatory T cells and immune tolerance. Cell. 133:775–787.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bennett CL, Christie J, Ramsdell F,
Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT,
Chance PF and Ochs HD: The immune dysregulation,
polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused
by mutations of FOXP3. Nat Genet. 27:20–21. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malik S and Awasthi A: Transcriptional
control of Th9 cells: Role of foxo1 in interleukin-9 induction.
Front Immunol. 9:9952018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wada M, Tsuchikawa T, Kyogoku N, Abiko T,
Miyauchi K, Takeuchi S, Kuwatani T, Shichinohe T, Miyahara Y,
Kageyama S, et al: Clinical implications of
CD4+CD25+Foxp3+Regulatory T cell
frequencies after CHP--MAGE-A4 cancer vaccination. Anticancer Res.
38:1435–1444. 2018.PubMed/NCBI
|
|
9
|
Lin SZ, Chen KJ, Xu ZY, Chen H, Zhou L,
Xie HY and Zheng SS: Prediction of recurrence and survival in
hepatocellular carcinoma based on two cox models mainly determined
by FoxP3+ regulatory T cells. Cancer Prev Res (Phila). 6:594–602.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Triulzi T, Tagliabue E, Balsari A and
Casalini P: FOXP3 expression in tumor cells and implications for
cancer progression. J Cell Physiol. 228:30–35. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McInnes N, Sadlon TJ, Brown CY, Pederson
S, Beyer M, Schultze JL, McColl S, Goodall GJ and Barry SC: FOXP3
and FOXP3-regulated microRNAs suppress SATB1 in breast cancer
cells. Oncogene. 31:1045–1054. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Merlo A, Casalini P, Carcangiu ML,
Malventano C, Triulzi T, Mènard S, Tagliabue E and Balsari A: FOXP3
expression and overall survival in breast cancer. J Clin Oncol.
27:1746–1752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ebert LM, Tan BS, Browning J, Svobodova S,
Russell SE, Kirkpatrick N, Gedye C, Moss D, Ng SP, MacGregor D, et
al: The regulatory T cell-associated transcription factor FoxP3 is
expressed by tumor cells. Cancer Res. 68:3001–3009. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen DJ, Li XS, Zhao H, Shi XL, Zhang HH,
Fan ZY, Yao YM and DU N: Inhibitory effect of lentiviral-mediated
RNA on the expression of Foxp3 protein in melanoma cells. Xi Bao Yu
Fen Zi Mian Yi Xue Za Zhi. 28:337–339. 2012.(In Chinese).
PubMed/NCBI
|
|
15
|
Kobayashi N, Hiraoka N, Yamagami W, Ojima
H, Kanai Y, Kosuge T, Nakajima A and Hirohashi S: FOXP3+ regulatory
T cells affect the development and progression of
hepatocarcinogenesis. Clin Cancer Res. 13:902–911. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang WH, Jiang CL, Yan W, Zhang YH, Yang
JT, Zhang C, Yan B, Zhang W, Han W, Wang JZ and Zhang YQ: FOXP3
expression and clinical characteristics of hepatocellular
carcinoma. World J Gastroenterol. 16:5502–5509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi JY, Ma LJ, Zhang JW, Duan M, Ding ZB,
Yang LX, Cao Y, Zhou J, Fan J, Zhang X, et al: FOXP3 Is a HCC
suppressor gene and Acts through regulating the TGF-β/Smad2/3
signaling pathway. BMC Cancer. 17:6482017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagasawa T: CXC chemokine ligand 12
(CXCL12) and its receptor CXCR4. J Mol Med. 92:433–439. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Dai LI and Zhou R: Association
between preeclampsia and the CXC chemokine family (Review). Exp
Ther Med. 9:1572–1576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou XM, He L, Hou G, Jiang B, Wang YH and
Zhao L: Clinicopathological significance of CXCR4 in non-small cell
lung cancer. Drug Des Devel Ther. 9:1349–1358. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Izumi D, Ishimoto T, Miyake K, Sugihara H,
Eto K, Sawayama H, Yasuda T, Kiyozumi Y, Kaida T, Kurashige J, et
al: CXCL12/CXCR4 activation by cancer-associated fibroblasts
promotes integrin β1 clustering and invasiveness in gastric cancer.
Int J Cancer. 138:1207–1219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rave-Frank M, Tehrany N, Kitz J, Leu M,
Weber HE, Burfeind P, Schliephake H, Canis M, Beissbarth T,
Reichardt HM and Wolff HA: Prognostic value of CXCL12 and CXCR4 in
inoperable head and neck squamous cell carcinoma. Strahlenther
Onkol. 192:47–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fontanella R, Pelagalli A, Nardelli A,
D'Alterio C, Ieranò C, Cerchia L, Lucarelli E, Scala S and Zannetti
A: A novel antagonist of CXCR4 prevents bone marrow-derived
mesenchymal stem cell-mediated osteosarcoma and hepatocellular
carcinoma cell migration and invasion. Cancer Lett. 370:100–107.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi JY, Yang LX, Wang ZC, Wang LY, Zhou J,
Wang XY, Shi GM, Ding ZB, Ke AW, Dai Z, et al: CC chemokine
receptor-like 1 functions as a tumour suppressor by impairing
CCR7-related chemotaxis in hepatocellular carcinoma. J Pathol.
235:546–558. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hao M, Zheng J, Hou K and Wang J, Chen X,
Lu X, Bo J, Xu C, Shen K and Wang J: Role of chemokine receptor
CXCR7 in bladder cancer progression. Biochem Pharmacol. 84:204–214.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benredjem B, Girard M, Rhainds D, St-Onge
G and Heveker N: Mutational analysis of atypical chemokine receptor
3 (ACKR3/CXCR7) interaction with its chemokine ligands CXCL11 and
CXCL12. J Biol Chem. 292:31–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burns JM, Summers BC, Wang Y, Melikian A,
Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ,
et al: A novel chemokine receptor for SDF-1 and I-TAC involved in
cell survival, cell adhesion, and tumor development. J Exp Med.
203:2201–2213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Lu Z, Liang Z, Ji D, Zhang P, Liu Q,
Zheng X and Yao Y: Metastasis-associated in colon cancer-1 is
associated with poor prognosis in hepatocellular carcinoma, partly
by promoting proliferation through enhanced glucose metabolism. Mol
Med Rep. 12:426–434. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wiedmann MW and Mossner J: Molecular
targeted therapy of hepatocellular carcinoma-results of the first
clinical studies. Curr Cancer Drug Targets. 11:714–733. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lopes JE, Torgerson TR, Schubert LA,
Anover SD, Ocheltree EL, Ochs HD and Ziegler SF: Analysis of FOXP3
reveals multiple domains required for its function as a
transcriptional repressor. J Immunol. 177:3133–3142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rudensky AY: Regulatory T cells and Foxp3.
Immunol Rev. 241:260–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grimmig T, Kim M, Germer CT, Gasser M and
Waaga-Gasser AM: The role of FOXP3 in disease progression in
colorectal cancer patients. Oncoimmunology. 2:e245212013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cunha LL, Morari EC, Nonogaki S, Soares
FA, Vassallo J and Ward LS: Foxp3 expression is associated with
aggressiveness in differentiated thyroid carcinomas. Clinics.
67:483–488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang L, Mei M, Sun X, et al: Expression
and significance of Foxp3 in hepatocellular carcinoma. Shandong Med
J. 48:8–11. 2010.(In Chinese).
|
|
37
|
Niu J, Jiang C, Li C, Liu L, Li K, Jian Z
and Gao T: Foxp3 expression in melanoma cells as a possible
mechanism of resistance to immune destruction. Cancer Immunol
Immunother. 60:1109–1118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Geng Y, Wang H, Lu C, Li Q, Xu B, Jiang J
and Wu C: Expression of costimulatory molecules B7-H1, B7-H4 and
Foxp3+ Tregs in gastric cancer and its clinical significance. Int J
Clin Oncol. 20:273–281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fu S, Zhang N, Yopp AC, Chen D, Mao M,
Chen D, Zhang H, Ding Y and Bromberg JS: TGF-beta induces Foxp3 +
T-regulatory cells from CD4+CD25-precursors. Am J Transplant.
4:1614–1627. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng K, Li HY, Su XL, Wang XY, Tian T, Li
F and Ren GS: Chemokine receptor CXCR7 regulates the invasion,
angiogenesis and tumor growth of human hepatocellular carcinoma
cells. J Exp Clin Cancer Res. 29:312010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xue TC, Chen RX, Han D, Chen J, Xue Q, Gao
DM, Sun RX, Tang ZY and Ye SL: Down-regulation of CXCR7 inhibits
the growth and lung metastasis of human hepatocellular carcinoma
cells with highly metastatic potential. Exp Ther Med. 3:117–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sutton A, Friand V, Brulé-Donneger S,
Chaigneau T, Ziol M, Sainte-Catherine O, Poiré A, Saffar L, Kraemer
M, Vassy J, et al: Stromal cell-derived factor-1/chemokine (C-X-C
motif) ligand 12 stimulates human hepatoma cell growth, migration,
and invasion. Mol Cancer Res. 5:21–33. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Monnier J, Boissan M, L'Helgoualc'h A,
Lacombe ML, Turlin B, Zucman-Rossi J, Théret N, Piquet-Pellorce C
and Samson M: CXCR7 is up-regulated in human and murine
hepatocellular carcinoma and is specifically expressed by
endothelial cells. Eur J Cancer. 48:138–148. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y and Rudensky AY: Foxp3 in control
of the regulatory T cell lineage. Nat Immunol. 8:457–462. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Overbeck-Zubrzycka D, Ali S, Kirby J and
Lennard T: FOXP3 transcription factor regulates metastatic spread
of breast cancer via control of expression of CXCR4 chemokine
receptor. Br J Surg. 98:842011.PubMed/NCBI
|
|
46
|
Duda DG, Kozin SV, Kirkpatrick ND, Xu L,
Fukumura D and Jain RK: CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway
inhibition: An emerging sensitizer for anticancer therapies? Clin
Cancer Res. 17:2074–2080. 2011. View Article : Google Scholar : PubMed/NCBI
|