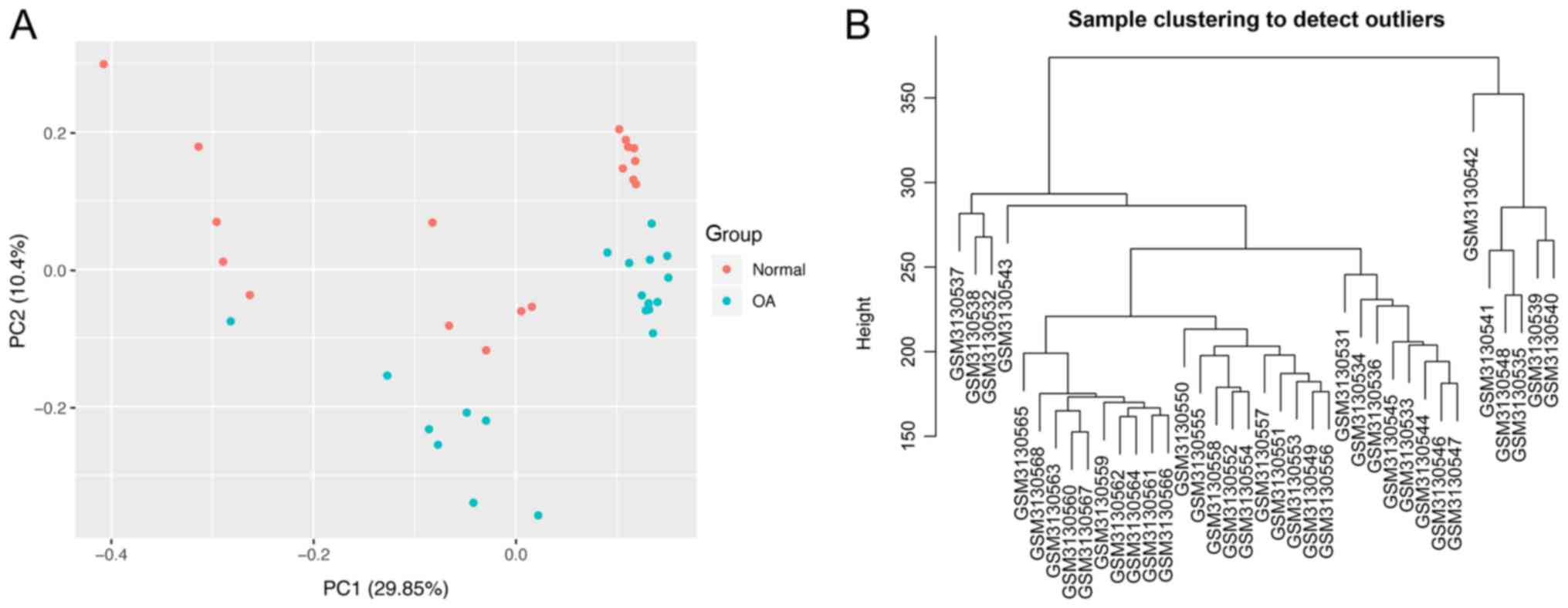
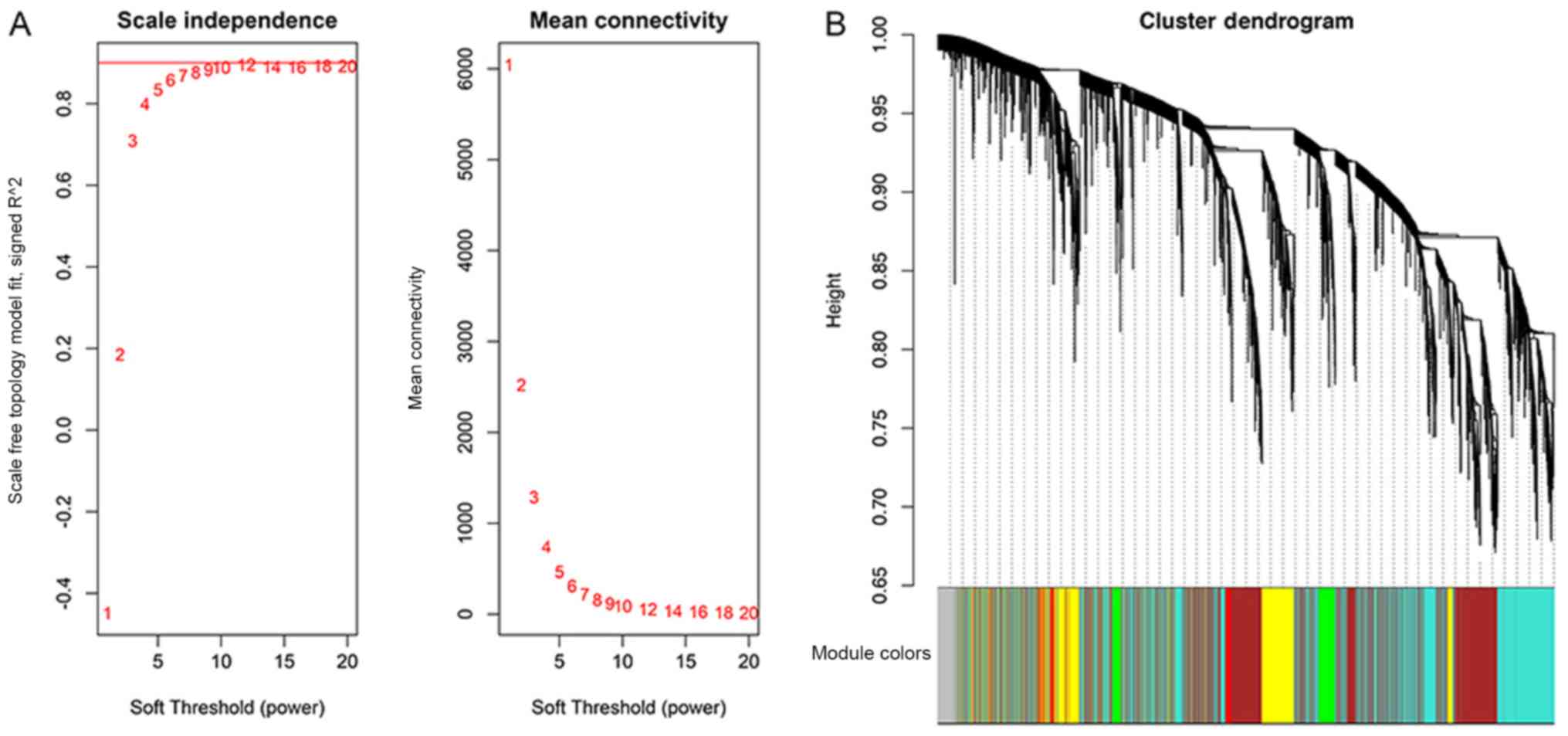
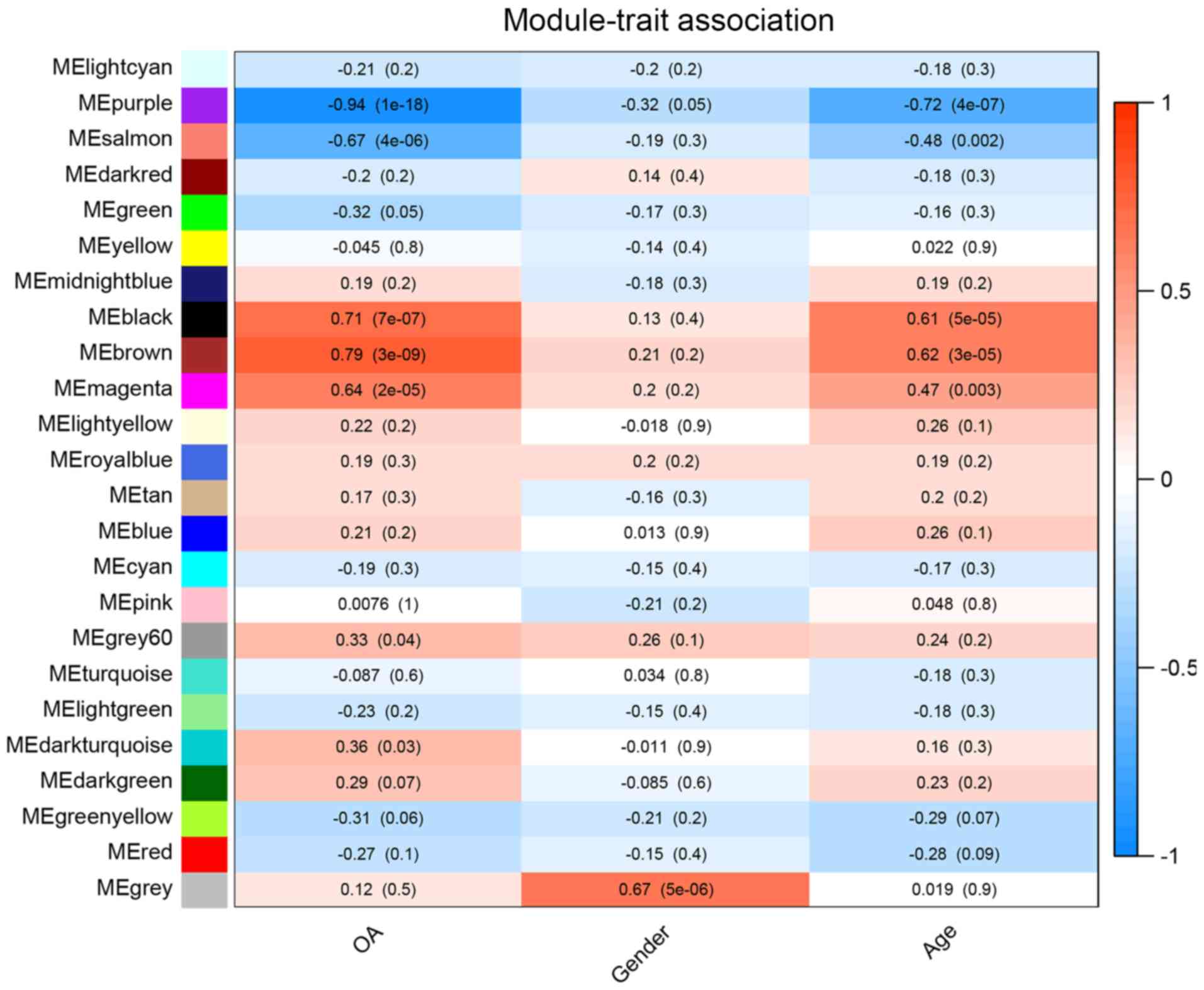
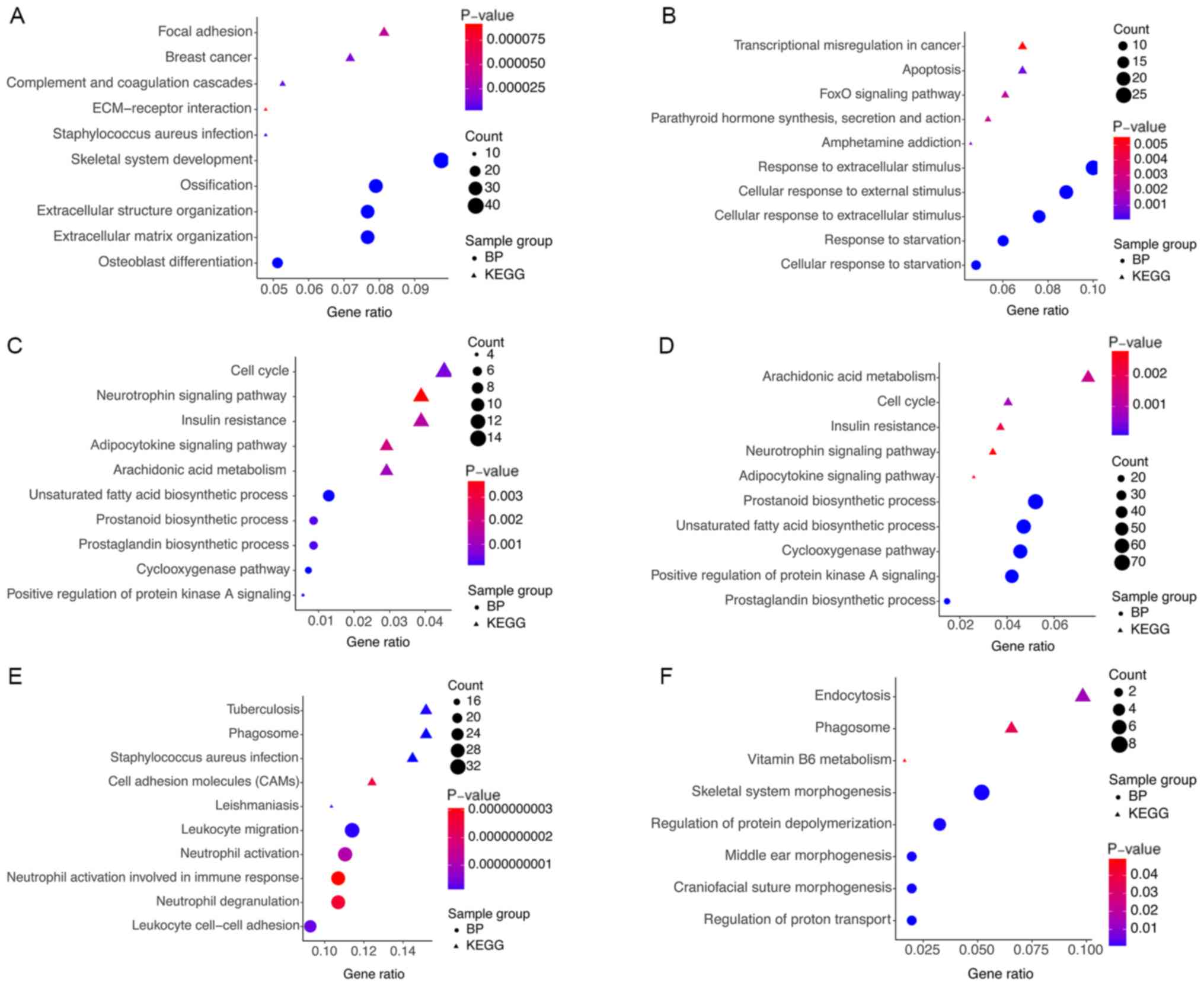
|
1
|
Thomas E, Peat G and Croft P: Defining and
mapping the person with osteoarthritis for population studies and
public health. Rheumatology (Oxford). 53:338–345. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xie F, Kovic B, Jin X, He X, Wang M and
Silvestre C: Economic and humanistic burden of osteoarthritis: A
systematic review of large sample studies. Pharmacoeconomics.
34:1087–1100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldring MB and Otero M: Inflammation in
osteoarthritis. Curr Opin Rheumatol. 23:471–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sellam J and Berenbaum F: The role of
synovitis in pathophysiology and clinical symptoms of
osteoarthritis. Nat Rev Rheumatol. 6:625–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tonge DP, Pearson MJ and Jones SW: The
hallmarks of osteoarthritis and the potential to develop
personalised disease-modifying pharmacological therapeutics.
Osteoarthritis Cartilage. 22:609–621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hunter DJ and Bierma-Zeinstra S:
Osteoarthritis. Lancet. 393:1745–1759. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren YM, Zhao X, Yang T, Duan YH, Sun YB,
Zhao WJ and Tian MQ: Exploring the key genes and pathways of
osteoarthritis in knee cartilage in a rat model using gene
expression profiling. Yonsei Med J. 59:760–768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu N, Hou J, Wu Y, Li G, Liu J, Ma G,
Chen B and Song Y: Identification of key genes in rheumatoid
arthritis and osteoarthritis based on bioinformatics analysis.
Medicine (Baltimore). 97:e109972018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fei Q, Lin J, Meng H, Wang B, Yang Y, Wang
Q, Su N, Li J and Li D: Identification of upstream regulators for
synovial expression signature genes in osteoarthritis. Joint Bone
Spine. 83:545–551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fisch KM, Gamini R, Alvarez-Garcia O,
Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI and Lotz
MK: Identification of transcription factors responsible for
dysregulated networks in human osteoarthritis cartilage by global
gene expression analysis. Osteoarthritis Cartilage. 26:1531–1538.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, et al: NCBI GEO: Archive for functional genomics data
sets-10 years on. Nucleic Acids Res. 39(Database Issue):
D1005–D1010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xin J, Mark A, Afrasiabi C, Tsueng G,
Juchler M, Gopal N, Stupp GS, Putman TE, Ainscough BJ, Griffith OL,
et al: High-performance web services for querying gene and variant
annotation. Genome Biol. 17:912016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Potier D, Atak ZK, Sanchez MN, Herrmann C
and Aerts S: Using cisTargetX to predict transcriptional targets
and networks in Drosophila. Methods Mol Biol. 786:291–314. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aerts S, Quan XJ, Claeys A, Naval Sanchez
M, Tate P, Yan J and Hassan BA: Robust target gene discovery
through transcriptome perturbations and genome-wide enhancer
predictions in Drosophila uncovers a regulatory basis for sensory
specification. PLoS Biol. 8:e10004352010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ringnér M: What is principal component
analysis? Nat Biotechnol. 26:303–304. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Connor MI and Hooten EG: Breakout
session: Gender disparities in knee osteoarthritis and TKA. Clin
Orthop Relat Res. 469:1883–1885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee TI and Young RA: Transcriptional
regulation and its misregulation in disease. Cell. 152:1237–1251.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abusarah J, Benabdoune H, Shi Q, Lussier
B, Martel-Pelletier J, Malo M, Fernandes JC, de Souza FP, Fahmi H
and Benderdour M: Elucidating the role of protandim and 6-Gingerol
in protection against osteoarthritis. J Cell Biochem.
118:1003–1013. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dunn SL, Soul J, Anand S, Schwartz JM,
Boot-Handford RP and Hardingham TE: Gene expression changes in
damaged osteoarthritic cartilage identify a signature of
non-chondrogenic and mechanical responses. Osteoarthritis
Cartilage. 24:1431–1440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu M, Huang G, Zhang Z, Liu J, Zhang Z,
Huang Z, Yu B and Meng F: Expression profile of long noncoding RNAs
in cartilage from knee osteoarthritis patients. Osteoarthritis
Cartilage. 23:423–432. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fujii W, Kawahito Y, Nagahara H, Kukida Y,
Seno T, Yamamoto A, Kohno M, Oda R, Taniguchi D, Fujiwara H, et al:
Monocarboxylate transporter 4, associated with the acidification of
synovial fluid, is a novel therapeutic target for inflammatory
arthritis. Arthritis Rheumatol. 67:2888–2896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu X, Yang S, Lin W, Wang L, Ying J, Ding
Y and Chen X: Roles of cell cyle regulators cyclin D1, CDK4, and
p53 in knee osteoarthritis. Genet Test Mol Biomarkers. 20:529–534.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Troeberg L and Nagase H: Proteases
involved in cartilage matrix degradation in osteoarthritis. Biochim
Biophys Acta 1824. 133–145. 2012.
|
|
30
|
Ahmed U, Thornalley PJ and Rabbani N:
Possible role of methylglyoxal and glyoxalase in arthritis. Biochem
Soc Trans. 42:538–542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo H, Yao L, Zhang Y and Li R: Liquid
chromatography-mass spectrometry-based quantitative proteomics
analysis reveals chondroprotective effects of astragaloside IV in
interleukin-1β-induced SW1353 chondrocyte-like cells. Biomed
Pharmacother. 91:796–802. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Andriacchi TP, Favre J, Erhart-Hledik JC
and Chu CR: A systems view of risk factors for knee osteoarthritis
reveals insights into the pathogenesis of the disease. Ann Biomed
Eng. 43:376–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scanzello CR, Plaas A and Crow MK: Innate
immune system activation in osteoarthritis: Is osteoarthritis a
chronic wound? Curr Opin Rheumatol. 20:565–572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marchev AS, Dimitrova PA, Burns AJ, Kostov
RV, Dinkova-Kostova AT and Georgiev MI: Oxidative stress and
chronic inflammation in osteoarthritis: Can NRF2 counteract these
partners in crime? Ann N Y Acad Sci 1401. 114–135. 2017. View Article : Google Scholar
|
|
35
|
Hess J, Angel P and Schorpp-Kistner M:
AP-1 subunits: Quarrel and harmony among siblings. J Cell Sci.
117:5965–5973. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ionescu AM, Schwarz EM, Vinson C, Puzas
JE, Rosier R, Reynolds PR and O'Keefe RJ: PTHrP modulates
chondrocyte differentiation through AP-1 and CREB signaling. J Biol
Chem. 276:11639–11647. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmucker AC, Wright JB, Cole MD and
Brinckerhoff CE: Distal interleukin-1β (IL-1β) response element of
human matrix metalloproteinase-13 (MMP-13) binds activator protein
1 (AP-1) transcription factors and regulates gene expression. J
Biol Chem. 287:1189–1197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moritani NH, Kubota S, Eguchi T, Fukunaga
T, Yamashiro T, Takano-Yamamoto T, Tahara H, Ohyama K, Sugahara T
and Takigawa M: Interaction of AP-1 and the ctgf gene: A possible
driver of chondrocyte hypertrophy in growth cartilage. J Bone Miner
Metab. 21:205–210. 2003.PubMed/NCBI
|
|
39
|
Papachristou D, Pirttiniemi P, Kantomaa T,
Agnantis N and Basdra EK: Fos- and Jun-related transcription
factors are involved in the signal transduction pathway of
mechanical loading in condylar chondrocytes. Eur J Orthod.
28:20–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim MG, Yang JH, Kim KM, Jang CH, Jung JY,
Cho IJ, Shin SM and Ki SH: Regulation of Toll-like
receptor-mediated Sestrin2 induction by AP-1, Nrf2, and the
ubiquitin-proteasome system in macrophages. Toxicol Sci.
144:425–435. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Briso EM, Guinea-Viniegra J, Bakiri L,
Rogon Z, Petzelbauer P, Eils R, Wolf R, Rincón M, Angel P and
Wagner EF: Inflammation-mediated skin tumorigenesis induced by
epidermal c-Fos. Genes Dev. 27:1959–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu YH, Liu W, Zhang L, Liu XY, Wang Y, Xue
B, Liu B, Duan R, Zhang B and Ji Y: Effects of microRNA-24
targeting C-myc on apoptosis, proliferation, and cytokine
expressions in chondrocytes of rats with osteoarthritis via MAPK
signaling pathway. J Cell Biochem. 119:7944–7958. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lemos DR, McMurdo M, Karaca G,
Wilflingseder J, Leaf IA, Gupta N, Miyoshi T, Susa K, Johnson BG,
Soliman K, et al: Interleukin-1β activates a MYC-dependent
metabolic switch in kidney stromal cells necessary for progressive
tubulointerstitial fibrosis. J Am Soc Nephrol. 29:1690–1705. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Anderton B, Camarda R, Balakrishnan S,
Balakrishnan A, Kohnz RA, Lim L, Evason KJ, Momcilovic O, Kruttwig
K, Huang Q, et al: MYC-driven inhibition of the glutamate-cysteine
ligase promotes glutathione depletion in liver cancer. EMBO Rep.
18:569–585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen D, Xiong XQ, Zang YH, Tong Y, Zhou B,
Chen Q, Li YH, Gao XY, Kang YM and Zhu GQ: BCL6 attenuates renal
inflammation via negative regulation of NLRP3 transcription. Cell
Death Dis. 8:e31562017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Iezaki T, Ozaki K, Fukasawa K, Inoue M,
Kitajima S, Muneta T, Takeda S, Fujita H, Onishi Y, Horie T, et al:
ATF3 deficiency in chondrocytes alleviates osteoarthritis
development. J Pathol. 239:426–437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen HH, Lai PF, Lan YF, Cheng CF, Zhong
WB, Lin YF, Chen TW and Lin H: Exosomal ATF3 RNA attenuates
pro-inflammatory gene MCP-1 transcription in renal
ischemia-reperfusion. J Cell Physiol. 229:1202–1211. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pernhorst K, Herms S, Hoffmann P, Cichon
S, Schulz H, Sander T, Schoch S, Becker AJ and Grote A: TLR4, ATF-3
and IL8 inflammation mediator expression correlates with seizure
frequency in human epileptic brain tissue. Seizure. 22:675–678.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Walsh DA, McWilliams DF, Turley MJ, Dixon
MR, Fransès RE, Mapp PI and Wilson D: Angiogenesis and nerve growth
factor at the osteochondral junction in rheumatoid arthritis and
osteoarthritis. Rheumatology (Oxford). 49:1852–1861. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G,
Cao L, Zhang X, Guan H, Yang Y, et al: Overexpression of HOXC10
promotes angiogenesis in human glioma via interaction with PRMT5
and upregulation of VEGFA expression. Theranostics. 8:5143–5158.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zan PF, Yao J, Wu Z, Yang Y, Hu S and Li
GD: Cyclin D1 gene silencing promotes IL-1β-induced apoptosis in
rat chondrocytes. J Cell Biochem. 119:290–299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lefebvre V and Dvir-Ginzberg M: SOX9 and
the many facets of its regulation in the chondrocyte lineage.
Connect Tissue Res. 58:2–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li Z, Wang Q, Chen G, Li X, Yang Q, Du Z,
Ren M, Song Y and Zhang G: Integration of gene expression profile
data to screen and verify hub genes involved in osteoarthritis.
Biomed Res Int 2018. 94827262018.
|
|
54
|
Moskalev AA, Smit-McBride Z, Shaposhnikov
MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R and Fraifeld
VE: Gadd45 proteins: Relevance to aging, longevity and age-related
pathologies. Ageing Res Rev. 11:51–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Amente S, Zhang J, Lavadera ML, Lania L,
Avvedimento EV and Majello B: Myc and PI3K/AKT signaling
cooperatively repress FOXO3a-dependent PUMA and GADD45a gene
expression. Nucleic Acids Res. 39:9498–9507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Novakofski KD, Torre CJ and Fortier LA:
Interleukin-1α, −6, and −8 decrease Cdc42 activity resulting in
loss of articular chondrocyte phenotype. J Orthop Res. 30:246–251.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fortier LA and Miller BJ: Signaling
through the small G-protein Cdc42 is involved in insulin-like
growth factor-I resistance in aging articular chondrocytes. J
Orthop Res. 24:1765–1772. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ushijima T, Okazaki K, Tsushima H and
Iwamoto Y: CCAAT/enhancer-binding protein β regulates the
repression of type II collagen expression during the
differentiation from proliferative to hypertrophic chondrocytes. J
Biol Chem. 289:2852–2863. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lepetsos P, Papavassiliou KA and
Papavassiliou AG: Redox and NF-κB signaling in osteoarthritis. Free
Radic Biol Med. 132:90–100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Morrovati F, Karimian Fathi N, Soleimani
Rad J and Montaseri A: Mummy prevents IL-1β-induced inflammatory
responses and cartilage matrix degradation via inhibition of NF-κB
subunits gene expression in pellet culture system. Adv Pharm Bull.
8:283–289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bradley EW, Carpio LR, Newton AC and
Westendorf JJ: Deletion of the PH-domain and leucine-rich repeat
protein phosphatase 1 (Phlpp1) increases fibroblast growth factor
(Fgf) 18 expression and promotes chondrocyte proliferation. J Biol
Chem. 290:16272–16280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Loeser RF, Goldring SR, Scanzello CR and
Goldring MB: Osteoarthritis: A disease of the joint as an organ.
Arthritis Rheum. 64:1697–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Guo SM, Wang JX, Li J, Xu FY, Wei Q, Wang
HM, Huang HQ, Zheng SL, Xie YJ and Zhang C: Identification of gene
expression profiles and key genes in subchondral bone of
osteoarthritis using weighted gene coexpression network analysis. J
Cell Biochem. 119:7687–7695. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Durand M, Komarova SV, Bhargava A,
Trebec-Reynolds DP, Li K, Fiorino C, Maria O, Nabavi N, Manolson
MF, Harrison RE, et al: Monocytes from patients with osteoarthritis
display increased osteoclastogenesis and bone resorption: The in
vitro osteoclast differentiation in arthritis study. Arthritis
Rheum. 65:148–158. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bertuglia A, Lacourt M, Girard C,
Beauchamp G, Richard H and Laverty S: Osteoclasts are recruited to
the subchondral bone in naturally occurring post-traumatic equine
carpal osteoarthritis and may contribute to cartilage degradation.
Osteoarthritis Cartilage. 24:555–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Botter SM, van Osch GJ, Waarsing JH, van
der Linden JC, Verhaar JA, Pols HA, van Leeuwen JP and Weinans H:
Cartilage damage pattern in relation to subchondral plate thickness
in a collagenase-induced model of osteoarthritis. Osteoarthritis
Cartilage. 16:506–514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Akasaki Y, Hasegawa A, Saito M, Asahara H,
Iwamoto Y and Lotz MK: Dysregulated FOXO transcription factors in
articular cartilage in aging and osteoarthritis. Osteoarthritis
Cartilage. 22:162–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Matsuzaki T, Alvarez-Garcia O, Mokuda S,
Nagira K, Olmer M, Gamini R, Miyata K, Akasaki Y, Su AI, Asahara H
and Lotz MK: FoxO transcription factors modulate autophagy and
proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci
Transl Med. 10(pii): eaan07462018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shen C, Cai GQ, Peng JP and Chen XD:
Autophagy protects chondrocytes from glucocorticoids-induced
apoptosis via ROS/Akt/FOXO3 signaling. Osteoarthritis Cartilage.
23:2279–2287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zeng GQ, Chen AB, Li W, Song JH and Gao
CY: High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis.
Genet Mol Res. 14:14811–14822. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sun HY, Hu KZ and Yin ZS: Inhibition of
the p38-MAPK signaling pathway suppresses the apoptosis and
expression of proinflammatory cytokines in human osteoarthritis
chondrocytes. Cytokine. 90:135–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chang L, Graham PH and Ni J: Targeting
PI3K/Akt/mTOR signaling pathway in the treatment of prostate cancer
radioresistance. Crit Rev Oncol Hematol. 96:507–517. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xue JF, Shi ZM, Zou J and Li XL:
Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of
articular chondrocytes and attenuates inflammatory response in rats
with osteoarthritis. Biomed Pharmacother. 89:1252–1261. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yu SM and Kim SJ: The thymoquinone-induced
production of reactive oxygen species promotes dedifferentiation
through the ERK pathway and inflammation through the p38 and PI3K
pathways in rabbit articular chondrocytes. Int J Mol Med.
35:325–332. 2015. View Article : Google Scholar : PubMed/NCBI
|