Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide, accounting for ~1.4 million incident cases and
600,000 mortalities every year (1).
The low early diagnosis rate, high recurrence and metastasis rates,
and lack of effective treatment are responsible for the
CRC-associated mortality statistics (2). Therefore, the identification of the
molecular mechanisms involved in the progression of CRC would be
useful in providing targets for cancer detection, prevention and
therapy, and ultimately improving patient prognosis.
MicroRNAs (miRNAs/miRs) are a small non-coding RNA
molecules present in plants, animals and certain viruses, and they
serve a role in RNA silencing and post-transcriptional regulation
of gene expression (3,4). In animal cells, the miRNA gene first
transcribes a longer primary miRNA in the nucleus, where it is
processed by the Drosha enzyme to form a 60–70 nucleotide hairpin
precursor miRNA. This is transported to the cytoplasm with the help
of the nuclear exportin-5 complex, and cleaved by the Dicer protein
to generate a mature miRNA of 18–25 nucleotides. Mature miRNAs and
a number of proteins form RNA silencing complexes, which inhibit
transcription or translation of the target gene through the
activity of sequences complementary to the 3′untranslated region of
the target gene mRNA that are contained within the complex
(5,6). It has been established that miRNAs are
important regulators of apoptosis, cell proliferation and
tumorigenesis (7–10).
miR-34a is considered to be a potential tumor
suppressor miRNA based on its frequent dysregulation in cancer
tissues (11) and its ability to
regulate the expression of numerous genes, including NOTCH1,
apoptosis regulator BCL2, MYC proto- oncogene, MET proto-oncogene,
cyclin-dependent kinases 4/6 and CD44 antigen (12–14).
miR-34a is frequently downregulated in multiple types of cancer and
is an independent prognostic indicator in various types of cancer,
including colon, breast, lung and gastric cancer (15–18). In
addition, it is significantly downregulated in the serum and tumor
tissue in CRC (19,20). However, the exact molecular mechanism
of miR-34a in the development of CRC has not been fully elucidated.
In the present study, miR-34a was inhibited or overexpressed in CRC
cells, revealing that it inhibited cell migration, invasion and
proliferation by regulating vimentin and early growth response
protein 1 (EGR1).
Materials and methods
Cell lines and culture
Human CRC cell lines SW480, SW620, RKO, LoVo and
SW1116 were provided by Sun Yat-sen University (Guangzhou, China)
and grown in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Sera Gold), 100 U/ml penicillin G
and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
The human CRC Caco-2 cell line and the human colonic epithelial
NCM460 cell line were provided by Sun Yat-sen University and grown
in Dulbecco's modified Eagle's medium (Gibco, Thermo Fisher
Scientific, Inc.) with 10% FBS. The cells were cultured at 37°C in
a 5% CO2 incubator. Detailed information on cell lines
is summarized in Table SI.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the cultured cancer cells was
extracted using TRIzol® reagent (Invitrogen, Thermo
Fisher Scientific, Inc.). cDNA was then synthesized from total RNA
or small RNA using the Prime-Script RT reagent kit (Takara Bio,
Inc.) or via the stem-loop method using the miRNA First Strand cDNA
Synthesis kit (Sangon Biotech Co., Ltd., Shanghai, China). The qPCR
was performed using 2X SG Fast qPCR Master Mix (Sangon Biotech Co.,
Ltd.) according to the manufacturer's protocol. The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95°C for 3 min, followed by 40 cycles at 95°C for 5 sec and at
60°C for 30 sec. The following primers were utilized for qPCR:
miR-34a forward, 5′-CCGCGTGGCAGTGTCTTAGCT-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′ (each, Sangon Biotech Co.,
Ltd.). For each case, ≥3 biological replicates were performed. The
qPCR analysis was performed on a PikoReal™ (Thermo Fisher
Scientific, Inc.). The miRNA was quantified with the
2−ΔΔCq method and normalized against U6 (21).
Oligonucleotide transient
transfection
SW480 and SW620 cells were seeded into 6-well plates
at a density of 3×105 cell per well. Prior to
transfection, cells were plated at 70–80% confluence. miR-34a
mimic, inhibitor and corresponding negative control were chemically
synthesized by Guangzhou RiboBio (Guangzhou RiboBio Co., Ltd.). The
mimic and inhibitor sequences were as follows: miR-34a mimic,
5′-UGGCAGUGUCUUAGCUGGUUGU-3′; control miR-34a mimic,
5′-UUUGUACUACACAAAAGUACUG-3′; miR-34a inhibitor,
5′-ACAACCAGCUAAGACACUGCCA-3′ and control miR-34a inhibitor,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′. The SW620 cells were transfected
with 50 nM miR-34a mimic (named miR-34a) or mimic negative control
(NC) (Guangzhou RiboBio Co., Ltd.) by nucleofection using
Lipofectamine® 3000 (Invitrogen, Thermo Fisher
Scientific, Inc.), and the SW480 cells were transfected with 100 nM
miR-34a inhibitor (inmiR-34a) or inhibitor NC, according to the
manufacturer's protocol. At 24 h post-transfection, RT-qPCR was
performed to verify the transfection efficiency.
Western blot analysis
SW480 and SW620 cells were collected 48 h after the
aforementioned transfections. Protein was extracted from cells
using a mixture of RIPA lysate (Beyotime Institute of Technology),
PMSF (Beyotime Institute of Technology) and 50× phosphatase
inhibitor (Applygen Technologies, Inc.) at a ratio of 100:1:2.
Protein concentrations were then quantified using a BCA Protein
Assay kit (Thermo Fisher Scientific, Inc.). A total of 40 µg
protein was separated by 10% SDS-PAGE (Beyotime Institute of
Technology) and subsequently transferred to polyvinylidene
difluoride membranes (Merck KGaA). The membranes were blocked with
low-fat milk (5%) at room temperature for 1 h, and then incubated
at 4°C overnight with primary antibodies against EGR1 (cat. no.
4153; 1:1,000; Cell Signaling Technology, Inc.), vimentin (cat. no.
5741; 1:1,000; Cell Signaling Technology, Inc.) and GAPDH (cat. no.
5174; 1:1,000; Cell Signaling Technology, Inc.). Subsequently, the
membranes were washed with TBST 3 times and incubated with a
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (cat. no. A0208; 1:1,000; Beyotime Institute of
Biotechnology) for 1 h at room temperature. Following 3 washes with
TBST, the results were detected using an enhanced chemiluminescence
system on a FluorChem R (ProteinSimple). Protein expression was
quantified using ImageJ software (version 1.47; National Institutes
of Health).
Wound healing assay
SW480 and SW620 cells were cultured in 6-well plate
following transfection until the confluence reached 90%. The cell
monolayers were scratched with micropipette tips to form a gap. The
cell culture surface was washed 3 times with PBS to remove cellular
debris and incubated in RPMI-1640 medium with 2% FBS for 48 h. The
wound closures were compared at 0 and 48 h. Images were captured
using an optical microscope (magnification, ×100; Nikon
Corporation) and analyzed with ImageJ software (version 1.47;
National Institutes of Health).
Transwell cell migration assay
Following 12 h of transfection, SW480 and SW620
cells were treated with 0.25% trypsin (Beijing Solarbio Science
& Technology Co., Ltd.) and suspended in serum-free RPMI-1640
medium at a density of 3×105 cells/ml. A total of 200 µl
cell suspension was placed in the top chamber of a 2-chamber Costar
Transwell 24-well plates (8-µm pores; Corning Inc.) and 600 µl
RPMI-1640 medium containing 15% FBS was added in the lower chamber.
The cells were cultured at 37°C for 48 h. The cells on the surface
of the upper chamber were swabbed and those under the surface of
the lower chamber were stained with crystal violet (0.1%) at room
temperature for 20 min. Cell migration was evaluated by counting
the cells that had migrated into the filters using an optical
microscope (magnification, ×100; Nikon Corporation).
Transwell cell invasion assay
The invasion assay was performed using the
aforementioned migration assay protocol, but following the addition
of 50 µl BD Matrigel™ (BD Biosciences) into each Transwell upper
chamber, which was then placed in a 37°C incubator for 2 h to
solidify. The tumor cell invasive capacity was then assessed as
aforementioned.
Colony formation assay
Following transfection for 12 h, the SW480 and SW620
cells were transferred to 6-well plates with RPMI-1640 medium with
10% FBS (300 cells per well). The cells were cultured at 37°C and
5% CO2 for 2 weeks. Subsequently, the cell colonies were
fixed with pure methanol for 15 min at room temperature and stained
with 0.1% crystal violet for 20 min at room temperature, and the
colonies were counted.
MTT assay
Following transfection for 12 h, SW480 and SW620
cells in each group were transferred to a 96-well plate and
cultured at 37°C, and 5% CO2 for 72 h. The MTT assay was
performed at 0, 24, 48 and 72 h. At each time point, the medium was
replaced and 20 µl MTT (5 mg/ml) was added to each well. Following
incubation at 37°C for 4 h, the medium was discarded and 150 µl
dimethyl sulfoxide (Beijing Solarbio Science & Technology Co.,
Ltd.) was added to each well. The optical density was measured at
490 nm.
Statistical analysis
Statistical analysis was performed using SPSS
version 17.0 (SPSS Inc.) and GraphPad Prism version 6.0 (GraphPad
Software, Inc.). The data are presented as mean ± standard
deviation. A Student's t-test was used for comparison between two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
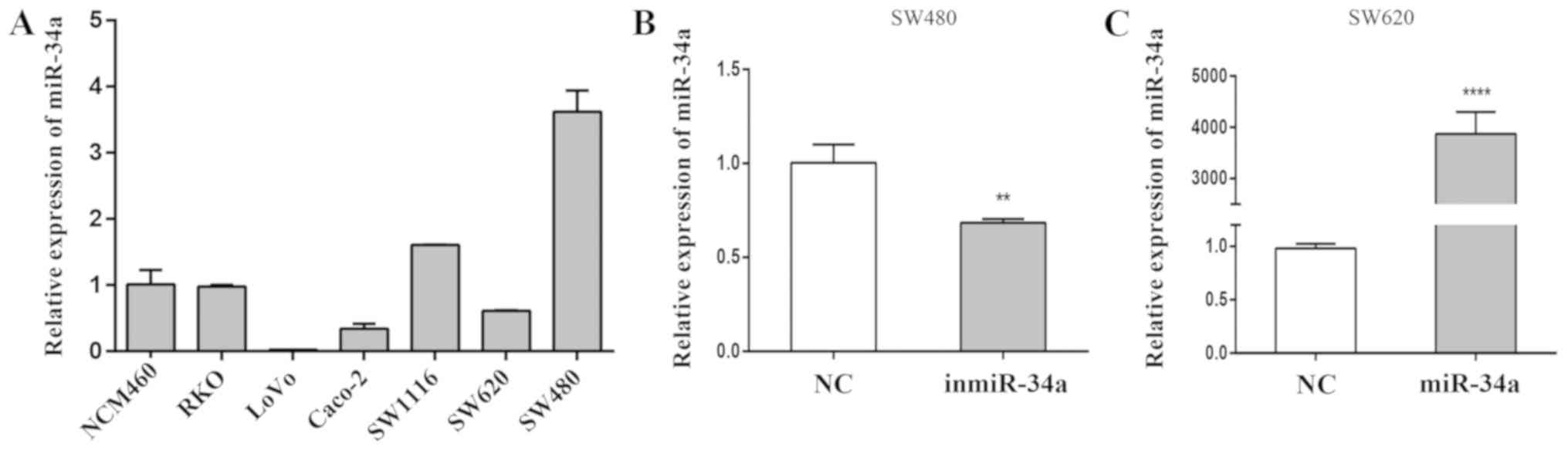
miR-34a is deregulated in highly
invasive cells, and may be successfully transfected into SW620 and
SW480 cells
RT-qPCR was used to detect the levels of miR-34a in
CRC and normal colonic epithelial cells. The expression of miR-34a
in highly invasive CRC SW620, RKO and LoVo cell lines is presented
in Fig. 1A. The SW620 cell line was
derived from a lymph node metastasis of a primary colon tumor,
whereas cells from the primary colon tumor in the same patient were
used to establish the SW480 cell line. These 2 cell lines were
selected for subsequent experiments to observe any notable
contrasts. At 24 h post-transfection, RT-qPCR was performed to
verify the transfection efficiency. Compared with the equivalent NC
group, differences were observed in the levels of relative miR-34a
expression following transfection of the inmiR-34a in the SW480
cells (P<0.01; Fig. 1B), and
miR-34a mimic into the SW620 cells (P<0.001; Fig. 1C).
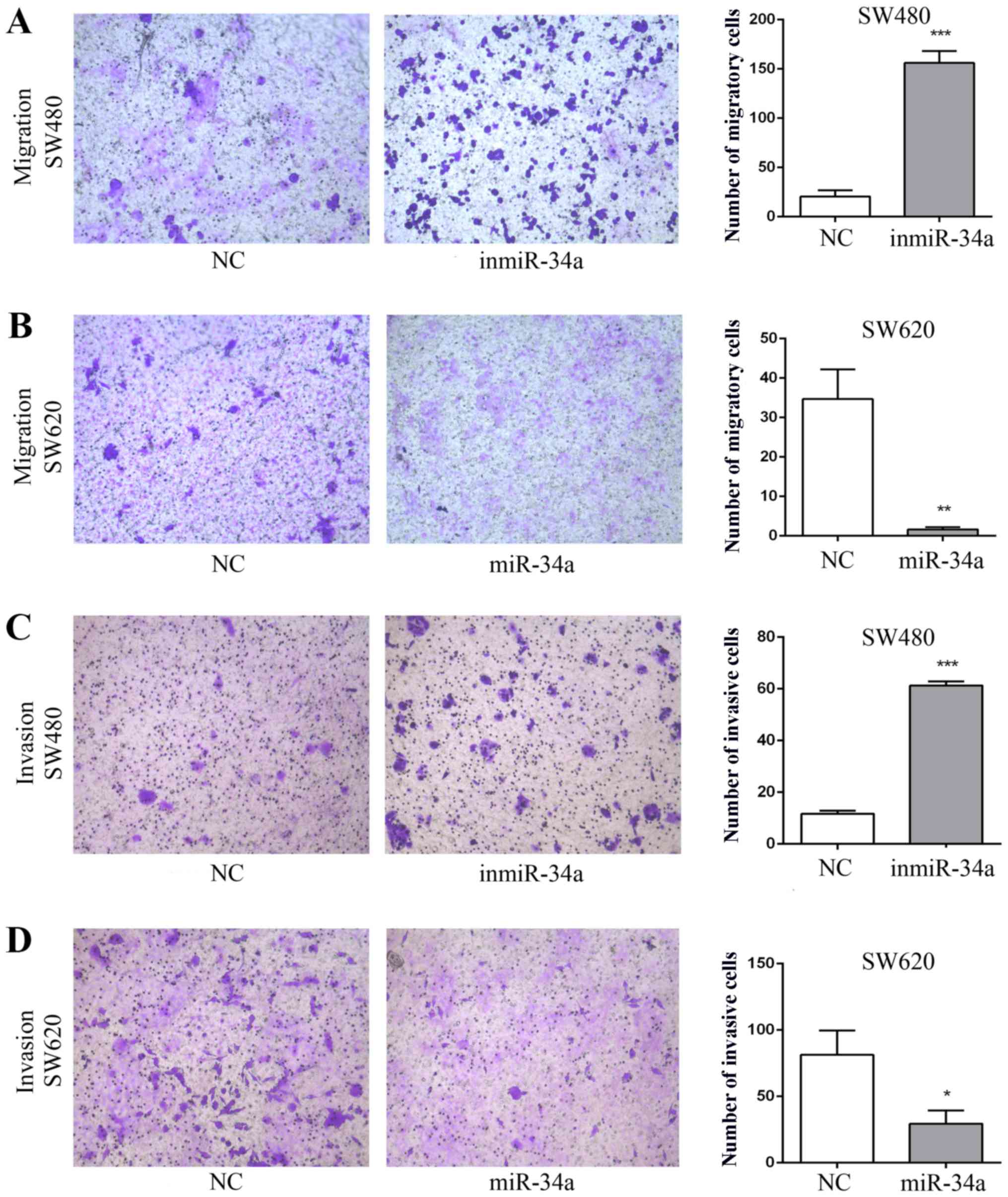
miR-34a suppresses the migration and
invasion of CRC cells
The effect of miR-34a on the migratory and invasive
abilities of SW480 and SW620 cell was assessed. The migratory
ability of the inmiR-34a group was significantly more marked
compared with that of the NC group (P<0.001; Fig. 2A). Furthermore, the overexpression of
miR-34a in SW620 cells attenuated cell migration (P<0.05;
Fig. 2B). The inmiR-34a group of the
SW480 cells exhibited a significant increase in cell invasion
(P<0.001; Fig. 2C), whereas the
increased levels of miR-34a in SW620 cells led to a decrease
(P<0.05; Fig. 2D). In addition,
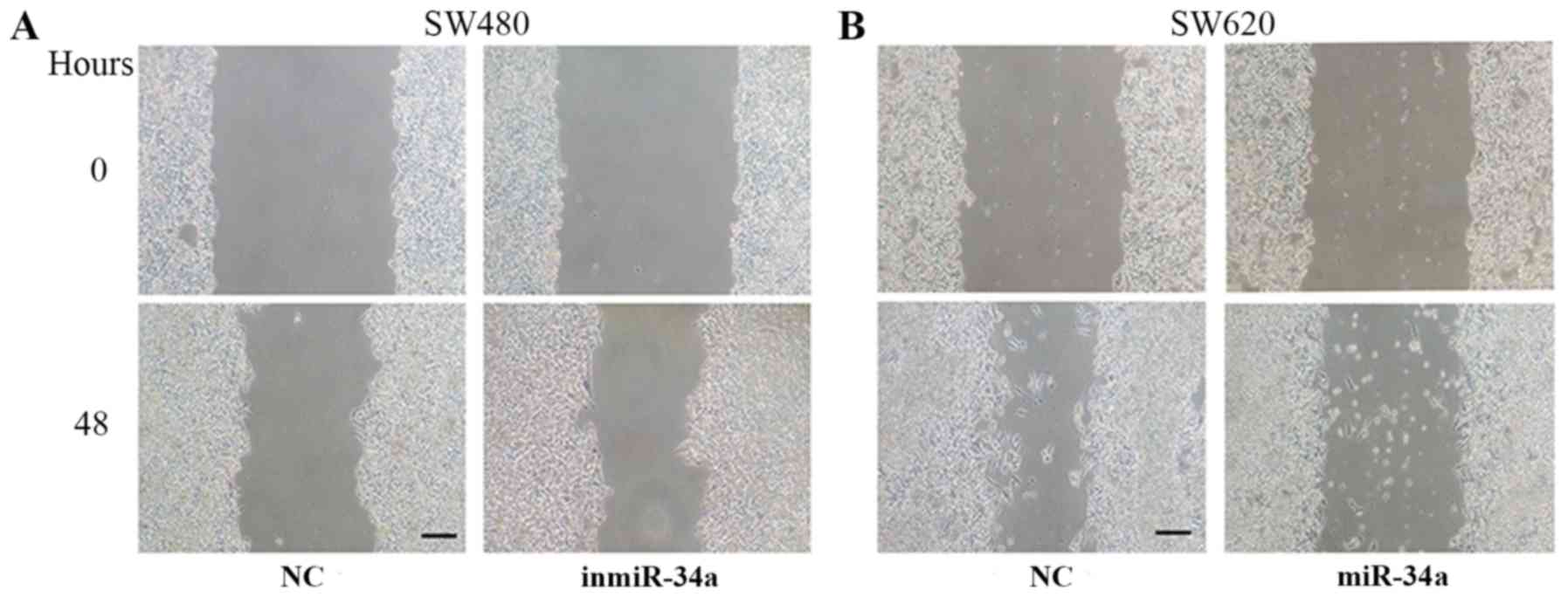
the wound healing assay was performed to evaluate the migration of
SW480 cells and SW620 cells. Compared with the equivalent control
groups, the degree of wound closure was increased in inmiR-34a
group (Fig. 3A), and decreased in
the SW620 mimic group (Fig. 3B).
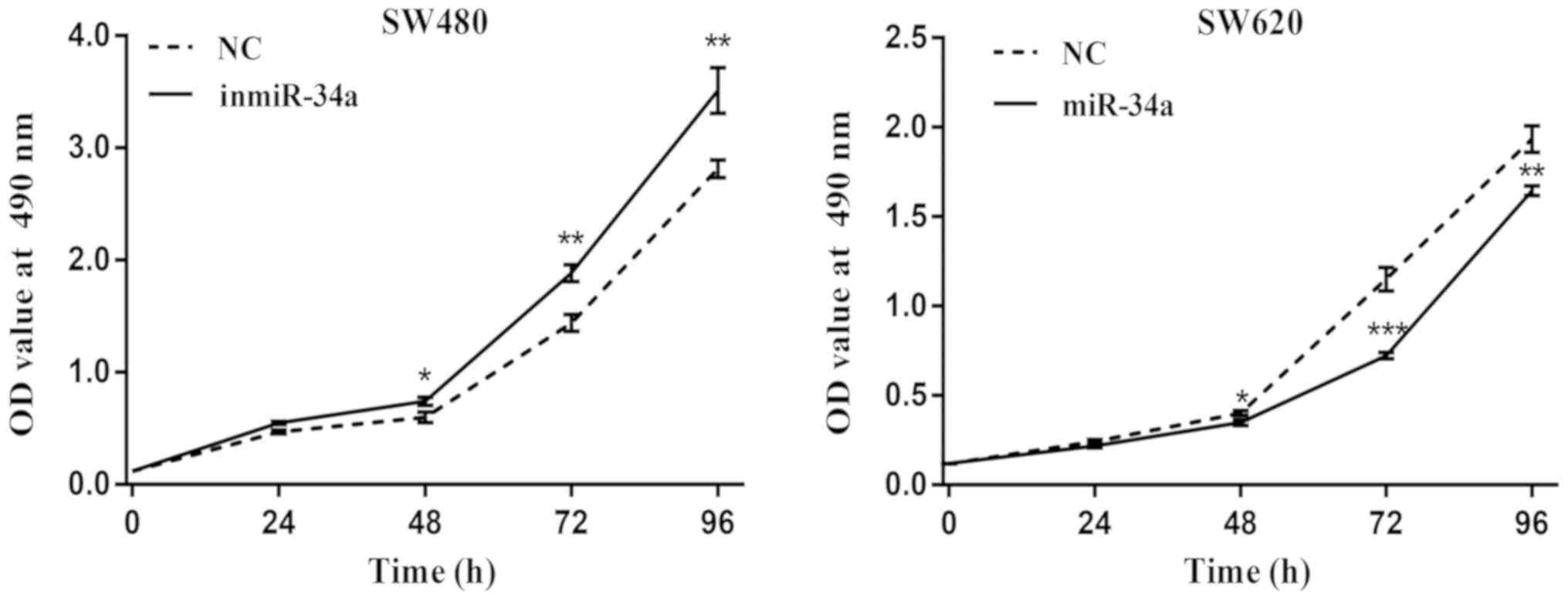
miR-34a inhibits the proliferation of
CRC cells
The proliferation of SW480 and SW620 cells following
transfection of miR-34a was examined. The results from the MTT
assay revealed an increased proliferative ability in the SW480
inmiR-34a group compared with the NC group, whereas a decrease in
the proliferation rate was observed in the SW620 cells with miR-34a
overexpression (Fig. 4). The
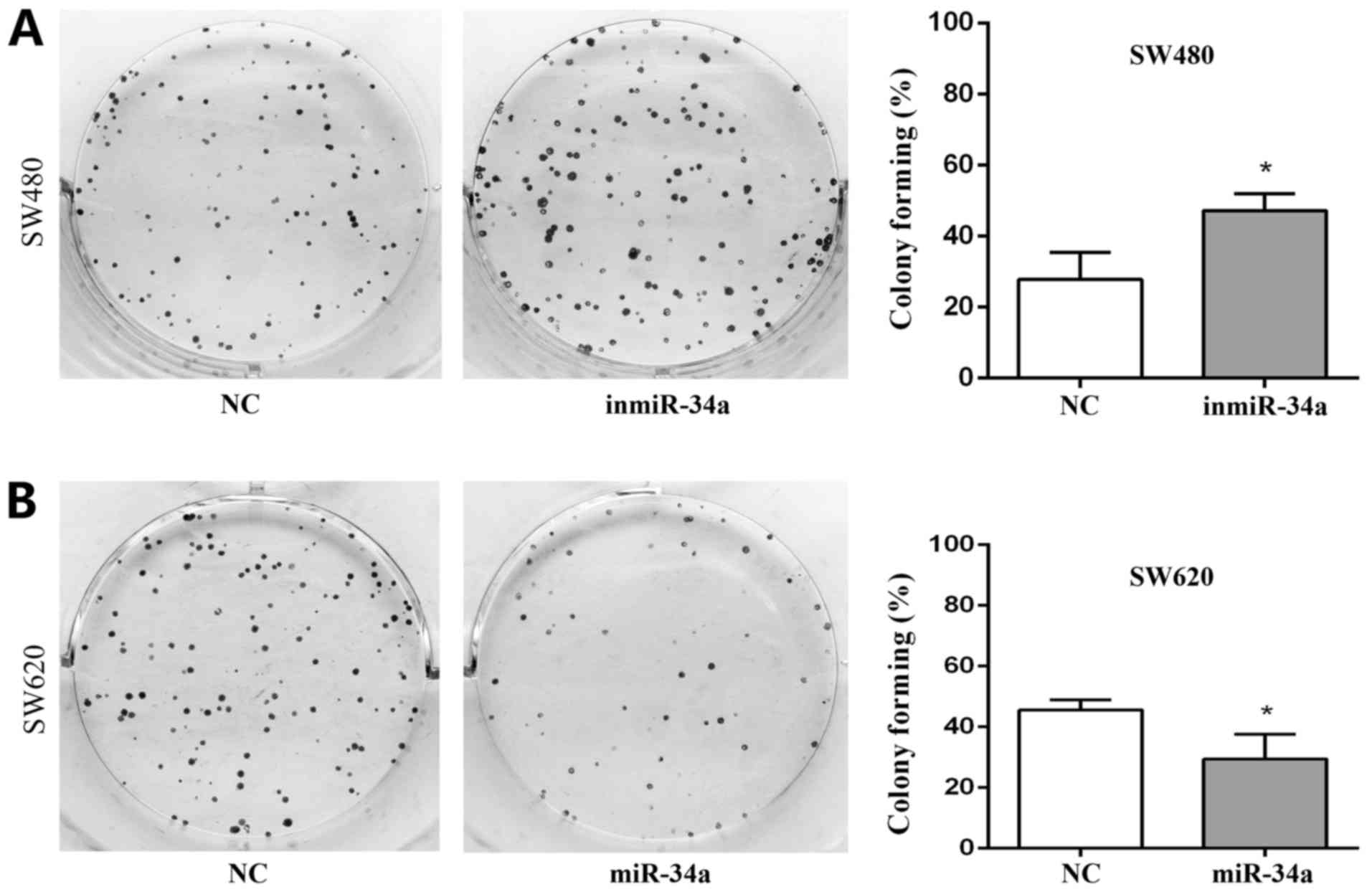
colony-formation rate for the SW480 inmiR-34a group was
47.11±2.75%, compared with 27.78±4.41% for the NC group (P<0.05;
Fig. 5A). In the SW620 cells, the
colony formation rate of miR-34a mimic group was 29.44±4.68%,
compared with 45.56±1.98% in the NC group (P<0.05; Fig. 5B). The colony formation ability of
the SW480 inmiR-34a and the SW620 NC groups was improved compared
with that of the groups to which they were compared, and their
colony sizes were larger.
miR-34a suppresses the expression of
vimentin and increases the expression of EGR1
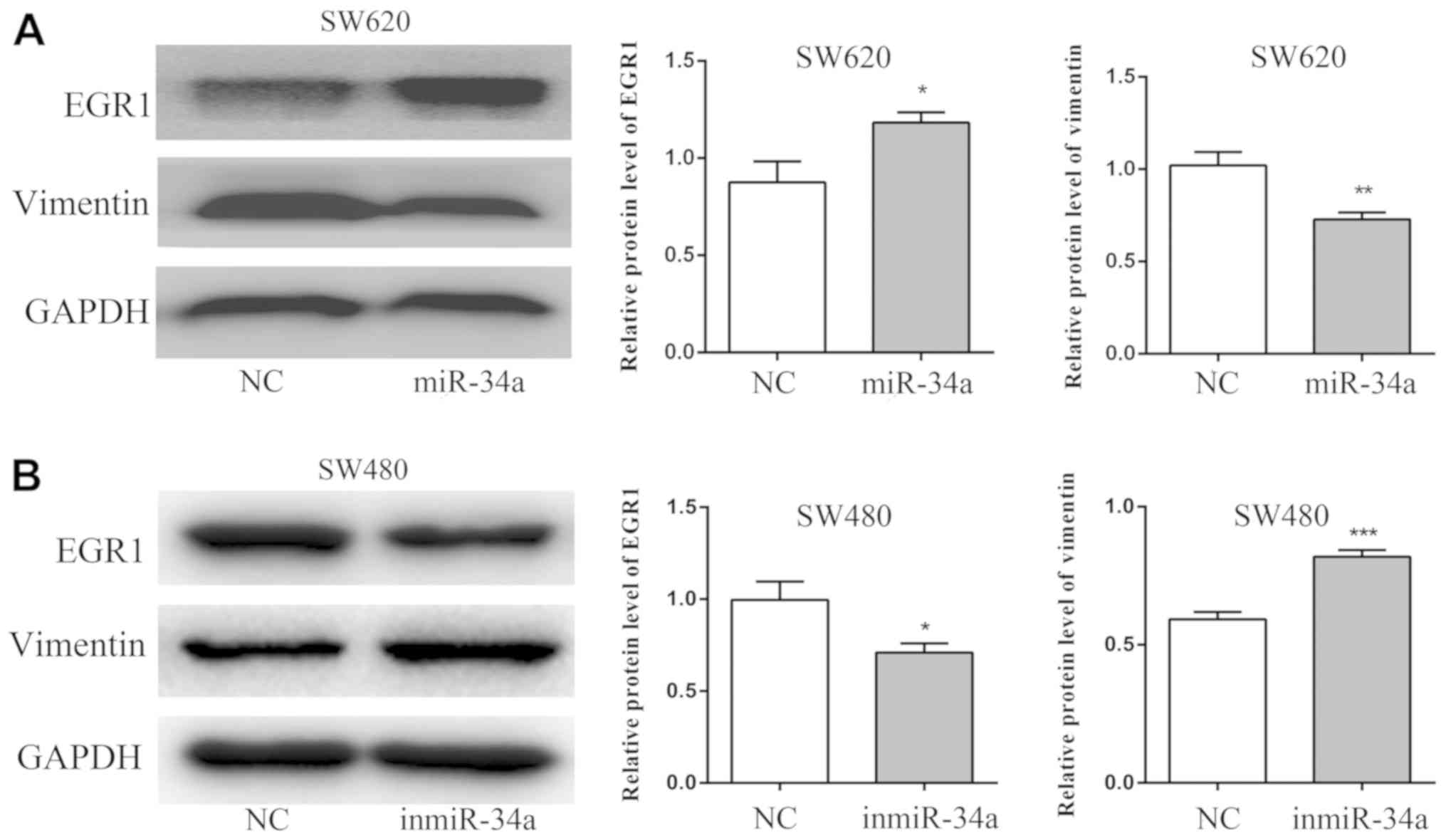
The results of the western blot analysis revealed
that the expression of EGR1 protein was increased in the miR-34a
group of SW620 cells compared with the NC group (P<0.05),
whereas the expression of vimentin was decreased (P<0.01;
Fig. 6A). By contrast, compared with
the NC group, the EGR1 level was decreased in the inmiR-34a group
of the SW480 cells (P<0.05), and the level of vimentin was
increased (P<0.001; Fig. 6B).
Discussion
CRC is the third most common cancer worldwide, and
its incidence and mortality are increasing globally (2). CRC is a heterogeneous disease; its
progression is caused by special genetic alterations. The
identification of the molecular mechanisms involved in the
progression of CRC would assist in providing targets for cancer
prevention and therapy, and improving patient prognosis. The
potential tumor suppressor miRNA miR-34a is frequently
downregulated in a number of types of cancer (12–14). The
results of the present study demonstrated that miR-34a is
deregulated in highly invasive CRC cell lines. Increasing the
expression of miR-34a in SW620 cells inhibited the cell
proliferative, migratory and invasive capabilities of these cells.
Furthermore, it suppressed the expression of vimentin and increased
the expression of EGR1. The opposite effects were observed when
miR-34a was inhibited in SW480 cells.
EGR1 is a 59-kDa zinc finger transcription factor
that appears to activate transcription by binding to DNA as a
monomer (22). EGR1 is also known to
exhibit a tumor suppressor function in a variety of human tumors,
including lung and breast cancer, glioblastoma and ovarian cancer
(23–25). Furthermore, previous studies have
indicated that EGR1 is a direct regulator of at least 4 major
suppressors: Transforming growth factor β-1 protein, PTEN, cellular
tumor antigen p53 family members and fibronectin, being involved in
the regulation of the invasive behavior of cancer cells (26–28). It
was also demonstrated that sustained EGR1 expression may lead to
preferential inhibition of tumor cell invasion and tumor growth
(29). However, few studies have
focused on EGR1 in CRC. In the present study, the addition of a
miR-34a mimic caused the expression of EGR1 to increase in CRC cell
lines. Therefore, EGR1 is considered to be involved in the
mechanism of miR-34a in suppressing tumorigenesis and development
of CRC. The results from the present study revealed that miR-34a
overexpression enhanced EGR1 levels in SW620 cells, whereas
inhibition of miR-34a in SW480 cells elicited the opposite effect.
However, the mechanism of how EGR1 participates in regulation of
the invasion and metastasis of CRC by miR-34a requires additional
investigation.
Vimentin, an intermediate silk protein, is involved
in cell structure and integrity (30). Previous studies have revealed that it
is abnormally expressed in numerous cancer types and that it
affects the shape and movement of cancer cells during the
epithelial-mesenchymal transition (EMT) process. Therefore, it is
considered to be a potential cancer therapeutic target (31–33). The
EMT is a biological phenomenon in which epithelial cells lose their
epithelial properties and acquire interstitial cell characteristics
under certain circumstances (34),
and it has been revealed that it is one of the mechanisms of tumor
cell invasion and metastasis (35).
Notably, vimentin is involved in the EMT (36). A previous study demonstrated that the
expression levels of vimentin in paraffin-embedded CRC specimens
were significantly increased compared with those in non-tumor
adjacent tissues, and that the high vimentin expression is
associated with various adverse clinicopathological factors
(37). Furthermore, the expression
of vimentin in CRC tissues is significantly increased in patients
with higher tumor stage, lymph node involvement, liver metastasis
and advanced tumor-node-metastasis stages (38). During the previous few years, miR-34a
has been revealed to be involved in the EMT regulation and,
therefore, to affect the invasion and metastasis of tumors
(39). The results of the present
study indicate that miR-34a overexpression inhibits the expression
of vimentin in SW620 cells, and miR-34a inhibition leads to an
increase in vimentin in SW480 cells.
In conclusion, the results of present study support
the hypothesis that miR-34a suppresses the cell migration, invasion
and proliferation of CRC cells by regulating vimentin and EGR1.
This suggests that miR-34a may prove to be a novel molecular target
for the treatment of CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81472275, 81702285
and 81702399) and the Natural Science Foundation of Guangdong
Province (grant nos. 2014A030313542 and 2017A030313644).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, YY and QH performed the experiments. WZ, CZ and
YS designed the current study. JL, YS and EJ analyzed the data. JL
and WZ wrote the manuscript. HG and YL provided technical
assistance and participated in critical revisions of important
intellectual content in the manuscript. WZ, JL, EJ and YS revised
the manuscript. All authors have read and approved the final
version of this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ung L, Lam AK, Morris DL and Chua TC:
Tissue-based biomarkers predicting outcomes in metastatic
colorectal cancer: A review. Clin Transl Oncol. 16:425–435. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meijer HA, Kong YW, Lu WT, Wilczynska A,
Spriggs RV, Robinson SW, Godfrey JD, Willis AE and Bushell M:
Translational repression and eIF4A2 activity are critical for
microRNA-mediated gene regulation. Science. 340:82–85. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S,
Song Z and He J: Downregulation of microRNA-27b-3p enhances
tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1
expression. Cell Death Dis. 7:e24542016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ao X, Nie P, Wu B, Xu W, Zhang T, Wang S,
Chang H and Zou Z: Decreased expression of microRNA-17 and
microRNA-20b promotes breast cancer resistance to taxol therapy by
upregulation of NCOA3. Cell Death Dis. 7:e24632016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rokavec M, Li H, Jiang L and Hermeking H:
The p53/miR-34 axis in development and disease. J Mol Cell Biol.
6:214–230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Slabáková E, Culig Z, Remšík J and Souček
K: Alternative mechanisms of miR-34a regulation in cancer. Cell
Death Dis. 8:e31002017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Ai F, Li X, Tian L, Wang X, Shen
S and Liu F: MicroRNA-34a suppresses colorectal cancer metastasis
by regulating Notch signaling. Oncol Lett. 14:2325–2333. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue
YF, Li G, Lu X, Sun Z and Tang KF: MicroRNA-34a inhibits migration
and invasion of colon cancer cells via targeting to Fra-1.
Carcinogenesis. 33:519–528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: MiR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franchina T, Amodeo V, Bronte G, Savio G,
Ricciardi GR, Picciotto M, Russo A, Giordano A and Adamo V:
Circulating miR-22, miR-24 and miR-34a as novel predictive
biomarkers to pemetrexed-based chemotherapy in advanced non-small
cell lung cancer. J Cell Physiol. 229:97–99. 2014.PubMed/NCBI
|
|
18
|
Cao W, Yang W, Fan R, Li H, Jiang J, Geng
M, Jin Y and Wu Y: miR-34a regulates cisplatin-induce gastric
cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour
Biol. 35:1287–1295. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nugent M, Miller N and Kerin MJ:
Circulating miR-34a levels are reduced in colorectal cancer. J Surg
Oncol. 106:947–952. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li
Y, Li Z, Ng SS, Sung JJ, et al: miR-34a-5p suppresses colorectal
cancer metastasis and predicts recurrence in patients with stage
II/III colorectal cancer. Oncogene. 34:4142–4152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Rangnekar VM, Adamson E and Mercola
D: Suppression of growth and transformation and induction of
apoptosis by EGR-1. Cancer Gene Ther. 5:3–28. 1998.PubMed/NCBI
|
|
23
|
Calogero A, Arcella A, De Gregorio G,
Porcellini A, Mercola D, Liu C, Lombari V, Zani M, Giannini G,
Gagliardi FM, et al: The early growth response gene EGR-1 behaves
as a suppressor gene that is down-regulated independent of ARF/Mdm2
but not p53 alterations in fresh human gliomas. Clin Cancer Res.
7:2788–2796. 2001.PubMed/NCBI
|
|
24
|
Levin WJ, Press MF, Gaynor RB, Sukhatme
VP, Boone TC, Reissmann PT, Figlin RA, Holmes EC, Souza LM and
Slamon DJ: Expression patterns of immediate early transcription
factors in human non-small cell lung cancer. The Lung Cancer Study
Group. Oncogene. 11:1261–1269. 1995.PubMed/NCBI
|
|
25
|
Ahmed MM: Regulation of radiation-induced
apoptosis by early growth response-1 gene in solid tumors. Curr
Cancer Drug Targets. 4:43–52. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Baron V, Adamson ED, Calogero A, Ragona G
and Mercola D: The transcription factor Egr1 is a direct regulator
of multiple tumor suppressors including TGFbeta1, PTEN, p53, and
fibronectin. Cancer Gene Ther. 13:115–124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Virolle T, Adamson ED, Baron V, Birle D,
Mercola D, Mustelin T and de Belle I: The Egr-1 transcription
factor directly activates PTEN during irradiation-induced
signalling. Nat Cell Biol. 3:1124–1128. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krones-Herzig A, Adamson E and Mercola D:
Early growth response 1 protein, an upstream gatekeeper of the p53
tumor suppressor, controls replicative senescence. Proc Natl Acad
Sci USA. 100:3233–3238. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim SO, Kwon JI, Jeong YK, Kim GY, Kim ND
and Choi YH: Induction of Egr-1 is associated with anti-metastatic
and anti-invasive ability of beta-lapachone in human
hepatocarcinoma cells. Biosci Biotechnol Biochem. 71:2169–2176.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katsumoto T, Mitsushima A and Kurimura T:
The role of the vimentin intermediate filaments in rat 3Y1 cells
elucidated by immunoelectron microscopy and computer-graphic
reconstruction. Biol Cell. 68:139–146. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McInroy L and Maatta A: Down-regulation of
vimentin expression inhibits carcinoma cell migration and adhesion.
Biochem Biophys Res Commun. 360:109–114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bargagna-Mohan P, Hamza A, Kim YE, Khuan
Abby Ho Y, Mor-Vaknin N, Wendschlag N, Liu J, Evans RM, Markovitz
DM, Zhan CG, et al: The tumor inhibitor and antiangiogenic agent
withaferin A targets the intermediate filament protein vimentin.
Chem Biol. 14:623–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nieto MA: Epithelial plasticity: A common
theme in embryonic and cancer cells. Science. 342:12348502013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wrighton KH: Cell migration: EMT promotes
contact inhibition of locomotion. Nat Rev Mol Cell Biol.
16:5182015. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiao S, Liu L, Lu X, Long J, Zhou X and
Fang M: The prognostic significance of bromodomain PHD-finger
transcription factor in colorectal carcinoma and association with
vimentin and E-cadherin. J Cancer Res Clin. 141:1465–1474. 2015.
View Article : Google Scholar
|
|
38
|
Toiyama Y, Yasuda H, Saigusa S, Tanaka K,
Inoue Y, Goel A and Kusunoki M: Increased expression of Slug and
Vimentin as novel predictive biomarkers for lymph node metastasis
and poor prognosis in colorectal cancer. Carcinogenesis.
34:2548–2557. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|