Introduction
Long non-coding RNAs (lncRNAs) are a major class of
non-coding RNAs with >200 base pairs (1,2). Recent
studies have revealed the important roles of lncRNAs in cancer
progression (3–6). A variety of lncRNAs have been observed
to be dysregulated in a number of human cancers, serve as oncogenes
or serve a tumor suppressor role (7,8). LncRNAs
are involved in the regulation of a variety of biological
processes, including cell growth, apoptosis and differentiation via
protein interaction, sponging microRNAs (miRNAs/miRs) and binding
to DNA (9–11). For example, lncRNA GATA binding
protein 6-antisense RNA (AS) has been indicated to interact with
lysyl oxidase like 2 to transcriptionally regulate gene expression
(12). LncRNA associated with BCBM
has been revealed to regulate interleukin (IL)-6-triggered signal
transducer and activator of transcription 3 phosphorylation by
binding to janus kinase 2 (13).
Furthermore, lncRNAs have been demonstrated to have a prognostic
value of in human cancers. The upregulation of serine peptidase
inhibitor kunitz type 1-antisense RNA 1 predicted a poor prognosis
in colorectal cancer (14), and the
overexpression of H19 imprinted maternally expressed transcript
predicted a poor prognosis in melanoma (15). These results demonstrated that
lncRNAs serves a crucial role in regulating human cancer
progression.
Colorectal cancer (CRC) is the third most common
type of cancer worldwide (16).
Worldwide, the morbidity and mortality of CRC has increased rapidly
in recent years, with 134,490 new cases and 49,190 cases of
CRC-associated mortality in 2016 (17). Previous studies have indicated that
lncRNAs are associated with CRC progression and prognosis. lncRNA
BRAF non-coding RNA was demonstrated to promote CRC tumorigenesis
and enhance adriamycin resistance, and LINC00312 was indicated to
repress CRC cell proliferation and metastasis via miR-21 (18,19).
However, the expression pattern and functional roles of a large
number of lncRNAs remain undetermined. Therefore, there is an
urgent requirement for the identification of CRC-associated lncRNAs
and for the generation of useful information that can be used to
understand their potential roles in CRC.
With the development of microarray profiles and RNA
sequencing, a series of public gene expression analyses were
published, including The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/gds) datasets. These
databases provide powerful tools that can be used to identify
cancer-associated mRNAs, miRNAs and lncRNAs. For example, Zhang
et al (20) identified 218
differently expressed genes in in papillary thyroid cancer using a
TCGA dataset. Yuan et al (21) identified 1,176 lncRNAs and 245 miRNAs
in colorectal cancer using the TCGA dataset. A number of lncRNAs,
which were predicted using bioinformatics, were identified to be
potential biomarkers in a variety of human cancer types. A total of
four lncRNAs (LINC01018, LOC553137, miR4435-2HG and TTTY14) were
revealed to be correlated with overall survival in patients with
gastric cancer (22). An increased
expression of miR100HG has also been associated with poor prognosis
in patients with breast cancer (23). Emerging evidence has indicated
bioinformatics analyses to be a powerful method that can be used to
understand the potential roles of novel lncRNAs in a variety of
human diseases, including cancer. Wang et al (24) demonstrated that dysregulated lncRNAs
in esophageal squamous cell carcinoma were associated with the
regulation of cancer metastasis, using co-expression analysis
(24). Cao et al (25) also demonstrated that dysregulated
lncRNAs in bladder cancer were associated with the regulation of
cell migration and the Ras signaling pathway.
In the present study, a GEO dataset GSE21510 was
re-annotated to identify CRC associated mRNAs and lncRNAs.
Bioinformatics analyses were also performed to understand the
potential roles of these lncRNAs in CRC. The current study aimed to
provide a novel insight into CRC-associated lncRNAs that could
serve as biomarkers in CRC.
Materials and methods
LncRNA classification pipeline
A pipeline, which was previously described by Zhang
et al (26), was used to
re-annotate microarray data.
Microarray data and data
preprocessing
In the present study, GSE21510 dataset (27) was downloaded from the GEO database to
identify differently expressed mRNAs and lncRNAs. GSE21510 included
25 normal and 123 CRC samples. A total of 104 patients were
assigned to the microarray study, including 13 patients with stage
I, 37 patients with stage II, 34 patients with stage III and 20
patients with stage IV CRC. Metastatic recurrence after surgery
occurred in 18 patients with stage I–III disease, and the median
follow-up time was 22 months for these patients. The raw data were
normalized using robust multi-array average (RMA) method (28) using R 2.6.2 statistical software
(29) with the affy package
(30) from BioConductor V3.9
(http://www.bioconductor.org/).
Normalization was separately performed for LCM dataset and
homogenized tissue dataset (31).
The normalized gene expression levels were presented as
log2-transformed values using the RMA method. LncRNAs or mRNAs with
fold changes (FC) ≥2 and P<0.05 were considered as
differentially expressed lncRNAs or mRNAs. Furthermore, gene
expression profile data in CRC (including 275 colon adenocarcinoma
samples and 92 rectum adenocarcinoma samples) and normal tissues
were downloaded from TCGA (https://portal.gdc.cancer.gov/) and used to examine
the expression pattern of key lncRNA in CRC and its association
with the patient clinicopathological features.
Functional group analysis
The Database for Annotation, Visualization and
Integrated Discovery v6.8 (http://david.ncifcrf.gov/) was used to perform Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analyses. GO analyses included biological
process, cellular component and molecular function. GO terms and
KEGG pathways with a value of P<0.05 were considered to be
significantly enriched functional annotations.
lncRNA-mRNA correlation network
The lncRNA-mRNA correlation network was established
according to lncRNA target predictions along with differentially
expressed lncRNA and mRNA profiles in GSE21510. A total of 3,647
lncRNA-mRNA pairs with an absolute Pearson value >0.85 were
selected to construct the co-expression network in CRC.
Statistical analysis
Statistical comparisons between groups were
performed using a t-test or Mann-Whitney U-test, as appropriate.
Kaplan-Meier survival curves were constructed with the log-rank
test and P-values were calculated. The cut-off threshold was set to
P<0.05. A log-rank test was conducted to examine the
significance of the difference between the curves. P<0.05 was
considered to indicate a statistically significant result, with a
95% confidence level.
Results
Identification of the differentially
expressed mRNAs and lncRNAs in CRC
A public GEO dataset, GSE21510, which included 25
normal and 123 CRC samples was analyzed to identify CRC-associated
mRNAs and lncRNAs. A total of 2,387 mRNAs were revealed to be
differentially expressed in CRC with a fold change of >1.5 and
P<0.01. Among these mRNAs, 1,874 were upregulated and 1,385 were
downregulated in CRC compared with normal samples. Hierarchical
clustering was used to identify differentially expressed mRNAs in
CRC (Fig. 1A).
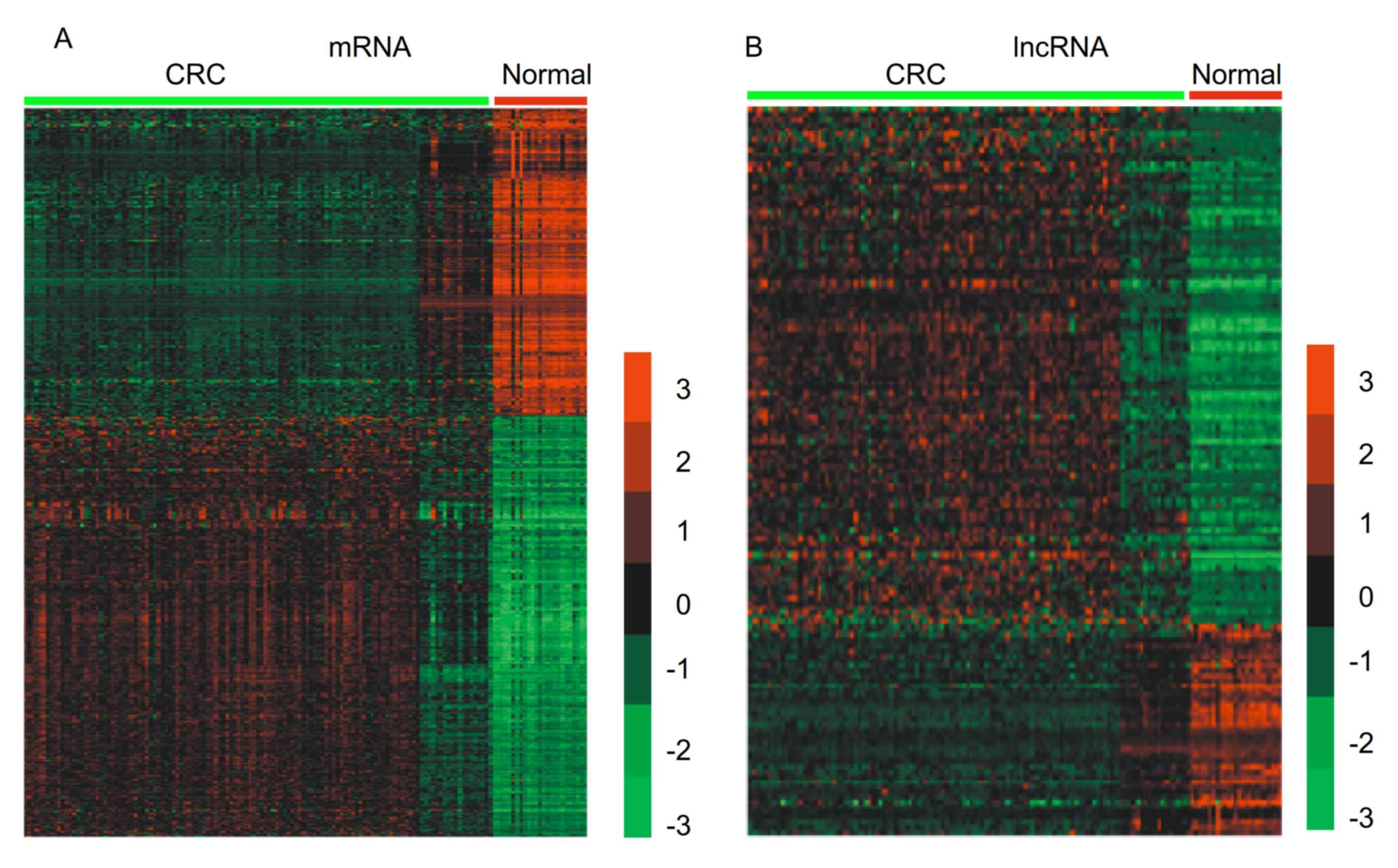 | Figure 1.Identification of the differentially
expressed mRNAs and lncRNAs in CRC. Hierarchical clustering
analysis indicating the differential (A) mRNA expression and (B)
lncRNA expression in CRC, using GSE21510. Red indicates high
expression and green indicates low expression. 3, 2, 1, 0, −1, −2,
and −3 are fold changes compared with normal samples. lncRNA, long
non-coding RNA; CRC, colorectal cancer; normal, normal tissue
samples. |
After re-annotating GSE21510 according to Zhang
et al (26) report, a total
of 107 upregulated lncRNAs and 43 downregulated lncRNAs were
identified in CRC compared with normal samples with a fold change
of >1.5 and P<0.01. Among these genes, a number of lncRNAs,
including colorectal neoplasia differentially expressed (CRNDE)
(32), growth arrest specific 5
(GAS5) (33), small nucleolar RNA
host gene 1 (SNHG1) (34) and, ZNFX1
antisense RNA 1 (ZFAS1) (35) have
been identified to be associated with CRC progression in previous
studies (36–38). However, most lncRNAs, including
LINC00525, LINC00869 and LINC01279, have not been previously
identified in human cancers and still need to be investigated
further. The present study identified that CRNDE, deleted in
lymphocytic leukemia 2 (DLEU2) and LOC101060264 were revealed to be
upregulated most, and LOC101929340, CDKN2B-AS1 and LOC101928405
lncRNAs were revealed to be downregulated most in CRC. Hierarchical
clustering was used to present differentially expressed lncRNAs in
CRC (Fig. 1B).
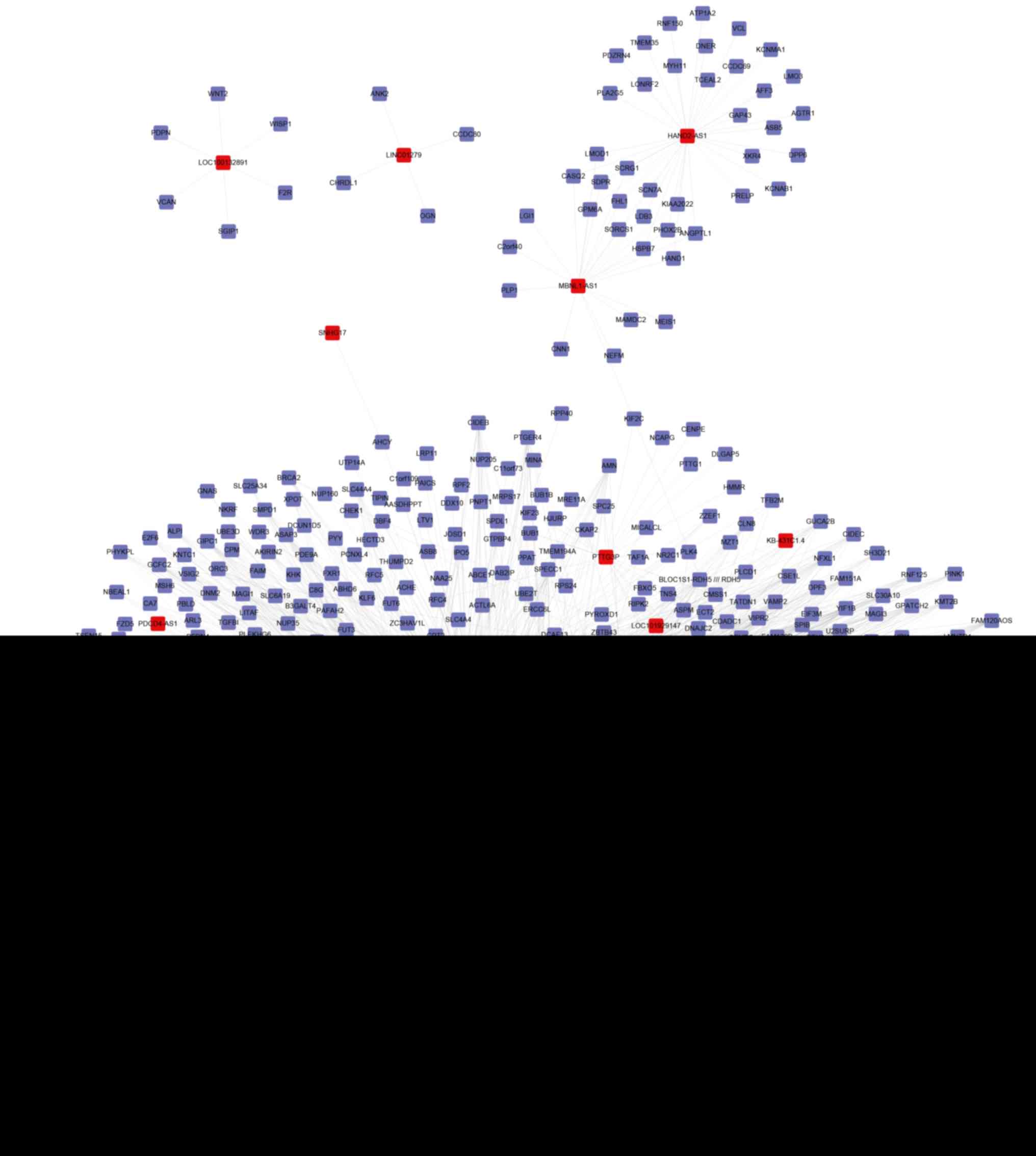
Construction of lncRNAs
co-expression network in CRC
The differentially expressed lncRNA co-expression
network in CRC was subsequently constructed according to expression
values in GSE21510. A total of 3,647 lncRNA-mRNA pairs with an
absolute Pearson value >0.85 were selected to construct the
co-expression network in CRC. This network included 47 lncRNAs and
636 mRNAs, as presented in Fig.
2.
A number of lncRNAs were identified as key
regulators in CRC. For example, lncRNA LOC389332 was co-expressed
with 328 mRNAs, LOC101928405 with 295 mRNAs, FLJ36848 with 260
mRNAs and EPB41L4A-AS2 with 251 mRNAs. A total of 4 mRNAs were
co-expressed with >20 lncRNAs in CRC, including yip1 interacting
factor homolog B (YIF1B), zinc finger and BTB domain containing 7B
(ZBTB7B), ruvb like AAA ATPase 2 (RUVBL2) and vesicle associated
membrane protein 2 (VAMP2) (Fig.
2).
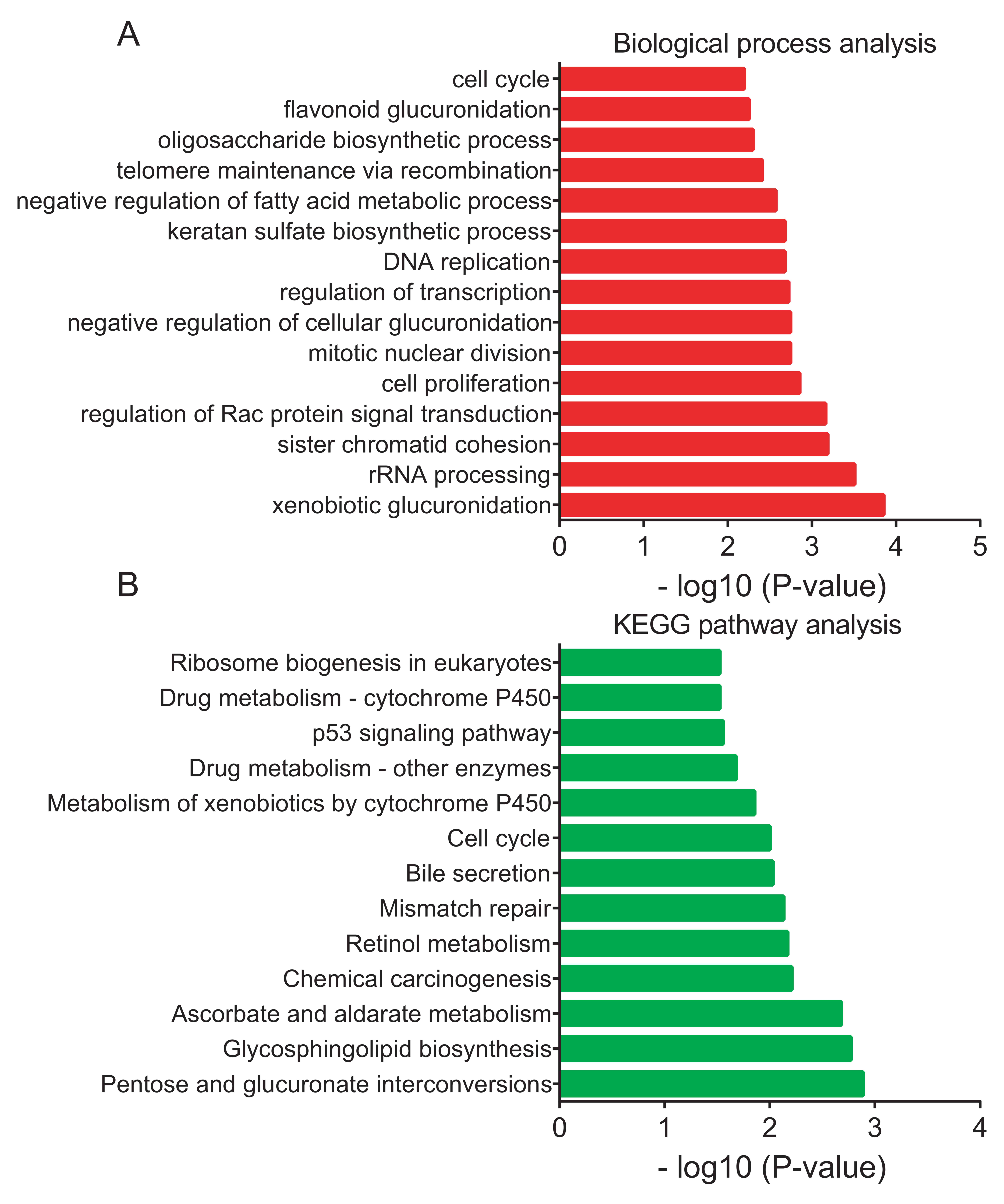
Bioinformatics analysis for
differentially expressed lncRNAs in CRC
Considering that lncRNAs serves a regulatory role in
a number of human cancers through downstream targets, GO and KEGG
analysis were subsequently performed to reveal the potential roles
of lncRNAs in CRC using their co-expressing mRNAs. GO analysis
indicated that differentially expressed lncRNAs were associated
with numerous biological processes, including ‘xenobiotic
glucuronidation’, ‘rRNA processing’, ‘sister chromatid cohesion’,
‘regulation of Rac protein signal transduction’, ‘cell
proliferation’, ‘mitotic nuclear division’, ‘negative regulation of
cellular glucuronidation’, ‘regulation of transcription’, ‘DNA
replication’, ‘keratan sulfate biosynthetic process’, ‘negative
regulation of fatty acid metabolic process’, ‘telomere maintenance
via recombination’, ‘oligosaccharide biosynthetic process’,
‘flavonoid glucuronidation’ and ‘cell cycle’ (all, P<0.001;
Fig. 3A).
KEGG pathway analysis revealed that differentially
expressed lncRNAs were associated with a series of pathways,
including ‘pentose and glucuronate interconversions’,
‘glycosphingolipid biosynthesis’, ‘ascorbate and aldarate
metabolism’, ‘chemical carcinogenesis’, ‘retinol metabolism’,
‘mismatch repair’, ‘bile secretion’, ‘cell cycle’, ‘metabolism of
xenobiotics by cytochrome P450’, ‘drug metabolism-other enzymes’,
p53 signaling pathway, ‘drug metabolism-cytochrome P450’ and
‘ibosome biogenesis in eukaryotes’ (all, P<0.001; Fig. 3B).
TCGA dataset validation of expression
of key lncRNAs in CRC
As presented in Fig.
4, a total of 16 key lncRNAs were identified to be dysregulated
in CRC when compared with normal samples using the TCGA dataset.
Among these lncRNAs, programmed cell death 4 (PDCD4)-AS1
(P<0.001), heart and neural crest derivatives expressed 2
(HAND2)-AS1 (P<0.001), erythrocyte membrane protein band 4.1
like 4A (EPB41L4A)-AS2 (P<0.001), LOC389332 (P<0.001), long
intergenic non-protein coding RNA 294 (LINC00294; P<0.001) and
PP14571 (P<0.001) were downregulated in CRC in comparison with
normal samples. However, LINC00202-2 (P<0.001), DLEU2
(P<0.001), GAS5 (P<0.001), SNHG1 (P<0.001), small
nucleolar RNA host gene 17 (SNHG17; P<0.001), pituitary
tumor-transforming 3 pseudogene (PTTG3P; P<0.001), muscle blind
like splicing regulator 1 (MBNL1)-AS1 (P<0.001), ZFAS1
(P<0.001), tetratricopeptide repeat domain 2B (TTC28)-AS1
(P<0.001) and cancer susceptibility 15 (CASC15; P<0.001) were
upregulated in CRC when compared with normal tissues.
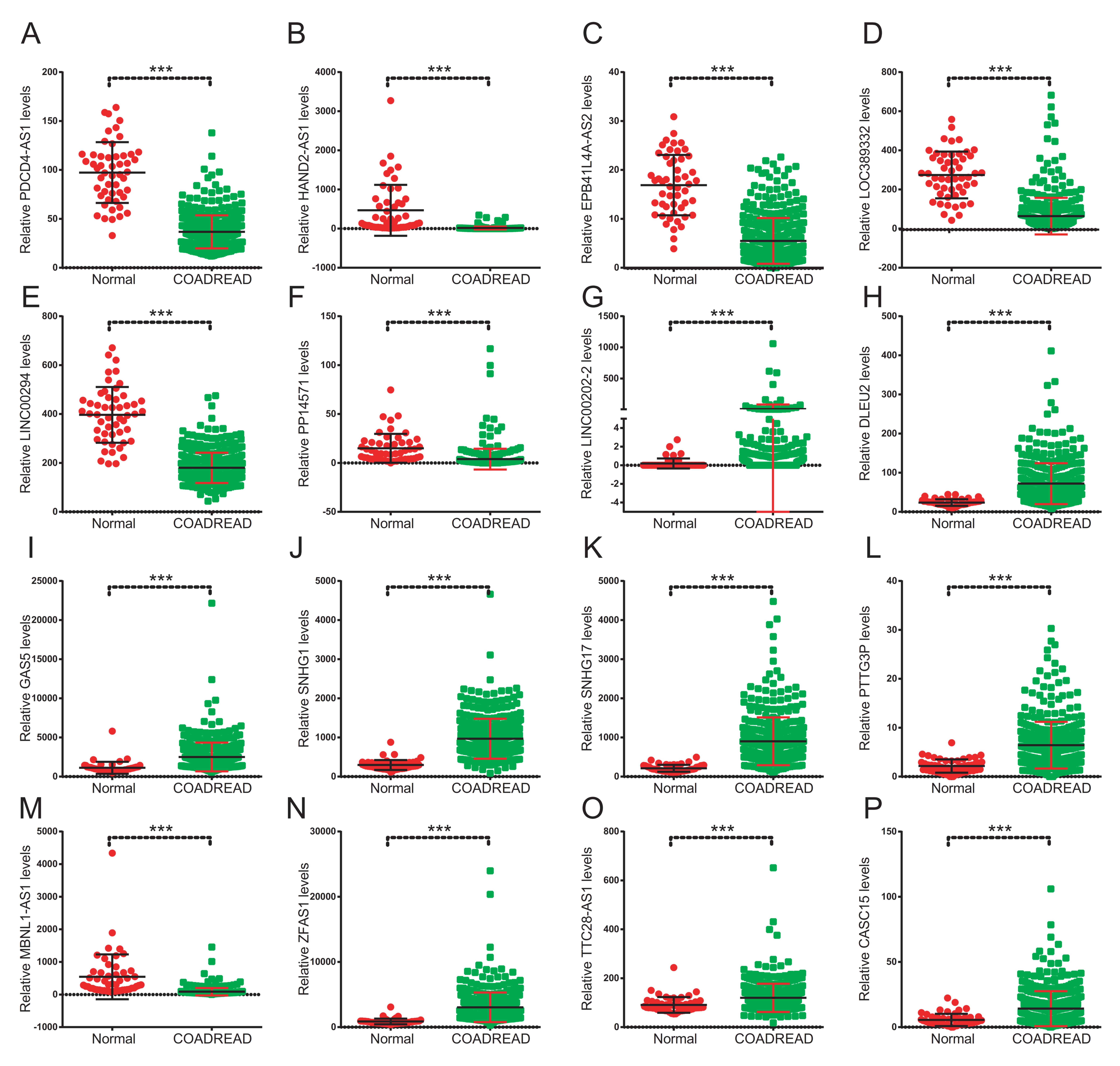 | Figure 4.Validation of expression of key
lncRNAs in CRC, using the TCGA dataset. The expression levels of
(A) PDCD4-AS1, (B) HAND2-AS1, (C) EPB41L4A-AS2, (D) LOC389332, (E)
LINC00294 and (F) PP14571 were decreased in CRC compared with
normal samples. The expression levels of (G) LINC00202-2, (H)
DLEU2, (I) GAS5, (J) SNHG1, (K) SNHG17, (L) PTTG3P, (M) MBNL1-AS1,
(N) ZFAS1, (O) TTC28-AS1 and (P) CASC15 were increased in CRC
compared with normal tissues. Statistical comparisons between
normal and CRC groups were performed using a Mann-Whitney U-test.
***P<0.001. COAD, colon adenocarcinoma; READ, rectum
adenocarcinoma; lncRNA, long non-coding RNA; CRC, colorectal
cancer; TCGA, The Cancer Genome Atlas; PDCD4, programmed cell death
4; AS, antisense RNA; HAND2, heart and neural crest derivatives
expressed 2; EPB41L4A, erythrocyte membrane protein band 4.1 like
4A; LINC00294, long intergenic non-protein coding RNA 294; DLEU2,
deleted in lymphocytic leukemia 2; GAS5, growth arrest specific 5;
SNHG1, small nucleolar RNA host gene 1; SNHG17, small nucleolar RNA
host gene 17; PTTG3P, pituitary tumor-transforming 3 pseudogene;
MBNL1, muscle blind like splicing regulator 1; ZFAS1, ZNFX1
antisense RNA 1; TTC28, tetratricopeptide repeat domain 2B; CASC15,
cancer susceptibility 15. |
Dysregulation of key lncRNAs is
associated with overall survival time in CRC
Kaplan-Meier analysis was subsequently performed to
explore whether key lncRNAs could serve as novel CRC biomarkers.
The online Cutoff Finder (39)
(http://molpath.charite.de/cutoff/output.jsp) was used
to determine the cut-off value to divide all patients with CRC into
the lncRNA low and high group. As presented in Fig. 5, higher expression levels of SNHG17
(P=0.0097), TTC28-AS1 (P=0.0065), EPB41L4A-AS2 (P=0.0075), DLEU2
(P=0.007) and SNHG1 (P=0.0033) were associated with shorter overall
survival time of patients with CRC compared with the respective low
expression groups, whereas the increased expression levels of
MBNL1-AS1 (P=0.0002) and LOC389332 (P=0.0468) were associated with
an improved overall survival in CRC. These results indicated that
the key lncRNAs identified in the current study may serve as
biomarkers for CRC.
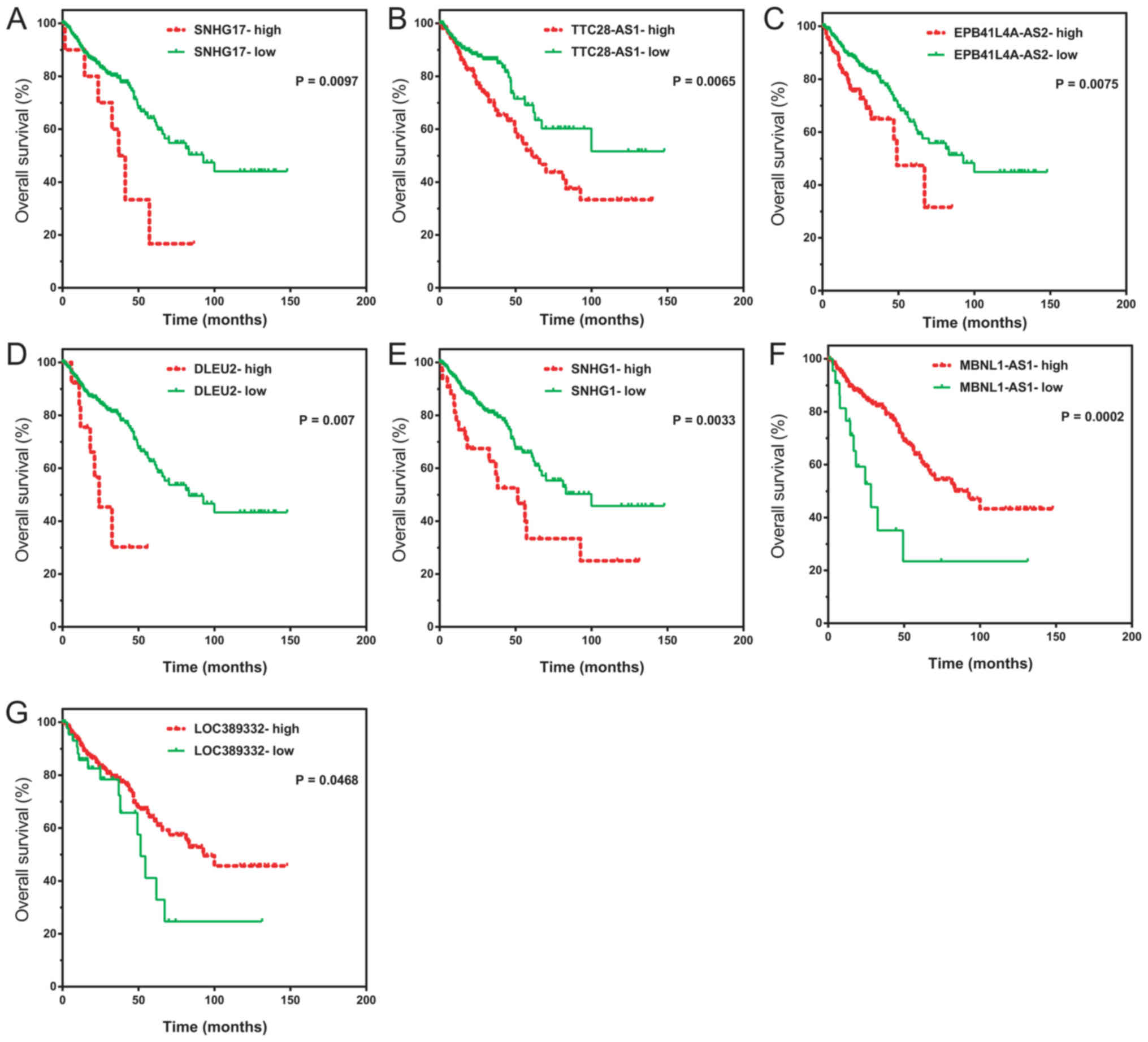 | Figure 5.Dysregulation of key lncRNAs is
associated with overall survival time in CRC. Kaplan-Meier analysis
of the overall survival rates in patients with high and low
expression levels of (A) SNHG17, (B) TTC28-AS1, (C) EPB41L4A-AS2,
(D) DLEU2, (E) SNHG1, (F) MBNL1-AS1 and (G) LOC389332. Log-rank
test was used to examine the significance of the difference between
the curves. lncRNA, long non-coding RNA; SNHG17, small nucleolar
RNA host gene 17; TTC28, tetratricopeptide repeat domain 2B; AS,
antisense RNA; EPB41L4A, erythrocyte membrane protein band 4.1 like
4A; DLEU2, deleted in lymphocytic leukemia 2; SNHG1, small
nucleolar RNA host gene 1; MBNL1, muscle blind like splicing
regulator 1. |
Discussion
LncRNAs are cancer progression regulators that are
associated with regulating cell proliferation, apoptosis and
migration. In CRC, a number of lncRNAs have been demonstrated to
serve crucial roles in cancer tumorigenesis. MEG3 has been
indicated to regulate the miR-141/PDCD4 axis to affect CRC
oxaliplatin resistance (40). DLEU1
has been indicated to promote CRC progression by activating KPNA3
(41). HAND2-AS1 has also been
observed to suppress CRC proliferation though sponging miR-1275
(42). However, the expression
pattern and functional roles of a large number of lncRNAs remain
unclear. In the present study, CRC-associated lncRNAs were
identified though re-annotating the GSE21510 dataset. A total of
1,125 upregulated lncRNAs and 1,245 downregulated lncRNAs were
identified in CRC compared with normal samples. Among these, a
number of lncRNAs, including CRNDE, GAS5, SNHG1 and ZFAS1, were
associated with CRC progression. CRNDE has previously been
indicated to promote CRC cell proliferation by epigenetically
silencing DUSP5/CDKN1A expression (43). GAS5, a well-known lncRNA in a variety
of cancer types, suppressed CRC cell proliferation via the
miR-182-5p/FOXO3a axis (44). SNHG1
has been demonstrated to serve as an oncogene in CRC though
activating the wnt pathway and sponging miR-145 (45). The upregulation of ZFAS1 was
associated with poor prognosis and promoted cancer proliferation
though sponging miR-484 in CRC (37). However, the functional roles of the
majority of lncRNAs remain unclear.
Bioinformatics analysis provides a powerful tool to
reveal the potential roles of unknown RNA transcripts in human
diseases. LncRNA co-expression networks are used to understand the
important roles of lncRNAs in a number of human diseases, including
diabetes (46) and major depressive
disorder (47). In the present
study, differentially expressed lncRNAs co-expression networks were
constructed in CRC, which included 47 lncRNAs and 636 mRNAs. The
current study indicated that a number of lncRNAs, including
LOC389332, LOC101928405, FLJ36848 and EPB41L4A-AS2 were identified
as key regulators in CRC and were co-expressed with >250 mRNAs
in the progression of CRC. LOC389332 was downregulated and
associated with favorable prognosis in clear cell renal cell
carcinoma (48). EPB41L4A-AS2 was
demonstrated to inhibit tumor proliferation and was associated with
favorable prognoses in breast cancer and other solid tumors
(49). Furthermore, a number of
mRNAs, including YIF1B, ZBTB7B, RUVBL2 and VAMP2, also served an
important role in this co-expression network. A previous study
indicated that YIF1B was associated with the anterograde traffic
pathway and the Golgi architecture (50). ZBTB7B is a transcription factor
associated with the regulation of lineage commitment of immature
T-cell precursors (51). VAMP2 is a
member of the vesicle-associated membrane protein, and VAMP2-NRG1
fusion is a novel oncogenic driver of non-small-cell lung
adenocarcinoma (52). Bioinformatics
analysis indicated that differentially expressed lncRNAs were
associated with ‘xenobiotic glucuronidation’, ‘rRNA processing’,
‘sister chromatid cohesion’, ‘regulation of Rac protein signal
transduction’, ‘cell proliferation’, ‘mitotic nuclear division’,
‘p53 signaling pathway’ and ‘cell cycle regulation’. Abnormal cell
cycle progression is considered a hallmark of human cancer
(53,54). Targeting cell cycle checkpoints is a
therapeutic approach against cancer (55). A number of compounds were reported as
potential anti-cancer drugs in CRC, including itraconazole, a drug
which targets cell cycle heterogeneity (56), and 5-Aminosalicylic acid, which
inhibits cell cycle progression in a phospholipase D dependent
manner (57). The present study
revealed that dysregulated lncRNAs in CRC were strongly associated
with the regulation of cancer cell cycle progression, indicating
that these lncRNAs may serve as novel therapeutic biomarkers for
this disease. Previous studies have indicated that the
dysregulation of lncRNAs may serve as novel biomarker for a variety
of cancers, including CRC. For example, upregulated SNHG6 has been
revealed to predict a poor prognosis in CRC (27). In the present study, the prognostic
value of key lncRNAs in CRC were evaluated using TCGA CRC dataset
PDCD4-AS1, HAND2-AS1, EPB41L4A-AS2, LOC389332, LINC00294 and
PP14571 were observed to be expressed at a low level, and
LINC00202-2, DLEU2, GAS5, SNHG1, SNHG17, PTTG3P, MBNL1-AS1, ZFAS1,
TTC28-AS1 and CASC15 were indicated to be highly expressed in CRC
compared with normal tissues. Furthermore, patients with CRC and
higher expression of SNHG17, TTC28-AS1, EPB41L4A-AS2, DLEU2 and
SNHG1 exhibited a shorter overall survival time. However, patients
with CRC with higher expression of MBNL1-AS1 and LOC389332
exhibited longer overall survival time compared with patients with
low expression levels of these genes. These results indicated that
high SNHG17, TTC28-AS1, EPB41L4A-AS2, DLEU2 and SNHG1 expression,
and low MBNL1-AS1 and LOC389332 expression could predict the
negative outcomes of patients with CRC. These analyses, to the best
of our knowledge, for the first time, indicated that these lncRNAs
may serve as biomarkers for CRC.
In conclusion, a total of 38 upregulated lncRNAs and
31 downregulated lncRNAs were observed in CRC. Bioinformatics
analysis indicated that differentially expressed lncRNAs were
associated with ‘xenobiotic glucuronidation’, ‘rRNA processing’,
‘sister chromatid cohesion’, ‘regulation of Rac protein signal
transduction’, ‘cell proliferation’, ‘mitotic nuclear division’ and
‘cell cycle regulation’. Furthermore, the results revealed that
high expression of SNHG17, TTC28-AS1, EPB41L4A-AS2, DLEU2 and
SNHG1, and low expression of MBNL1-AS1 and LOC389332 were
associated with shorter overall survival time of patients with CRC.
The current study provides useful information for identifying novel
biomarkers in CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Youth Research
Fund Project of the Health and Family Planning Commission of
Nantong (grant no. WQ2015052), the Project for Medical Key Youth
Talent of Nantong (grant no. Youth 005), and Nantong Science and
Technology Project (grant no. YYZ16043).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XC and BZ conceived and designed the study. XuZ, XiZ
and LSh developed the methodology. JW, GC, LSh and LSo analyzed and
interpreted the data. XuZ and XiZ wrote, reviewed and revised the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Song YX and Wang ZN: Non-coding
RNAs in gastric cancer. Gene. 560:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI
|
|
4
|
Iguchi T, Uchi R, Nambara S, Saito T,
Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et
al: A long noncoding RNA, lncRNA-ATB, is involved in the
progression and prognosis of colorectal cancer. Anticancer Res.
35:1385–1388. 2015.PubMed/NCBI
|
|
5
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:11480–11484. 2015.PubMed/NCBI
|
|
6
|
Cui Y, Zhang F, Zhu C, Geng L, Tian T and
Liu H: Upregulated lncRNA SNHG1 contributes to progression of
non-small cell lung cancer through inhibition of miR-101-3p and
activation of Wnt/β-catenin signaling pathway. Oncotarget.
8:17785–17794. 2017.PubMed/NCBI
|
|
7
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: LncRNA loc285194 is a p53-regulated tumor
suppressor. Nucleic Acids Res. 41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi XF, Sun M, Liu HB, Yao YW and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neumann P, Jaé N, Knau A, Glaser SF,
Fouani Y, Rossbach O, Krüger M, John D, Bindereif A, Grote P, et
al: The lncRNA GATA6-AS epigenetically regulates endothelial gene
expression via interaction with LOXL2. Nature Commun. 9:2372018.
View Article : Google Scholar
|
|
13
|
Wang S, Liang K, Hu Q, Li P, Song J, Yang
Y, Yao J, Mangala LS, Li C, Yang W, et al: JAK2-binding long
noncoding RNA promotes breast cancer brain metastasis. J Clin
Invest. 127:4498–4515. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li C, Li W, Zhang Y, Zhang X, Liu T, Zhang
Y, Yang Y, Wang L, Pan H, Ji J and Wang C: Increased expression of
antisense lncrna SPINT1-AS1 predicts a poor prognosis in colorectal
cancer and is negatively correlated with its sense transcript. Onco
Targets Ther. 11:3969–3978. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang E, Li W, Yin D, De W, Zhu L, Sun S
and Han L: c-Myc-regulated long non-coding RNA H19 indicates a poor
prognosis and affects cell proliferation in non-small-cell lung
cancer. Tumor Biol. 37:4007–4015. 2016. View Article : Google Scholar
|
|
16
|
Sostres C, Gargallo CJ and Lanas A:
Aspirin, cyclooxygenase inhibition and colorectal cancer. World
World J Gastrointest Pharmacol Ther. 5:40–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marley AR and Nan H: Epidemiology of
colorectal cancer. Int J Mol Epidemiol Genet. 7:105–114.
2016.PubMed/NCBI
|
|
18
|
Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y,
Yang S, Zhang W and Zhang R: LncRNA BANCR promotes tumorigenesis
and enhances adriamycin resistance in colorectal cancer. Aging
(Albany NY). 10:2062–2078. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li G, Wang C, Wang Y, Xu B and Zhang W:
LINC 00312 represses proliferation and metastasis of colorectal
cancer cells by regulation of miR-21. J Cell Mol Med. 22:5565–5572.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang K, Liu J, Li C, Peng X, Li H and Li
Z: Identification and validation of potential target genes in
papillary thyroid cancer. Eur J Pharmacol. 843:217–225. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan W, Li X, Liu L, Wei C, Sun D, Peng S
and Jiang L: Comprehensive analysis of lncRNA-associated ceRNA
network in colorectal cancer. Biochem Biophys Res Commun.
508:374–379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miao Y, Sui J, Xu SY, Liang GY, Pu YP and
Yin LH: Comprehensive analysis of a novel four-lncRNA signature as
a prognostic biomarker for human gastric cancer. Oncotarget.
8:75007–75024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L,
Yang Q, Zhu H, Nie J, Wu C and Jiao B: LncRNA MIR100HG promotes
cell proliferation in triple-negative breast cancer through triplex
formation with p27 loci. Cell Death Dis. 9:8052018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao Q, Wang N, Qi J, Gu Z and Shen HJ:
Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder
transitional cell carcinoma via regulation of chemokine (C-C motif)
ligand 1 expression. Mol Med Rep. 13:27–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsukamoto S, Ishikawa T, Iida S, Ishiguro
M, Mogushi K, Mizushima H, Uetake H, Tanaka H and Sugihara K:
Clinical significance of osteoprotegerin expression in human
colorectal cancer. Clin Cancer Res. 17:2444–2450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Binefa G, Rodríguez-Moranta F, Teule A and
Medina-Hayas M: Colorectal cancer: From prevention to personalized
medicine. World J Gastroenterol. 20:6786–6808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
R Core Team, . R: A language and
environment for statistical computingR Foundation for Statistical
Computing; Vienna, Austria: 2013
|
|
30
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Golubeva Y and Rogers K: Collection and
preparation of rodent tissue samples for histopathological and
molecular studies in carcinogenesis. Methods Mol Biol. 511:3–60.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Zhou D, Ying M, Chen M, Chen P,
Chen Z and Zhang F: Expression of long non-coding RNA (lncRNA)
small nucleolar RNA Host Gene 1 (SNHG1) exacerbates hepatocellular
carcinoma through suppressing miR-195. Med Sci Monit. 22:4820–4829.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ying Z, Sun L, Xuan L, Pan Z, Li K, Liu S,
Huang Y, Zhao X, Huang L, Wang Z, et al: Reciprocal changes of
circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute
myocardial infarction. Sci Rep. 6:223842016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Y, Qin ZS, Feng Y, Tang XJ, Zhang T
and Yang L: Long non-coding RNA (lncRNA) small nucleolar RNA host
gene 1 (SNHG1) promote cell proliferation in colorectal cancer by
affecting P53. Eur Rev Med Pharmacol Sci. 22:976–984.
2018.PubMed/NCBI
|
|
37
|
Xie S, Ge Q, Wang X, Sun X and Kang Y:
Long non-coding RNA ZFAS1 sponges miR-484 to promote cell
proliferation and invasion in colorectal cancer. Cell Cycle.
17:154–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu F, Gao H, Li S, Ni X and Zhu Z: Long
non-coding RNA ZFAS1 correlates with clinical progression and
prognosis in cancer patients. Oncotarget. 8:61561–61569.
2017.PubMed/NCBI
|
|
39
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff finder: A
Comprehensive and straightforward web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li M, Bian Z, Yao S, Zhang J, Jin G, Wang
X, Yin Y and Huang Z: Up-regulated expression of SNHG6 predicts
poor prognosis in colorectal cancer. Pathol Res Pract. 214:784–789.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu T, Han Z, Li H, Zhu Y, Sun Z and Zhu
A: LncRNA DLEU1 contributes to colorectal cancer progression via
activation of KPNA3. Mol Cancer. 17:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou J, Lin J, Zhang H, Zhu F and Xie R:
LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer
progression by upregulating KLF14. Biochem Biophys Res Commun.
503:1848–1853. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ding J, Li J, Wang H, Tian Y, Xie M, He X,
Ji H, Ma Z, Hui B, Wang K and Ji G: Long noncoding RNA CRNDE
promotes colorectal cancer cell proliferation via epigenetically
silencing DUSP5/CDKN1A expression. Cell Death Dis. 8:e29972017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheng K, Zhao Z, Wang G, Wang J and Zhu W:
lncRNA GAS5 inhibits colorectal cancer cell proliferation via the
miR-182-5p/FOXO3a axis. Oncol Rep. 40:2371–2380. 2018.PubMed/NCBI
|
|
45
|
Tian T, Qiu R and Qiu X: SNHG1 promotes
cell proliferation by acting as a sponge of miR-145 in colorectal
cancer. Oncotarget. 9:2128–2139. 2017.PubMed/NCBI
|
|
46
|
Wang P, Fu H, Cui J and Chen X:
Differential lncRNA-mRNA co-expression network analysis revealing
the potential regulatory roles of lncRNAs in myocardial infarction.
Mol Med Rep. 13:1195–1203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Egede LE and Zheng D: Independent factors
associated with major depressive disorder in a national sample of
individuals with diabetes. Diabetes Care. 26:104–111. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jin P, Wang J and Liu Y: Downregulation of
a novel long non-coding RNA, LOC389332, is associated with poor
prognosis and tumor progression in clear cell renal cell carcinoma.
Exp Ther Med. 13:1137–1142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu S, Wang P, You Z, Meng H, Mu G, Bai X,
Zhang G, Zhang J and Pang D: The long non-coding RNA EPB41L4A-AS2
inhibits tumor proliferation and is associated with favorable
prognoses in breast cancer and other solid tumors. Oncotarget.
7:20704–20717. 2016.PubMed/NCBI
|
|
50
|
Alterio J, Masson J, Diaz J, Chachlaki K,
Salman H, Areias J, Al Awabdh S, Emerit MB and Darmon M: Yif1B is
involved in the Anterograde traffic pathway and the Golgi
Architecture. Traffic. 16:978–993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang L, Wildt KF, Castro E, Xiong Y,
Feigenbaum L, Tessarollo L and Bosselut R: The zinc finger
transcription factor Zbtb7b represses CD8-lineage gene expression
in peripheral CD4+ T cells. Immunity. 29:876–887. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jung Y, Yong S, Kim P, Lee HY, Jung Y,
Keum J, Lee S and Kim J and Kim J: VAMP2-NRG1 fusion gene is a
novel oncogenic driver of non-small-cell lung adenocarcinoma. J
Thorac Oncol. 10:1107–1111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Scaggiante B, Dapas B, Bonin S, Grassi M,
Zennaro C, Farra R, Cristiano L, Siracusano S, Zanconati F,
Giansante C and Grassi G: Dissecting the expression of EEF1A1/2
genes in human prostate cancer cells: The potential of EEF1A2 as a
hallmark for prostate transformation and progression. Br J Cancer.
106:166–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Visconti R, Della Monica R and Grieco D:
Cell cycle checkpoint in cancer: A therapeutically targetable
double-edged sword. J Exp Clin Cancer Res. 35:1532016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Buczacki SJA, Popova S, Biggs E, Koukorava
C, Buzzelli J, Vermeulen L, Hazelwood L, Francies H, Garnett MJ and
Winton DJ: Itraconazole targets cell cycle heterogeneity in
colorectal cancer. J Exp Med. 215:1891–1912. 2018.PubMed/NCBI
|
|
57
|
Baan B, Dihal AA, Hoff E, Bos CL,
Voorneveld PW, Koelink PJ, Wildenberg ME, Muncan V, Heijmans J,
Verspaget HW, et al: 5-Aminosalicylic acid inhibits cell cycle
progression in a phospholipase D dependent manner in colorectal
cancer. Gut. 61:1708–1715. 2012. View Article : Google Scholar : PubMed/NCBI
|