Introduction
Psoriasis is a chronic, systemic, recurrent
inflammatory disease, which is characterized by erythema, papules
and scaling (1). This condition is
caused by genetic and environmental factors, including gene
mutations, infection and stress (2).
The pathological manifestations of psoriasis include abnormal
keratinocyte differentiation/proliferation and inflammatory cell
infiltration (3). It is currently
recognized that the pathogenesis of psoriasis is associated with
the interleukin (IL)-23/type 17 T-helper cell immune axis, leading
to abnormalities in immune cells and associated cytokines, which
further induces excessive proliferation of keratinocytes (4). Although extensive research has been
performed on the pathogenesis of psoriasis, the mechanisms of
psoriasis have remained to be fully elucidated.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNA with a length of >200 nt (5,6). It is
speculated that lncRNAs are involved in numerous important
biological processes, including cellular homeostasis, genomic
imprinting, immunity and development (7). In addition, certain lncRNAs have a role
in cardiovascular, neurological and developmental diseases, as well
as cancers (5,8). Recently, lncRNAs were reported to have
pivotal roles in psoriasis. For instance, the lncRNA psoriasis
susceptibility-associated RNA gene induced by stress (PRINS) is the
most frequent transcript detected in the non-defective epidermis of
patients with psoriasis, and it is thought to contribute to the
pathogenesis of psoriasis (9,10).
Certain studies have reported that PRINS is capable of regulating
G1P3, which is expressed at high levels in psoriatic lesions and
has an anti-apoptotic role in keratinocytes (9,11).
Furthermore, stressors including microbial components and
ultraviolet-B radiation are capable of inducing high expression of
PRINS in keratinocytes (9). Msh
homeobox 2 pseudogene 1 (MSX2P1), which is another lncRNA,
facilitates the proliferation of IL-22-stimulated keratinocytes by
suppressing microRNA (miRNA/miR)-6731-5p and upregulating the
expression of S100A7 (12). In
addition, lncRNAs have been indicated to be an important type of
competing endogenous RNA (ceRNA) (13). lncRNAs inhibit miRNA-mediated target
repression by competing for miRNA-binding sites with mRNAs.
However, the regulatory mechanisms involving ceRNAs in psoriasis
have remained elusive. In the present study, psoriasis-associated
RNA networks were reconstructed based on the ceRNA theory, and the
biological functions of the networks were also investigated.
Materials and methods
Processing of raw data
High-throughput sequencing data for
psoriasis-associated lncRNAs, mRNAs and miRNAs were downloaded from
the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/; lncRNA and mRNA
expression profile accession numbers, GSE54456 and GSE74697; miRNA
expression profile expression number, GSE31037). For the
high-throughput raw sequencing data for lncRNAs and mRNAs, quality
assessment was performed with FastQC (v0.11.7) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/);
further, sequencing reads were trimmed by removing adapters and
low-quality sequences were trimmed by using Trimmomatic (v0.38)
(14). Hisat2 (v2.1.0) (15) was used to map the trimmed reads to
the human GRCh38 reference genome (http://useast.ensembl.org/info/data/ftp/index.html),
and the HTSeq-count (16) was then
used to quantify the genes. Furthermore, microarray data (accession
no. GSE13355) for comparing the difference in the expression of
lncRNAs were also downloaded from the GEO database. After the probe
was set, re-annotation was performed to identify specific lncRNAs.
The method of the re-annotation method was performed as previously
described by Shen et al (17). For the miRNA high-throughput raw
sequencing data, miRDeep2 (18) was
used to remove adapters, map the trimmed reads to the human GRCh38
reference genome and quantify the expression of miRNAs.
Screening for differentially expressed
lncRNAs, miRNAs and mRNAs
The differentially expressed lncRNAs, mRNAs and
miRNAs between psoriatic lesions involving the skin and healthy
controls were identified with the DESeq2 package (19). The differentially expressed lncRNAs,
mRNAs and miRNAs were selected according to the adjusted P-values
(P<0.05 and |log2 fold change|>1).
Functional enrichment analysis
DAVID 6.8 (20) was
used to perform the Gene Ontology (GO) functional enrichment
analysis in the category Biological Processes (BP), and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway functional
enrichment analysis of differentially expressed genes was also
performed. P<0.05 was considered to indicate statistical
significance.
Construction of the
psoriasis-associated lncRNA-miRNA-mRNA network
miRNA-RNA interactions were downloaded from the
experimentally verified databases DIANA-TarBase V8 (21) and miRTarBase (V7.0) (22), and miRNA-lncRNA interactions were
downloaded from starBase v2.0 (23)
and LncBase V2 (24). To construct
the psoriasis-associated ceRNA network, the differentially
expressed miRNAs, mRNAs and lncRNAs in psoriasis vs. normal samples
were integrated with the miRNA-mRNA and miRNA-lncRNA pairs
downloaded from the databases. Subsequently, miRNA-mRNA and
miRNA-lncRNA pairs that shared common miRNAs were integrated into
the psoriasis-associated ceRNA network. In the present study, to
create the psoriasis-associated ceRNA network, only upregulated
lncRNAs and downregulated miRNAs were integrated with upregulated
mRNAs, while only downregulated lncRNAs and upregulated miRNAs were
integrated with downregulated mRNAs (25). The degree of interaction - a
topological property that indicates the number of edges that
connect to a node-was also calculated. lncRNAs in the ceRNA network
that interacted with >5 different miRNAs were identified as key
lncRNAs in the present study. The networks were visualized using
Cytoscape 3.6.1 (26).
Results
Identification of differentially
expressed lncRNAs, mRNAs and miRNAs
Analysis of the high-throughput data deposited under
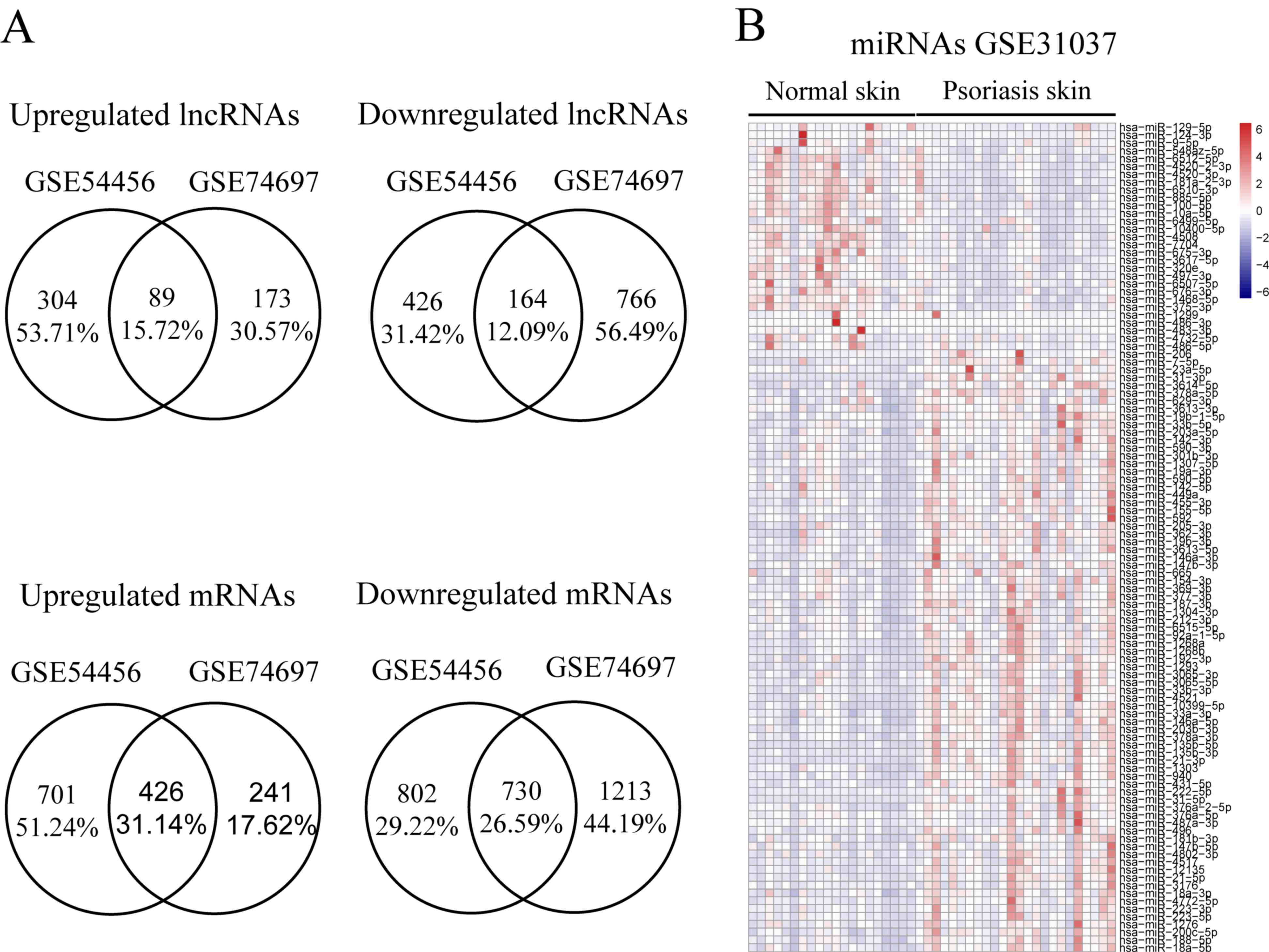
accession numbers GSE54456 and GSE74697 revealed that a total of 89
upregulated lncRNAs and 164 downregulated lncRNAs were shared
between the datasets for psoriatic lesions involving the skin and
healthy controls, 426 upregulated mRNAs and 730 downregulated mRNAs
were shared between the two datasets (Fig. 1A). The sRNA sequencing data
(GSE31037) were obtained from biopsy samples of 24 psoriasis
lesions from skin biopsies and 20 normal skin biopsies from healthy
controls. The expression profiles of miRNAs between
psoriasis-affected skin and healthy control skin were compared with
the R Deseq2 package, and 77 upregulated miRNAs and 29
downregulated miRNAs were identified (Fig. 1B).
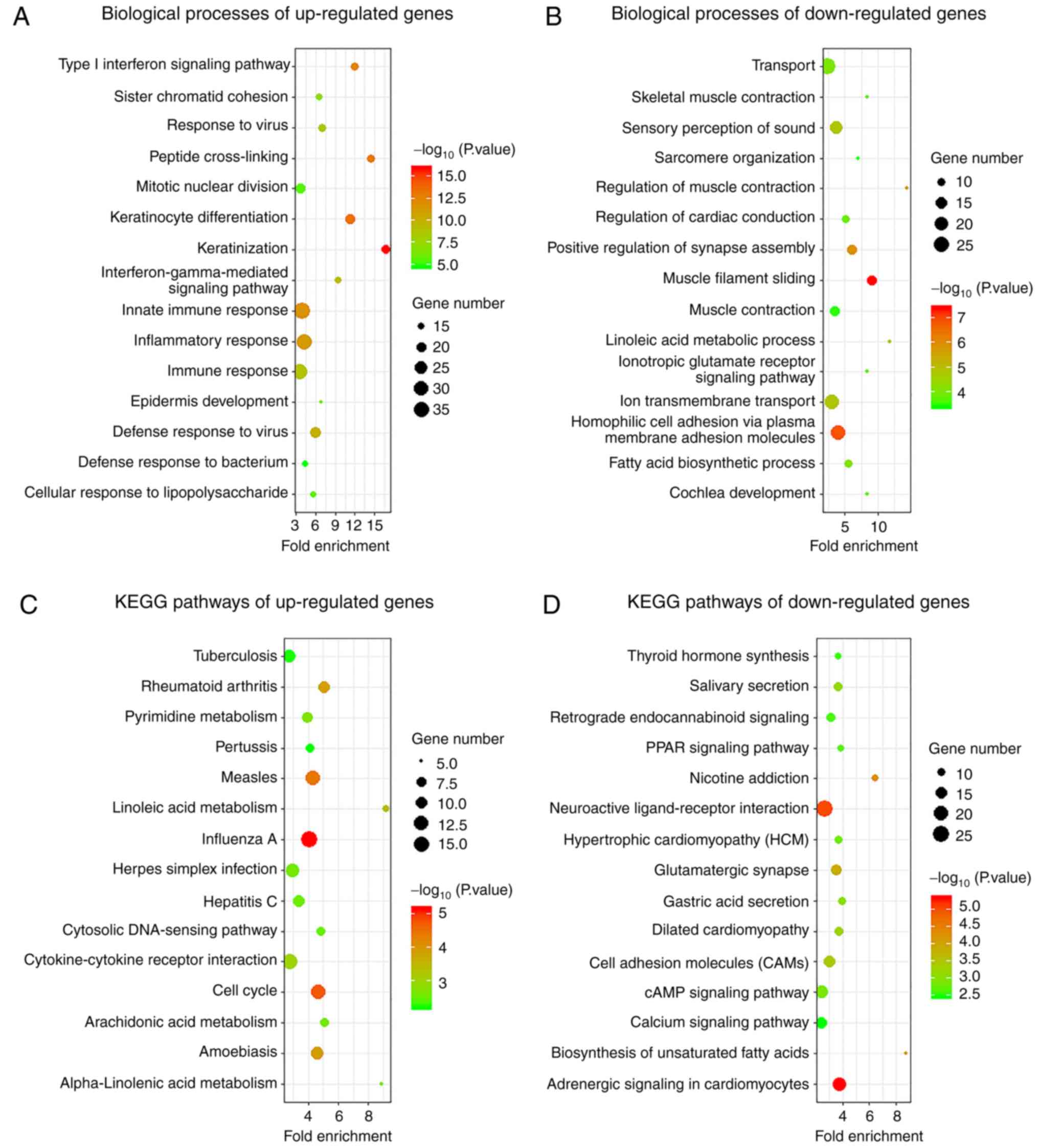
Functional enrichment analysis of
differentially expressed mRNAs in psoriasis
To investigate the biological mechanisms of lncRNAs
in the development of psoriasis, GO functional and KEGG pathway
enrichment analyses were performed for differentially expressed
genes (DEGs) in psoriasis. The results indicated that the
upregulated mRNAs were enriched for the following GO terms:
Keratinization, keratinocyte differentiation, peptide
cross-linking, type I interferon signaling pathway, innate immune
response, inflammatory response, defence response to virus,
interferon-γ-mediated response, immune response, response to virus,
sister chromatid cohesion, epidermis development, cellular response
to lipopolysaccharide, mitotic nuclear division and defence
response to bacterium (Fig. 2A). By
contrast, the downregulated genes were enriched in the following GO
terms: Muscle filament sliding, homophilic cell adhesion via plasma
membrane adhesion molecules, positive regulation of synapse
assembly, regulation of muscle contraction, ion transmembrane
transport, linoleic acid metabolic process, sensory perception of
sound, fatty acid biosynthesis process, transport, regulation of
cardiac conduction, cochlear development, ionotropic glutamate
receptor signaling pathway, skeletal muscle contraction, muscle
contraction and sarcomere organization (Fig. 2B).
The most significant KEGG pathway terms for the
upregulated mRNAs are provided in Fig.
2C: Influenza A, cell cycle, measles, amoebiasis, rheumatoid
arthritis, linoleic acid metabolism, cytokine-cytokine receptor
interaction, pyrimidine metabolism, α-linolenic acid metabolism,
arachidonic acid metabolism, herpes simplex infection, hepatitis C,
cytosolic DNA-sensing pathway, tuberculosis and pertussis. The
downregulated genes were mainly involved in adrenergic signaling in
cardiomyocytes, neuroactive ligand-receptor interaction, nicotine
addiction, biosynthesis of unsaturated fatty acids, glutamatergic
synapse, cell adhesion, dilated cardiomyopathy, salivary secretion,
gastric acid secretion, cAMP signaling pathway, hypertrophic
cardiomyopathy, peroxisome proliferator activated receptor-α
signaling pathway, retrograde endocannabinoid signaling, thyroid
hormone synthesis and calcium signaling pathway (Fig. 2D).
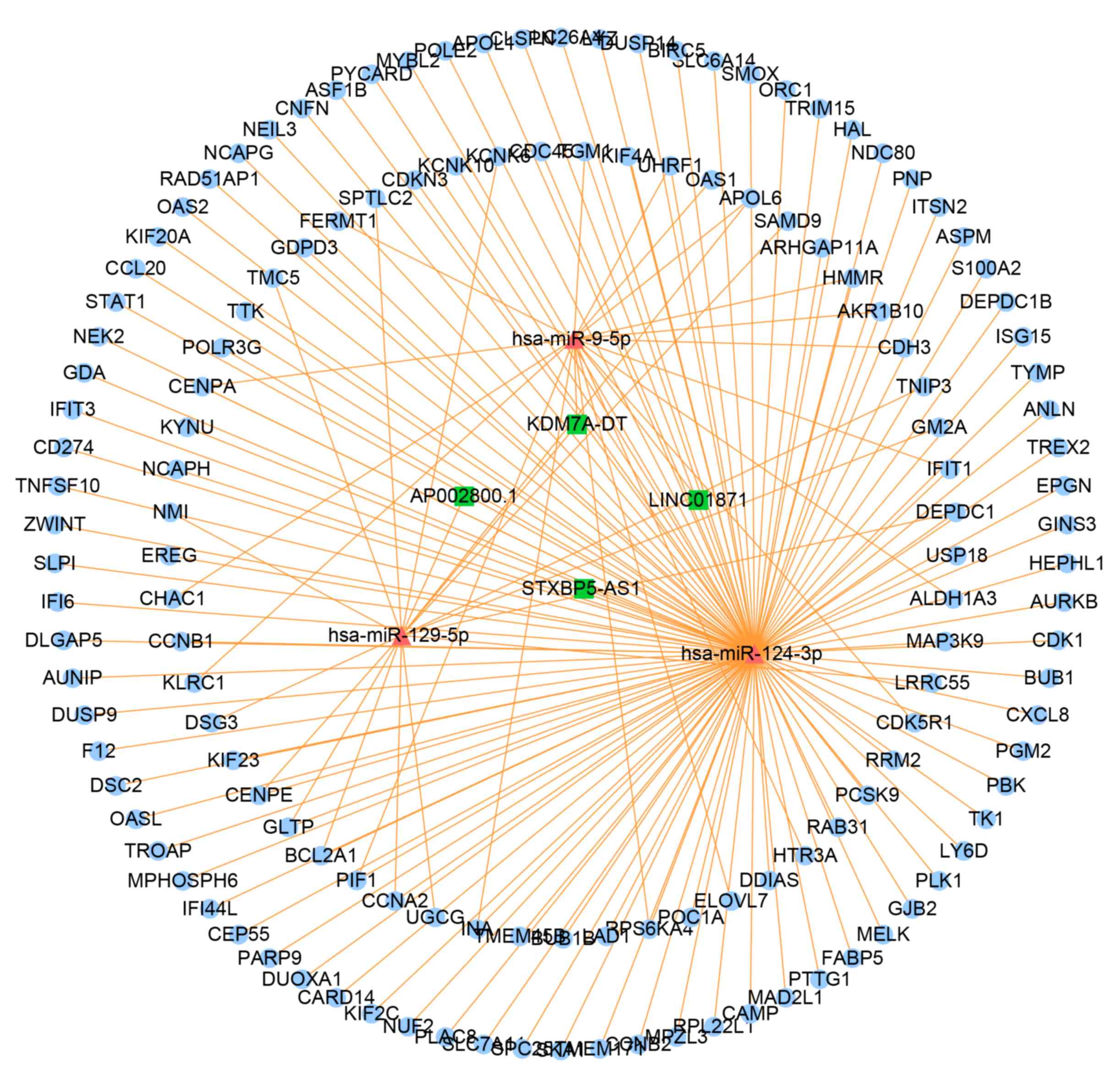
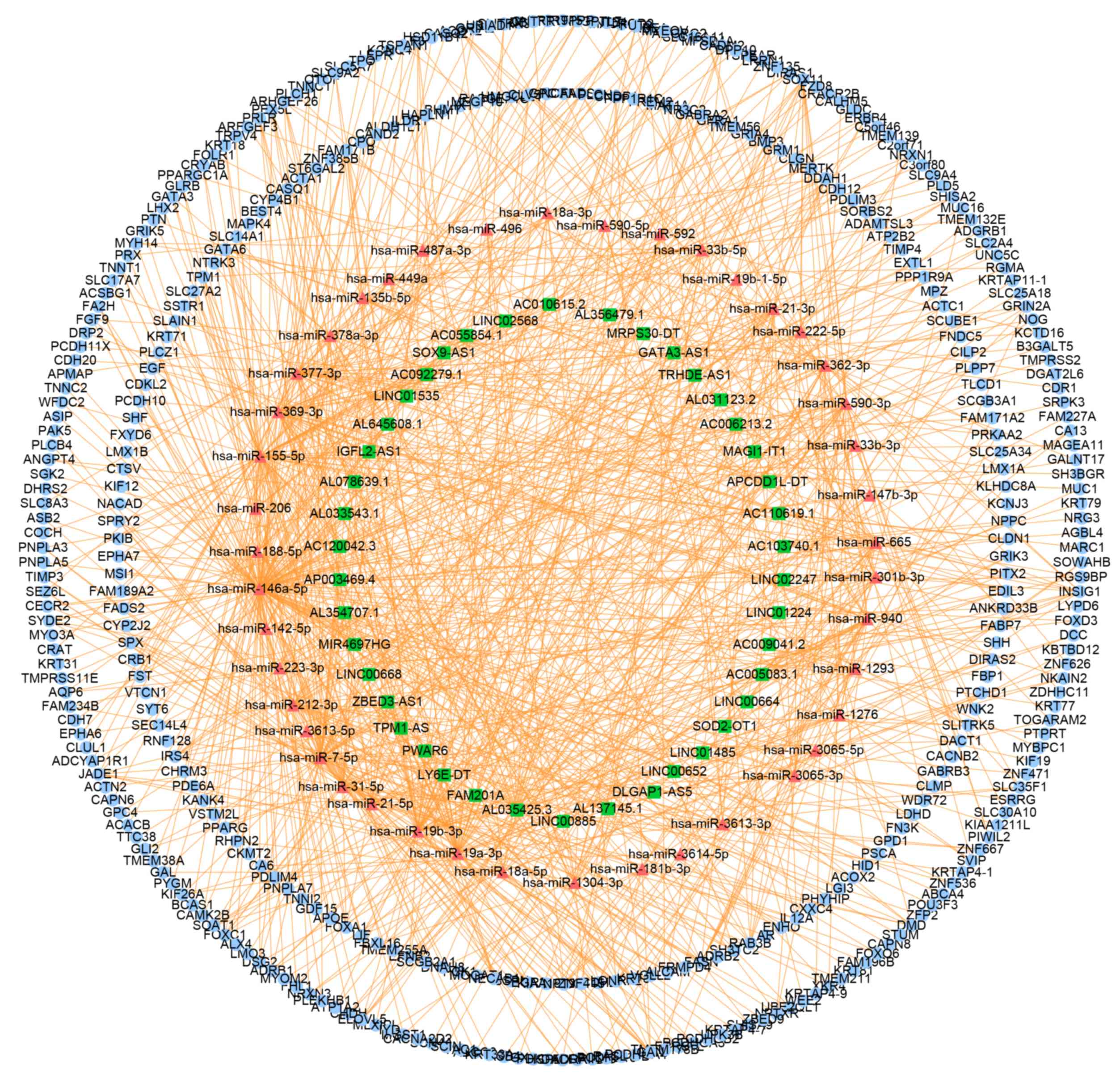
Construction of the
psoriasis-associated ceRNA network
As presented in Figs.
3 and 4, the upregulated
lncRNA-mediated ceRNA network contained 4 lncRNA nodes, 3 miRNA
nodes and 139 mRNA nodes, and the downregulated lncRNA-mediated
ceRNA network contained 42 lncRNA nodes, 43 miRNA nodes and 382
mRNA nodes.
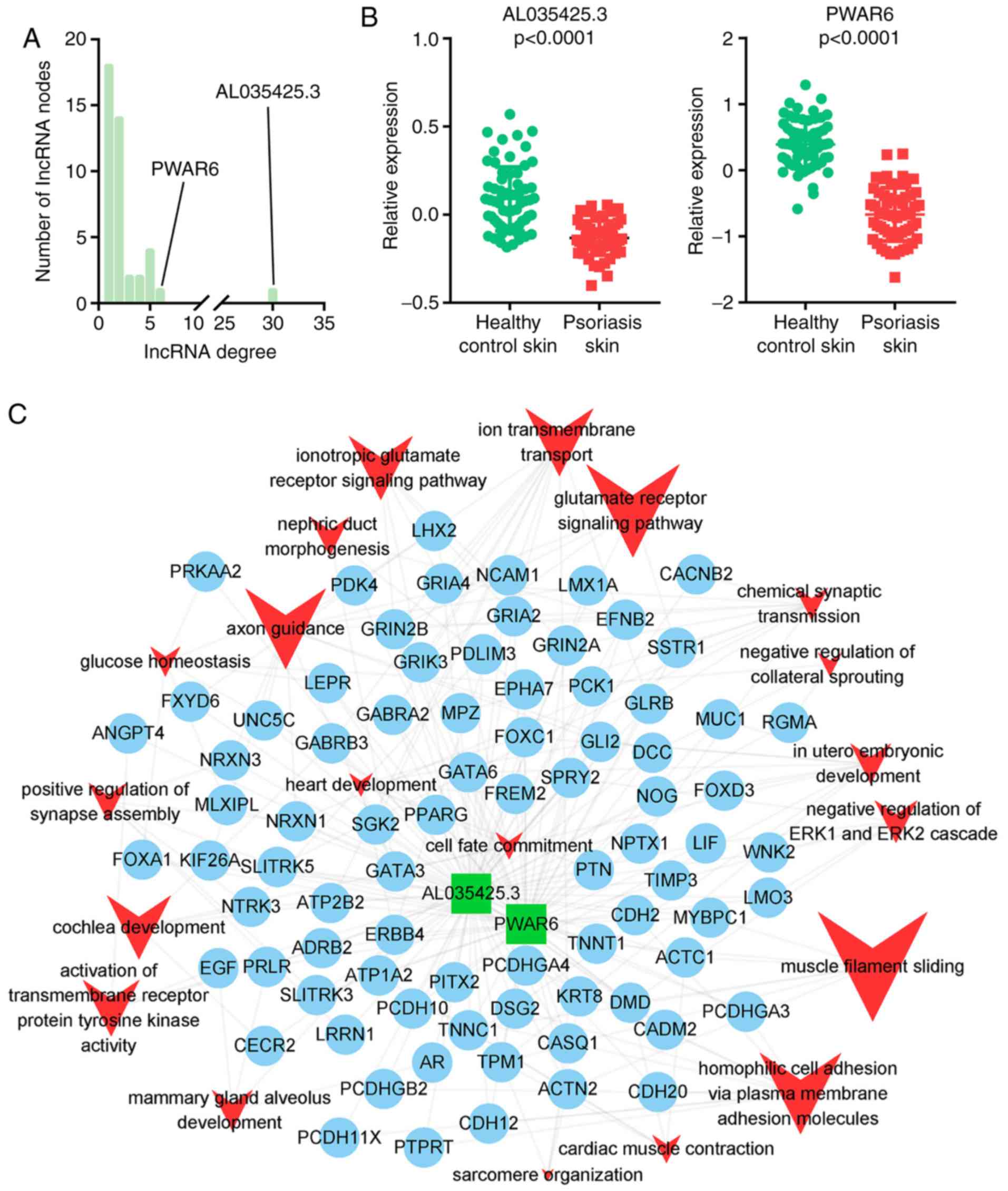
Identification of key lncRNAs
lncRNA nodes that interacted with >5 different
miRNAs were identified as key lncRNAs, which may have critical
roles in biological networks. Of note, in the downregulated
lncRNA-mediated ceRNA network, only two lncRNAs, AL035425.3 and
Prader Willi/Angelman region RNA 6 (PWAR6), interacted with >5
different miRNAs (Fig. 5A), while in
the upregulated lncRNA-mediated ceRNA network, none of the lncRNAs
interacted with >5 different miRNAs. To further validate the
differentially expressed key lncRNAs, a t-test was used to compare
their expression between psoriasis lesions involving the skin and
healthy controls. As presented in Fig.
5B, the key lncRNAs were downregulated in psoriasis-affected
skin. To better understand the mechanism of those key lncRNAs in
psoriasis, sub-ceRNA networks were extracted and GO functional
enrichment analysis for the DEGs associated with those key lncRNAs
was performed (Fig. 5C). In the GO
enrichment analysis, the DEGs associated with lncRNA AL035425.3 and
PWAR6 were involved in the biological processes of muscle filament
sliding, glutamate receptor signaling pathway, homophilic cell
adhesion via plasma membrane adhesion molecules, axon guidance, ion
transmembrane transport, cochlear development and ionotropic
glutamate receptor signaling pathway.
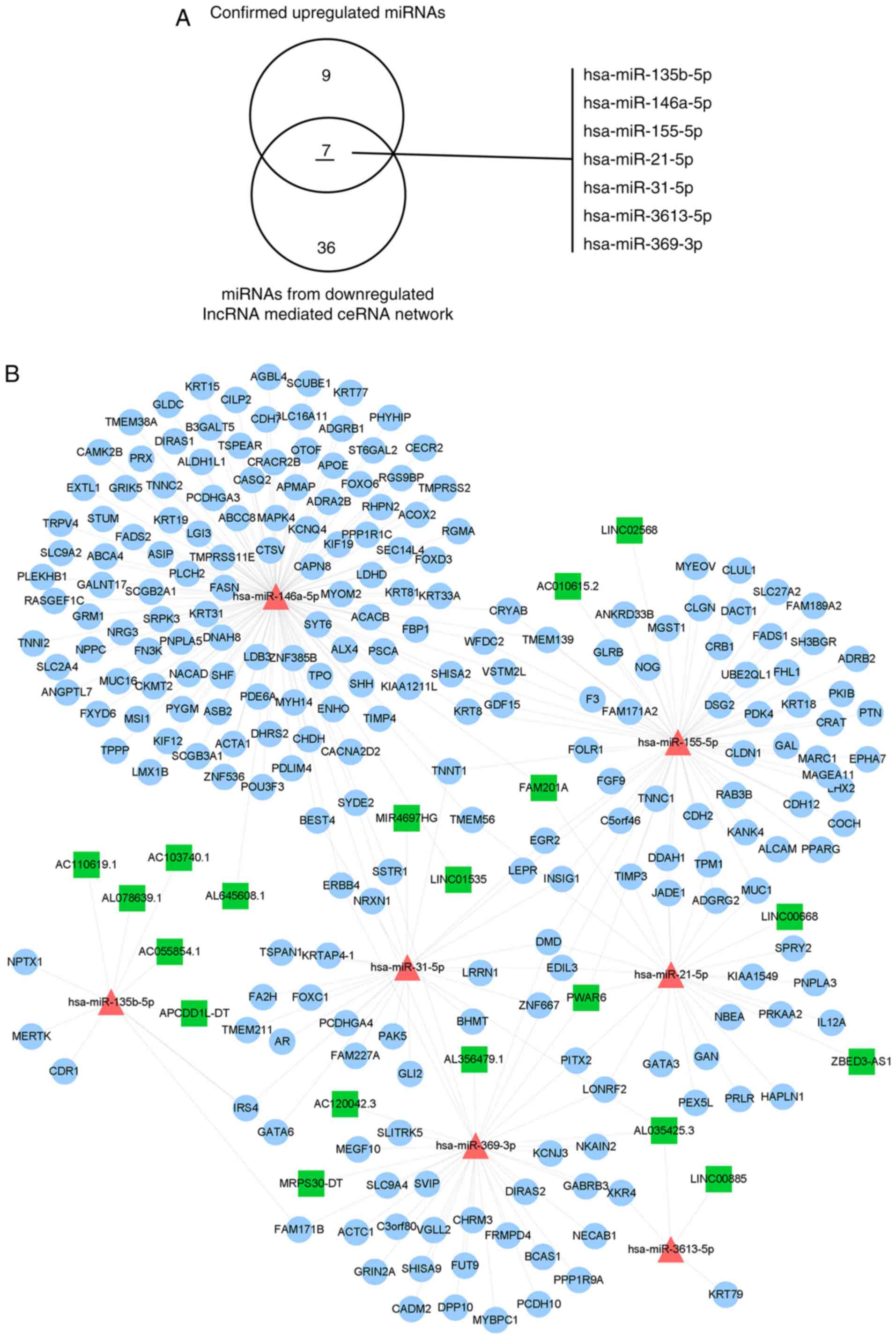
Construction of a specific validated
miRNA-associated lncRNA-miRNA-mRNA sub-network
A systematic literature search in the PubMed and Web
of Science databases was performed to identify real-time
PCR-validated differentially expressed microRNAs, using the
following key words: ‘MicroRNA’, ‘microRNAs’, ‘miRNA’, ‘miRs’ and
‘Psoriasis’. A total of 37 miRNAs were identified, including 20
downregulated miRNAs and 17 upregulated miRNAs (Table I) (27–48). Of
the identified miRNAs, 7 validated upregulated miRNAs were also
present in the downregulated lncRNA-mediated ceRNA network, but
none of the validated downregulated miRNAs were present in the
upregulated lncRNA-mediated ceRNA network (Fig. 6A). The validated differentially
expressed miRNA-associated lncRNA-miRNA-mRNA sub-networks were
extracted (Fig. 6B). Finally, DEGs
were identified from the literature. The miRNAs associated with the
lncRNA-miRNA-mRNA sub-network were subsequently extracted and
functional enrichment analysis was performed (Tables II and III). These genes were mainly enriched in
the following GO terms: Muscle filament sliding, regulation of
muscle contraction, in utero embryonic development, and enriched in
the following KEGG pathways: AMPK signalling pathway, neuroactive
ligand-receptor interaction, hypertrophic cardiomyopathy, calcium
signalling pathway, insulin signalling pathway, adipocytokine
signalling pathway, dilated cardiomyopathy, adrenergic signalling
in cardiomyocytes and insulin resistance.
 | Table I.Differentially expressed miRNAs in
psoriasis skin identified by the literature search. |
Table I.
Differentially expressed miRNAs in
psoriasis skin identified by the literature search.
| A, Downregulated
miRNAs |
|---|
|
|---|
|
| miRNA fold-change
in GSE31037 |
|
|---|
|
|
|
|
|---|
| miRNA name | log2
fold-change | P-value | (Refs.) |
|---|
| miR-10b-5p | −0.84 |
2.27×10−5 | (27) |
| miR-125b | −0.91 |
1.12×10−15 | (28) |
| miR-138 | −0.22 |
2.98×10−2 | (29) |
| miR-143-3p | −0.33 |
4.32×10−2 | (27) |
| miR-145-5p | −0.05 |
7.84×10−1 | (27) |
| miR-181b-5p | −0.58 |
9.74×10−3 | (30) |
| miR-194 | −0.28 |
1.17×10−1 | (31) |
| miR-196b-5p | −0.62 |
4.00×10−5 | (27) |
| miR-197 | −0.28 |
2.11×10−2 | (32) |
| miR-20a-3p | 0.61 |
1.26×10−4 | (33) |
| miR-217 | 0.17 |
5.51×10−1 | (34) |
| miR-320b | −0.54 |
5.42×10−4 | (35) |
| miR-338-3p | −0.48 |
5.24×10−3 | (27) |
| miR-423 | 0.05 |
8.52×10−1 | (32) |
| miR-424 | 0.83 |
1.99×10−6 | (36) |
| miR-4516 | −0.90 |
1.03×10−2 | (37) |
| miR-486-3p | −1.09 |
4.17×10−3 | (38) |
| miR-876-5p | 0.06 |
9.37×10−1 | (39) |
| miR-99a | −0.99 |
2.47×10−11 | (32) |
| miR-675-3p | −2.05 |
2.56×10−11 | (40) |
|
| B, Upregulated
miRNAs |
|
|
| miRNA
fold-change in GSE31037 |
|
|
|
|
|
| miRNA
name | log2
fold-change | P-value | (Refs.) |
|
| miR-122-5p | −0.48 |
6.17×10−1 | (41) |
| miR-130a-3p | 0.01 |
9.55×10−1 | (42) |
| miR-135b-5p | 2.56 |
2.46×10−33 | (40) |
| miR-146a-5p | 1.28 |
1.67×10−12 | (27) |
| miR-146b | 0.36 |
9.03×10−2 | (43) |
| miR-155-5p | 1.53 |
1.60×10−12 | (44) |
| miR-203 | −0.12 |
5.77×10−1 | (28) |
| miR-205 | 0.56 |
8.04×10−5 | (45) |
| miR-21-5p | 2.07 |
2.40×10−26 | (27) |
| miR-210 | −0.54 |
2.03×10−2 | (46) |
| miR-221 | −0.07 |
6.02×10−1 | (45) |
| miR-222-3p | 0.11 |
4.26×10−1 | (45) |
| miR-31-5p | 5.46 |
7.08×10−49 | (27) |
| miR-3613-5p | 1.68 |
2.16×10−11 | (40) |
| miR-369-3p | 1.30 |
3.78×10−11 | (47) |
| miR-431 | 2.20 |
1.67×10−7 | (40) |
| miR-744-3p | 0.38 |
2.78×10−2 | (40) |
 | Table II.Gene Ontology enrichment analysis of
differentially expressed mRNAs from the specific miRNA-associated
long non-coding RNA-miRNA-mRNA sub-networks (top 20). |
Table II.
Gene Ontology enrichment analysis of
differentially expressed mRNAs from the specific miRNA-associated
long non-coding RNA-miRNA-mRNA sub-networks (top 20).
| Term | n (%) | P-value | Genes |
|---|
| GO:0030049~muscle
filament sliding | 9 (3.93) |
1.17×10−8 | ACTC1, TNNT1,
TNNC2, ACTA1, TNNC1, MYBPC1, DMD, TPM1, TNNI2 |
|
GO:0006937~regulation of muscle
contraction | 5 (2.18) |
1.71×10−5 | TNNT1, TNNC2,
TNNC1, TPM1, TNNI2 |
| GO:0001701~in utero
embryonic development | 11 (4.80) |
7.68×10−5 | MUC1, AR, NOG,
LMX1B, GATA6, GATA3, FOXC1, GLI2, TPM1, PITX2, FOXD3 |
| GO:0003009~skeletal
muscle contraction | 5 (2.18) |
1.66×10−4 | TNNT1, TNNC2,
TNNC1, MYH14, TNNI2 |
| GO:0060048~cardiac
muscle contraction | 6 (2.62) |
1.81×10−4 | ACTC1, TNNC1, DMD,
TPM1, CASQ2, TNNI2 |
|
GO:0045214~sarcomere organization | 5 (2.18) |
3.54×10−4 | KRT19, KRT8, LDB3,
TPM1, CASQ2 |
|
GO:0007267~cell-cell signaling | 11 (4.80) |
8.84×10−4 | AR, ADRB2, CRB1,
SSTR1, FGF9, FADS1, ADRA2B, GDF15, MERTK, ASIP, SHH |
| GO:0042493~response
to drug | 12 (5.24) |
9.89×10−4 | ACTC1, GATA6,
GATA3, PPARG, NPPC, GRIN2A, PTN, TIMP4, ACACB, GAL, ABCC8,
MGST1 |
| GO:0048645~organ
formation | 3 (1.31) |
1.35×10−3 | AR, GATA6, SHH |
| GO:0045165~cell
fate commitment | 5 (2.18) |
2.08×10−3 | SPRY2, ERBB4,
GATA6, GATA3, PPARG |
|
GO:0007156~homophilic cell adhesion via
plasma membrane adhesion molecules | 8 (3.49) |
2.72×10−3 | CDH12, CDH7, DSG2,
CADM2, PCDH10, CDH2, PCDHGA4, PCDHGA3 |
|
GO:0007010~cytoskeleton organization | 8 (3.49) |
3.03×10−3 | APOE, DMD, KRT15,
PAK5, KRT31, CECR2, GAN, TPM1 |
| GO:0042472~inner
ear morphogenesis | 5 (2.18) |
3.26×10−3 | SPRY2, KCNQ4, FGF9,
GATA3, INSIG1 |
| GO:0007605~sensory
perception of sound | 7 (3.06) |
4.96×10−3 | COCH, SPRY2, KCNQ4,
TSPEAR, GABRB3, OTOF, MYH14 |
|
GO:0009953~dorsal/ventral pattern
formation | 4 (1.75) |
6.22×10−3 | NOG, LMX1B, LHX2,
SHH |
| GO:0042593~glucose
homeostasis | 6 (2.62) |
6.89×10−3 | SLC2A4, LEPR, PDK4,
PPARG, TRPV4, PRKAA2 |
| GO:0043627~response
to estrogen | 5 (2.18) |
7.25×10−3 | KRT19, GATA6,
GATA3, PPARG, GAL |
| GO:0032868~response
to insulin | 5 (2.18) |
8.06×10−3 | EGR2, FADS1, TRPV4,
GAL, ABCC8 |
|
GO:0007171~activation of transmembrane
receptor protein tyrosine kinase activity | 3 (1.31) |
8.45×10−3 | ADRB2, NRG3,
PRLR |
| GO:0034332~adherens
junction organization | 4 (1.75) |
9.35×10−3 | CDH12, CDH7, CADM2,
CDH2 |
 | Table III.Kyoto Encyclopedia of Genes and
Genomes signaling pathway enrichment analysis of differentially
expressed mRNAs from the specific miRNAs-associated long non-coding
RNA-miRNA-mRNA sub-network. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes signaling pathway enrichment analysis of differentially
expressed mRNAs from the specific miRNAs-associated long non-coding
RNA-miRNA-mRNA sub-network.
| Term | n (%) | P-value | Genes |
|---|
| hsa04152:AMPK
signaling pathway | 8 (3.49) |
8.22×10−4 | IRS4, SLC2A4, LEPR,
PPARG, FASN, FBP1, PRKAA2, ACACB |
|
hsa04080:Neuroactive ligand-receptor
interaction | 11 (4.80) |
2.25×10−3 | GLRB, ADRB2, PRLR,
GABRB3, CHRM3, SSTR1, LEPR, GRIK5, GRIN2A, ADRA2B, GRM1 |
|
hsa05410:Hypertrophic cardiomyopathy | 6 (2.62) |
2.80×10−3 | ACTC1, TNNC1, DMD,
PRKAA2, CACNA2D2, TPM1 |
| hsa04020:Calcium
signaling pathway | 8 (3.49) |
6.85×10−3 | ADRB2, TNNC2,
CHRM3, ERBB4, TNNC1, GRIN2A, CAMK2B, GRM1 |
| hsa04910:Insulin
signaling pathway | 7 (3.06) |
7.39×10−3 | IRS4, PYGM, SLC2A4,
FASN, FBP1, PRKAA2, ACACB |
|
hsa04920:Adipocytokine signaling
pathway | 5 (2.18) |
1.12×10−2 | IRS4, SLC2A4, LEPR,
PRKAA2, ACACB |
| hsa05414:Dilated
cardiomyopathy | 5 (2.18) |
2.06×10−2 | ACTC1, TNNC1, DMD,
CACNA2D2, TPM1 |
| hsa04261:Adrenergic
signaling in cardiomyocytes | 6 (2.62) |
2.88×10−2 | ACTC1, ADRB2,
TNNC1, CAMK2B, CACNA2D2, TPM1 |
| hsa04931:Insulin
resistance | 5 (2.18) |
4.58×10−2 | PYGM, SLC2A4,
PRKAA2, ACACB, SLC27A2 |
Discussion
Psoriasis is a chronic immune-mediated inflammatory
disease that affects 3.2% of the adult population in the US
(49). Over the past decades, the
role of different cytokines in psoriasis has been studied widely
(50), and therefore,
cytokine-targeting drugs have been developed, including adalimumab
(which targets tumor necrosis factor-α), secukinumab (which targets
IL-17A) and ustekinumab (which targets IL-12 and IL-23 p40)
(51). However, the roles of lncRNAs
in psoriasis have remained largely elusive.
lncRNAs have important roles in immune and
inflammatory pathways by regulating gene expression through
multiple mechanisms (52). Among
these mechanisms, the ceRNA theory is commonly studied. According
to this theory, lncRNAs act as ‘sponges’ for miRNAs and decrease
the effects of miRNAs on their target genes, thus promoting the
expression of target genes (53).
For instance, Qiao et al (12) reported that lncRNA-MSX2P1 activates
S100A71 and facilitates the growth of IL-22-stimulated
keratinocytes by inhibiting miR-6731-5p. Furthermore, Li et
al (54) demonstrated that
lncRNA H19 regulates the differentiation of keratinocytes by
increasing desmoglein 1 expression through sponging
miR-130b-3p.
In the present study, psoriasis-associated
lncRNA-miRNA-mRNA networks were constructed based on the ceRNA
theory, and the DEGs were also subjected to GO BP enrichment
analysis and KEGG pathway enrichment analysis. GO BP enrichment
analysis of the upregulated mRNAs indicated that the significantly
enriched GO BP terms were psoriasis-associated biological
processes, including keratinization (55), keratinocyte differentiation (3), inflammatory response (56) and immune response (56). Furthermore, the downregulated mRNAs
were mainly enriched in the BP terms muscle filament sliding,
homophilic cell adhesion via plasma membrane adhesion molecules,
positive regulation of synapse assembly and regulation of muscle
contraction.
In the present study, the downregulated lncRNAs
AL035425.3 and PWAR6 were identified as key lncRNAs. GO BP analysis
of the key lncRNAs revealed that AL035425.3 and PWAR6 were involved
in muscle filament sliding, the glutamate receptor signaling
pathway, homophilic cell adhesion via plasma membrane adhesion
molecules, axon guidance, ion transmembrane transport, cochlear
development and ionotropic glutamate receptor signaling pathway. Of
the two key lncRNAs, PWAR6 was previously reported to be a tumor
suppressor lncRNA in glioma, and high expression of PWAR6 was
reported to be an indicator of better survival in glioma patients
(57). In addition, PWAR6 is
functionally important in Prader-Willi syndrome, and the disruption
of its expression is associated with the pathogenesis of the
disease (58). Two potential target
miRNAs of PWAR6 were identified from the validated miRNA-associated
lncRNA-miRNA-mRNA sub-network: miR-155-5p and miR-369-3p. Previous
studies have demonstrated that knockdown of miR-155-5p suppresses
psoriasis-associated inflammatory responses through regulation of
the NLR family pyrin domain containing 3 inflammasome (44); furthermore, the expression levels of
miR-369-3p in skin had a positive correlation with the Psoriasis
Area and Severity Index, which is used to assess disease severity
(47). Taken together, these results
indicate that PWAR6 may be involved in the progress of psoriasis
via regulation of the expression of miR-155-5p and miR-369-3p.
The present study has certain limitations that
should be considered: First, although the expression profiles of
lncRNA and mRNA were determined from the same specimens, the miRNA
expression profile is from an independent dataset. Thus, combining
these profiles into a network may introduce selection bias, and
further experiments using the same specimens are required. In
addition, further validation of key lncRNA expression levels in
psoriasis samples is required. Finally, additional functional
investigations of these lncRNAs in the context of psoriasis
progression are required.
In summary, the present study presented a novel and
important psoriasis-associated ceRNA network based on the ceRNA
theory. The results indicate that certain lncRNAs have key roles in
the development of psoriasis. In addition, the present study
identified two lncRNAs, AL035425.3 and PWAR6, that were indicated
to have a central role in psoriasis. The present study provides
insight into the molecular mechanisms of psoriasis, and this may
help to identify novel therapeutic targets for the treatment of
psoriasis in the future.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant nos. 81673050 and
81872522), the Program of Science and Technology Commission of
Shanghai Municipality (grant no. 18140901800) and the Excellent
Subject Leader Program of Shanghai Municipal Commission of Health
and Family Planning (grant no. 2018BR30).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ and YS designed the current study, performed data
analysis and wrote the manuscript. QY and YG performed the
literature review regarding microRNAs in psoriasis. ZL, HX and YW
analyzed the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greb JE, Goldminz AM, Elder JT, Lebwohl
MG, Gladman DD, Wu JJ, Mehta NN, Finlay AY and Gottlieb AB:
Psoriasis. Nat Rev Dis Primers. 2:160822016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ogawa E, Sato Y, Minagawa A and Okuyama R:
Pathogenesis of psoriasis and development of treatment. J Dermatol.
45:264–272. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Cesare A, Di Meglio P and Nestle FO:
The IL-23/Th17 Axis in the Immunopathogenesis of Psoriasis. J
Invest Dermatol. 129:1339–1350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szell M, Danis J, Bata-Csorgo Z and Kemeny
L: PRINS, a primate-specific long non-coding RNA, plays a role in
the keratinocyte stress response and psoriasis pathogenesis.
Pflugers Arch. 468:935–943. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Danis J, Goblos A, Bata-Csorgo Z, Kemeny L
and Szell M: PRINS Non-Coding RNA regulates nucleic acid-induced
innate immune responses of human keratinocytes. Front Immunol.
8:10532017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szegedi K, Sonkoly E, Nagy N, Németh IB,
Bata-Csörgo Z, Kemény L, Dobozy A and Széll M: The anti-apoptotic
protein G1P3 is overexpressed in psoriasis and regulated by the
non-coding RNA, PRINS. Exp Dermatol. 19:269–278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiao M, Li R, Zhao X, Yan J and Sun Q:
Up-regulated lncRNA-MSX2P1 promotes the growth of IL-22-stimulated
keratinocytes by inhibiting miR-6731-5p and activating S100A7. Exp
Cell Res. 363:243–254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolger AM, Marc L and Bjoern U:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Daehwan K, Ben L and Salzberg SL: HISAT: A
fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Simon A, Paul Theodor P and Wolfgang H:
HTSeq-a Python framework to work with high-throughput sequencing
data. Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen L, Liu W, Cui J, Li J and Li C:
Analysis of long non-coding RNA expression profiles in ovarian
cancer. Oncol Lett. 14:1526–1530. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Friedl?Nder MR, Mackowiak SD, Na L, Wei C
and Nikolaus R: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–d302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian L, Hu X, He Y, Wu Z, Li D and Zhang
H: Construction of lncRNA-miRNA-mRNA networks reveals functional
lncRNAs in abdominal aortic aneurysm. Exp Ther Med. 16:3978–3986.
2018.PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan JJ, Qiao M, Li RH, Zhao XT, Wang XY
and Sun Q: Downregulation of miR-145-5p contributes to
hyperproliferation of keratinocytes and skin inflammation in
psoriasis. Br J Dermatol. 180:365–372. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sonkoly E, Wei T, Janson PC, Sääf A,
Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B,
Scheynius A, et al: MicroRNAs: Novel regulators involved in the
pathogenesis of psoriasis? PLoS One. 2:e6102007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu D, Yu W, Li M, Wang H, Liu D, Song X,
Li Z and Tian Z: MicroRNA-138 regulates the balance of Th1/Th2 via
targeting RUNX3 in psoriasis. Immunol Lett. 166:55–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng C, Bai M, Yu NZ, Wang XJ and Liu Z:
MicroRNA-181b negatively regulates the proliferation of human
epidermal keratinocytes in psoriasis through targeting TLR4. J Cell
Mol Med. 21:278–285. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu X, An J, Hua Y, Li Z, Yan N, Fan W and
Su C: MicroRNA-194 regulates keratinocyte proliferation and
differentiation by targeting Grainyhead-like 2 in psoriasis. Pathol
Res Pract. 213:89–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lerman G, Avivi C, Mardoukh C, Barzilai A,
Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S,
Orenstein A, et al: MiRNA expression in psoriatic skin: Reciprocal
regulation of hsa-miR-99a and IGF-1R. PLoS One. 6:e209162011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li R, Qiao M, Zhao X, Yan J, Wang X and
Sun Q: MiR-20a-3p regulates TGF-β1/Survivin pathway to affect
keratinocytes proliferation and apoptosis by targeting SFMBT1 in
vitro. Cell Signal. 49:95–104. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu H, Hou L, Liu J and Li Z: MiR-217 is
down-regulated in psoriasis and promotes keratinocyte
differentiation via targeting GRHL2. Biochem Biophys Res Commun.
471:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Yu X, Wang L, Ma W and Sun Q:
miR-320b is down-regulated in psoriasis and modulates keratinocyte
proliferation by targeting AKT3. Inflammation. 41:2160–2170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ichihara A, Jinnin M, Yamane K, Fujisawa
A, Sakai K, Masuguchi S, Fukushima S, Maruo K and Ihn H:
microRNA-mediated keratinocyte hyperproliferation in psoriasis
vulgaris. Br J Dermatol. 165:1003–1010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chowdhari S, Sardana K and Saini N:
miR-4516, a microRNA downregulated in psoriasis inhibits
keratinocyte motility by targeting fibronectin/integrin α9
signaling. Biochim Biophys Acta Mol Basis Dis. 1863:3142–3152.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang M, Sun Z, Dang E, Li B, Fang H, Li
J, Gao L, Zhang K and Wang G: TGFβ/SMAD/microRNA-486-3p signaling
axis mediates keratin 17 expression and keratinocyte
hyperproliferation in psoriasis. J Invest Dermatol. 137:2177–2186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
A R, Yu P, Hao S and Li Y: MiR-876-5p
suppresses cell proliferation by targeting Angiopoietin-1 in the
psoriasis. Biomed Pharmacother. 103:1163–1169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Joyce CE, Zhou X, Xia J, Ryan C, Thrash B,
Menter A, Zhang W and Bowcock AM: Deep sequencing of small RNAs
from human skin reveals major alterations in the psoriasis
miRNAome. Hum Mol Genet. 20:4025–4040. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang M, Ma W, Gao Y, Jia K, Zhang Y, Liu
H and Sun Q: IL-22-induced miR-122-5p promotes keratinocyte
proliferation by targeting Sprouty2. Exp Dermatol. 26:368–374.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong Y, Chen H, Liu L, Wang Z, Tian F and
Zhao Y: microRNA-130a promotes human keratinocyte viability and
migration and inhibits apoptosis through direct regulation of
STK40-mediated NF-κB pathway and indirect regulation of
SOX9-meditated JNK/MAPK pathway: A potential role in psoriasis. DNA
Cell Biol. 36:219–226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hermann H, Runnel T, Aab A, Baurecht H,
Rodriguez E, Magilnick N, Urgard E, Šahmatova L, Prans E,
Maslovskaja J, et al: miR-146b probably assists miRNA-146a in the
suppression of keratinocyte proliferation and inflammatory
responses in psoriasis. J Invest Dermatol. 137:1945–1954. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo Q, Zeng J, Li W, Lin L, Zhou X, Tian
X, Liu W, Zhang L and Zhang X: Silencing of miR155 suppresses
inflammatory responses in psoriasis through inflammasome NLRP3
regulation. Int J Mol Med. 42:1086–1095. 2018.PubMed/NCBI
|
|
45
|
Zibert JR, Lovendorf MB, Litman T, Olsen
J, Kaczkowski B and Skov L: MicroRNAs and potential target
interactions in psoriasis. J Dermatol Sci. 58:177–185. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu R, Zeng J, Yuan J, Deng X, Huang Y,
Chen L, Zhang P, Feng H, Liu Z, Wang Z, et al: MicroRNA-210
overexpression promotes psoriasis-like inflammation by inducing Th1
and Th17 cell differentiation. J Clin Invest. 128:2551–2568. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guo S, Zhang W, Wei C, Wang L, Zhu G, Shi
Q, Li S, Ge R, Li K, Gao L, et al: Serum and skin levels of
miR-369-3p in patients with psoriasis and their correlation with
disease severity. Eur J Dermatol. 23:608–613. 2013.PubMed/NCBI
|
|
48
|
Wang C, Zong J, Li Y, Wang X, Du W and Li
L: MiR-744-3p regulates keratinocyte proliferation and
differentiation via targeting KLLN in psoriasis. Exp Dermatol.
28:283–291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rachakonda TD, Schupp CW and Armstrong AW:
Psoriasis prevalence among adults in the United States. J Am Acad
Dermatol. 70:512–516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Baliwag J, Barnes DH and Johnston A:
Cytokines in psoriasis. Cytokine. 73:342–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nograles KE and Krueger JG: Anti-cytokine
therapies for psoriasis. Exp Cell Res. 317:1293–1300. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li
XM and Ye DQ: Emerging role of long noncoding RNAs in autoimmune
diseases. Autoimmun Rev. 14:798–805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li CX, Li HG, Huang LT, Kong YW, Chen FY,
Liang JY, Yu H and Yao ZR: H19 lncRNA regulates keratinocyte
differentiation by targeting miR-130b-3p. Cell Death Dis.
8:e31742017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Akiyama M: Early-onset generalized
pustular psoriasis is representative of autoinflammatory
keratinization diseases. J Allergy Clin Immunol. 143:809–810. 2018.
View Article : Google Scholar
|
|
56
|
Deng Y, Chang C and Lu Q: The inflammatory
response in psoriasis: A Comprehensive review. Clin Rev Allergy
Immunol. 50:377–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lin X, Jiang T, Bai J, Li J, Wang T, Xiao
J, Tian Y, Jin X, Shao T, Xu J, et al: Characterization of
transcriptome transition associates long noncoding RNAs with glioma
progression. Mol Ther Nucleic Acids. 13:620–632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lei M, Mitsuhashi S, Miyake N, Ohta T,
Liang D, Wu L and Matsumoto N: Translocation breakpoint disrupting
the host SNHG14 gene but not coding genes or snoRNAs in typical
Prader-Willi syndrome. J Hum Genet. 64:647–652. 2019. View Article : Google Scholar : PubMed/NCBI
|