Introduction
Esophageal carcinoma is the eighth most common
cancer worldwide, and the sixth leading cause of cancer-associated
deaths, with an estimated 455,800 new cases and 400,200 mortalities
per year globally (1,2). Based on histopathological analysis,
esophageal carcinoma is divided into two subtypes, esophageal
squamous cell carcinoma (ESCC) and esophageal adenocarcinoma
(3). ESCC, accounts for about 90% of
esophageal cancer cases and is characterized by invasiveness,
recurrence and ability to metastasize (4). Despite the advances in diagnostic and
surgical approaches, the clinical outcomes of patients with ESCC
remain unsatisfactory with a 5 year survival rate of 26.2–49.4%
(5). Genetic and epigenetic
alterations are closely associated with the progression of ESCC;
however, the detailed mechanisms underlying the pathogenesis of
ESCC remain to be determined (6,7).
Therefore, a detailed investigation into the formation and
progression of ESCC is required to advance the identification of
novel therapeutic targets for the treatment of patients with
ESCC.
MicroRNAs (miRNAs) are a group of single stranded,
non-coding RNA molecules of 18–24 nucleotides in length (8). miRNAs are expressed in plants, animals
and some viruses, and have been considered as critical regulators
of gene expression (9). miRNAs
negatively regulate gene expression through base-pairing with the
complementary sites in the 3′-untranslated regions (3′-UTRs) of
their target genes (10) resulting
in inhibition of translation and/or degradation of mRNAs (11). To date, miRNA molecules have been
reported to be aberrantly expressed in almost all types of cancer,
including ESCC (12), lung (13), colorectal (14), gastric (15) and prostate (16) cancer. Dysregulated miRNAs are
implicated in the tumorigenesis and tumor development of ESCC via
the regulation of cell proliferation, cell cycle, apoptosis,
metastasis, angiogenesis, and resistance to radiation and
chemotherapeutic treatment (17–19).
miRNAs may function as tumor suppressors or oncogenes depending on
the specific roles of their target genes (20,21).
Therefore, miRNAs may serve as therapeutic targets for the
treatment of patients with ESCC.
miR-652 has been recognized as a cancer-associated
miRNA in endometrial cancer (22),
non-small cell lung cancer (23),
pediatric acute lymphoblastic leukemia (24) and pancreatic cancer (25). However, the expression status and
roles of miR-652 in ESCC as well as the molecular mechanisms
involved remain largely unknown. Therefore, in the current study,
miR-652 expression in ESCC and the biological roles of miR-652 in
the development of ESCC were investigated. In addition, the
potential mechanisms underlying these functions were explored. The
present study aimed to determine the role of miR-652 in ESCC.
Materials and methods
Patients and specimens
The present study was approved by The Ethics
Committee of Shengjing Hospital of China Medical University
(Liaoning, China). Written informed consent was obtained from all
patients recruited in this research. In total, 37 pairs of ESCC
tissues and adjacent non-tumor tissues (2 cm away from tumor
tissues) were collected from patients (25 males, 12 females; age
range, 51–73 years) who underwent surgery at the Shengjing Hospital
of China Medical University between February 2015 and September
2017. None of the patients had been treated for cancer prior to
surgical resection. All tissues were obtained and stored in liquid
nitrogen until required for RNA and protein extraction.
Cell culture
A normal human esophageal epithelial cell line
(HET-1A) was purchased from American Type Culture Collection. In
total, four human ESCC cell lines, including KYSE70, KYSE150, TE-1,
and Eca109, were purchased from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences. All cell lines were
grown in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS) and 1% streptomycin/penicillin
mixture (all from Invitrogen; Thermo Fisher Scientific, Inc.).
Cells were maintained at 37°C in a humidified atmosphere containing
5% CO2.
Cell transfection
miR-652 mimics (5′-AAUGGCGCCACUAGGGUUGUG-3′) and
miRNA mimic negative control (miR-NC; 5′-UUCUCCGAACGUGUCACGUTT-3′)
were synthesized by Shanghai GenePharma Co., Ltd. miR-652 mimics
were used to increase the expression of miR-652, while miR-NC
served as the negative control. Human FGFR1 overexpression vector
lacking the 3′-UTR pCMV-FGFR1 and empty pCMV plasmid were
constructed by the Chinese Academy of Sciences. Cells in the
mid-log phase were collected and plated into six-well plates with a
density of 5×105 cells/well. The aforementioned
oligonucleotides (100 pmol) and vectors (4 µg) were transiently
transfected into cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and 48 h after transfection, reverse
transcription-quantitative PCR (RT-qPCR) and transwell invasion
assays were performed. Cell Counting Kit-8 (CCK-8) assay and
western blot analysis were conducted at 24 and 72 h after
transfection, respectively.
RNA extraction and RT-qPCR
Total cellular RNA was extracted from tissue samples
or cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). For quantification of miR-652 expression,
first-strand complementary DNA (cDNA) was prepared from total
cellular RNA using a TaqMan MicroRNA Reverse Transcription kit
using the following temperature protocol: 16°C for 30 min, 42°C for
30 min and 85°C for 5 min. Samples were then subjected to
quantitative PCR using a TaqMan MicroRNA PCR kit (both from Applied
Biosystems; Thermo Fisher Scientific, Inc.). The temperature
protocol for qPCR was as follows: 50°C for 2 min, 95°C for 10 min;
40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec. For FGFR1 mRNA detection,
total RNA was reverse-transcribed into cDNA using a PrimeScript RT1
reagent kit using the following temperature protocol: 37°C for 15
min and 85°C for 5 sec. Subsequently, qPCR was conducted using a
SYBR Premix Ex Taq™ (both from Takara Biotechnology Co., Ltd.). The
thermocycling conditions were as follows: 5 min at 95°C, followed
by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. The miR-652
and FGFR1 mRNA levels were normalized to that of U6 small nuclear
RNA and GAPDH, respectively. The primers were designed as follows:
miR-652 forward, 5′-ACACTCCAGCTGGGCAACCCTAGGAGAGGGTGC-3′ and
reverse, 5′-GTGTCGTGGAGTCGGCAATTC-3′; U6 forward,
5′-TGGAACGCTTCACGAATTTGCG-3′ and reverse
5′-GGAACGATACAGAGAAGATTAGC-3′; FGFR1 forward,
5′-CTGGTGACAGAGGACAATG-3′ and reverse 5′-AGATCCGGTCAAATAATGCC-3′;
and GAPDH forward, 5′-CCTGGTATGACAACGAATTTG-3′ and reverse
5′-CAGTGAGGGTCTCTCTCTTCC-3′. All data were quantified using the
2−ΔΔCq method (26).
RT-qPCR was performed in 3 replicate wells per group and repeated
three times.
CCK-8 assay
CCK-8 assay was used to evaluate cell proliferation.
After 24 h incubation, transfected cells were collected and plated
into 96-well plates at a density of 2×103 cells/well. A
total of 10 µl CCK-8 solution (Dojindo Molecular Technologies,
Inc.) was added into each well at different time points (0, 1, 2
and 3 days). Transfected cells were then incubated at 37°C with 5%
CO2 for an additional 2 h. The absorbance was determined
at an optical density of 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Transwell invasion assay
Transfected cells were collected after 48 h of
incubation and suspended in FBS-free DMEM. A total of
5×104 transfected cells resuspended in DMEM without FBS
were added to the top chamber of the transwell apparatus pre-coated
with Matrigel (both from BD Biosciences) at 37°C for 4 h. The
bottom chamber was filled with 600 µl DMEM supplemented with 10%
FBS. After incubating for 24 h, a cotton swab was used to carefully
wipe the cells which had not invaded. The invaded cells were fixed
with 95% methanol at room temperature for 30 min and stained with
0.5% crystal violet at room temperature for 30 min. The mean number
of cells in five randomly selected fields of view was counted under
an inverted light microscope (magnification ×200; IX83; Olympus
Corporation).
Bioinformatics prediction
TargetScan (Release 7.2, March 2018; www.targetscan.org) and microRNA.org
(Release, August 2010; last updated, 2010-11-01; www.microrna.org/microrna/) were used to search
for the potential targets of miR-652.
Luciferase reporter assay
The 3′-UTR fragments of FGFR1 with wild-type (wt) or
mutant (mut) miR-652 binding site were amplified by Shanghai
GenePharma Co., Ltd. (Shanghai, China) and inserted into the pGL3
vector (Promega Corporation). The chemically generated luciferase
reporter plasmids were designated as pGL3-FGFR1-3′-UTR wt and
pGL3-FGFR1-3′-UTR mut, respectively. Cells were seeded into 24-well
plates, and co-transfected with miR-652 mimics or miR-NC and
pGL3-FGFR1-3′-UTR wt or pGL3-FGFR1-3′-UTR mut using
Lipofectamine® 2000 reagent according to the protocol
specified by the manufacturer. The transfected cells were harvested
48 h after co-transfection and assayed using a dual-luciferase
reporter assay system (Promega Corporation) in accordance with the
manufacturer's protocol. Firefly luciferase activity was normalized
to that of the Renilla luciferase activity.
Western blot analysis
Total protein was isolated from tissue samples or
cells using a Protein Extraction Reagent (Pierce; Thermo Fisher
Scientific, Inc.). The concentration of the total protein was
detected using a bicinchoninic acid assay kit (Beyotime Institute
of Biotechnology). After centrifugation (14,000 × g) at 4°C for 15
min, equal amounts of proteins were subjected to SDS-PAGE (10% gel)
and transferred onto polyvinylidene difluoride membranes (EMD
Millipore). After the transfer, the membranes were blocked with 5%
non-fat dried milk in Tris-buffered saline containing 0.1% Tween-20
(TBST) at room temperature for 1 h, and then incubated with primary
antibodies overnight at 4°C. The primary antibodies used included
rabbit anti-human FGFR1 antibody (cat no. ab173305; 1:1,000
dilution; Abcam) and rabbit anti-human GAPDH antibody (cat no.
ab181602; 1:1,000 dilution; Abcam). After extensive washing with
TBST, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no.
ab205718; 1:5,000 dilution; Abcam) for 2 h at room temperature. The
protein signals were visualised using an Enhanced Chemiluminescence
Plus reagent (GE Healthcare). Quantity One software version 4.62
(Bio-Rad Laboratories, Inc.) was utilized for densitometry.
Statistical analysis
All assays were repeated at least three times. The
results are presented as the mean ± standard deviation and were
analyzed with SPSS statistical software (version 11.0; SPSS, Inc.).
The differences between groups were examined using a paired
Student's t-test, Student's t-test or one-way analysis of variance.
A post-hoc Student-Newman-Keuls test was used to test for
significance between multiple groups. The correlation between
miR-652 and FGFR1 mRNA levels in ESCC tissues was determined using
Spearman's rank correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-652 is significantly downregulated
in ESCC tissues and cell lines
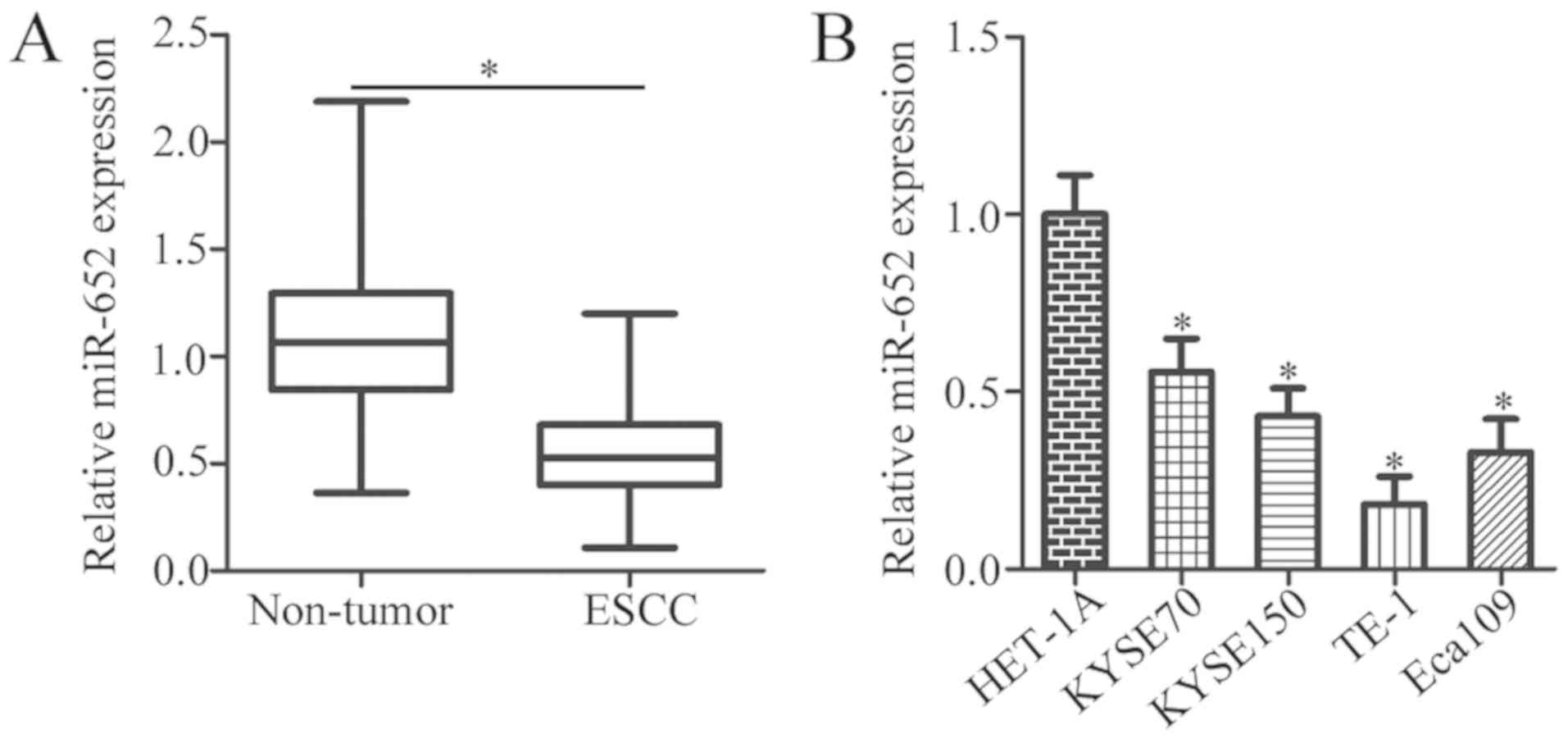
To illustrate the expression status of miR-652 in
ESCC, 37 pairs of ESCC tissues and adjacent non-tumor tissues were
collected. miR-652 expression levels were significantly decreased
in ESCC tissues compared with that of the adjacent non-tumor
tissues (Fig. 1A; P<0.05).
Additionally, the expression level of miR-652 was determined in
four ESCC cell lines (KYSE70, KYSE150, TE-1 and Eca109) and a
normal human esophageal epithelial cell line (HET-1A). The
expression levels of miR-652 were lower in the aforementioned ESCC
cell lines relative to HET-1A cells (Fig. 1B; P<0.05).
miR-652 suppresses the proliferation
and invasiveness of ESCC cells
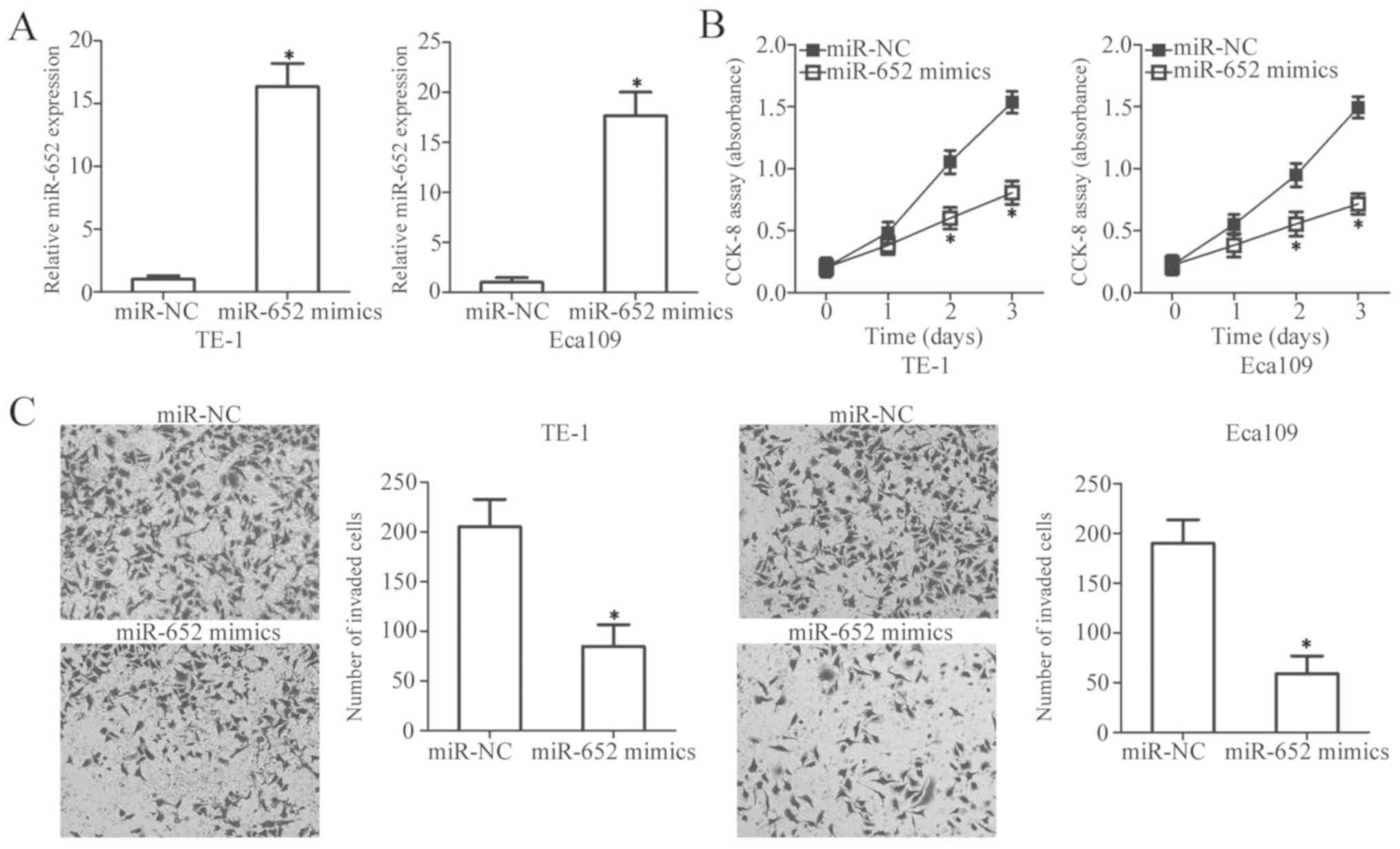
To explore the biological functions of miR-652 in
the development of ESCC, miR-652 mimics and miR-NC were chemically
synthesized, and then transiently transfected into TE-1 and Eca109
cells which expressed the lowest levels of miR-652 of the four ESCC
cell lines. Following transfection, miR-652 was significantly
upregulated in TE-1 and Eca109 cells transfected with miR-652
mimics compared with the miR-NC-transfected cells (Fig. 2A; P<0.05). The regulatory effect
of miR-652 overexpression on ESCC cell proliferation was evaluated
by a CCK-8 assay. miR-652 expression significantly decreased cell
proliferation after 2 days compared with the miR-NC-transfected
cells in both cell lines (Fig. 2B;
P<0.05). Additionally, miR-652 upregulation significantly
decreased the invasiveness of TE-1 and Eca109 cells (Fig. 2C; P<0.05). These data thus suggest
that miR-652 may serve an inhibitory role in the proliferation and
invasion of ESCC cells.
FGFR1 is a direct target gene of
miR-652 in ESCC cells
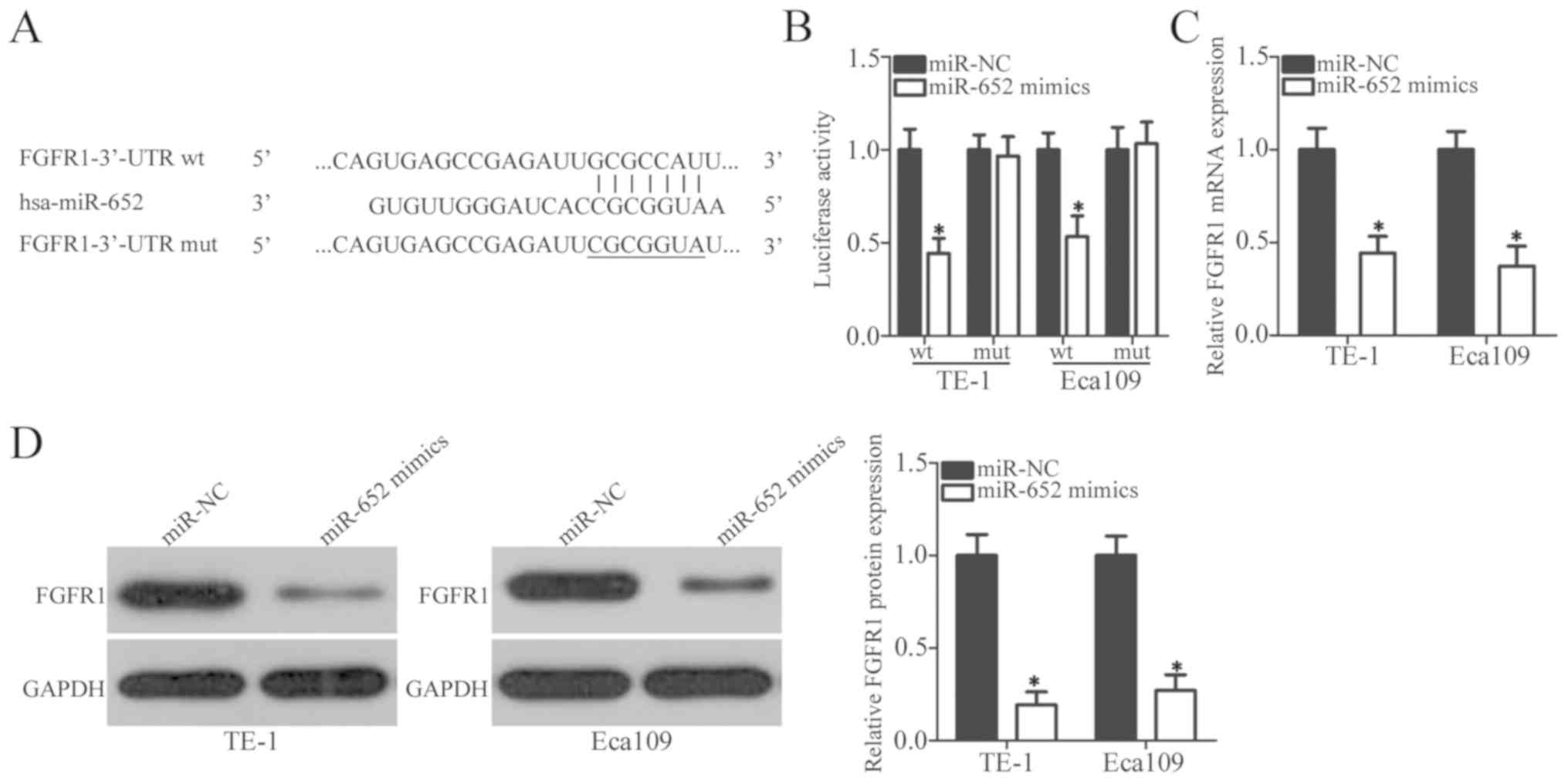
To explore the direct target genes of miR-652 in
ESCC cells, online target exploratory programs, TargetScan and
microRNA.org, were used to search for the putative
targets of miR-652. The 3′-UTR of FGFR1 contains a 7-bp specific
complementary sequence which may directly bind miR-652 (Fig. 3A). A total of 398 human genes were
predicted as potential targets of miR-652, and FGFR1 was selected
for further identification as it was previously reported to be
closely associated with ESCC progression (27–31).
Luciferase reporter assay was performed to determine whether
miR-652 could directly interact with the 3′-UTR of FGFR1.
Luciferase activity of the reporter plasmid containing wt FGFR1
3′-UTR was significantly downregulated in TE-1 and Eca109 cells
after co-transfection with miR-652 mimics (P<0.05); however, the
luciferase activity of the mut FGFR1 3′-UTR was unaltered (Fig. 3B). To further clarify the regulatory
effect of miR-652 on FGFR1, the level of FGFR1 was detected in TE-1
and Eca109 cells transfected with miR-652 mimics or miR-NC. FGFR1
expression was significantly decreased in both ESCC cell lines when
transfected with miR-652 mimics compared with the
miR-NC-transfected cells at both the mRNA level (Fig. 3C; P<0.05) and protein level
(Fig. 3D; P<0.05). Together,
these results suggest that miR-652 may inhibit FGFR1 expression in
ESCC cells by directly binding to its 3′-UTR.
FGFR1 expression is increased in ESCC
tissues, and its expression is inversely correlated with miR-652
expression
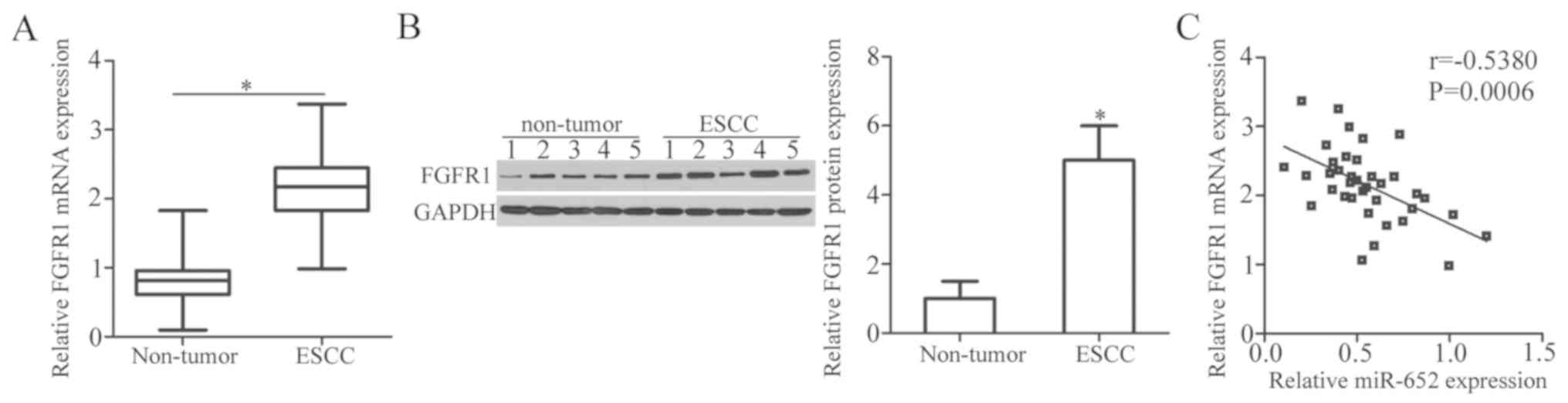
To further explore the association between miR-652
and FGFR1 in ESCC, FGFR1 and miR-652 expression was measured in 37
pairs of ESCC tissues and adjacent non-tumor tissues. The results
of RT-qPCR analysis revealed that the expression levels of FGFR1
mRNA were significantly higher in ESCC tissues compared in adjacent
non-tumor tissues (Fig. 4A;
P<0.05). Additionally, western blot analysis verified that the
protein expression levels of FGFR1 were increased in ESCC tissues
relative to that in adjacent non-tumor tissues (Fig. 4B; P<0.05). Furthermore, Spearman's
rank correlation analysis was used to examine the relationship
between miR-652 and FGFR1 mRNA levels in ESCC tissues. The
expression level of FGFR1 mRNA was inversely correlated with that
of miR-652 in ESCC tissues (Fig. 4C;
r=−0.5380, P=0.0006).
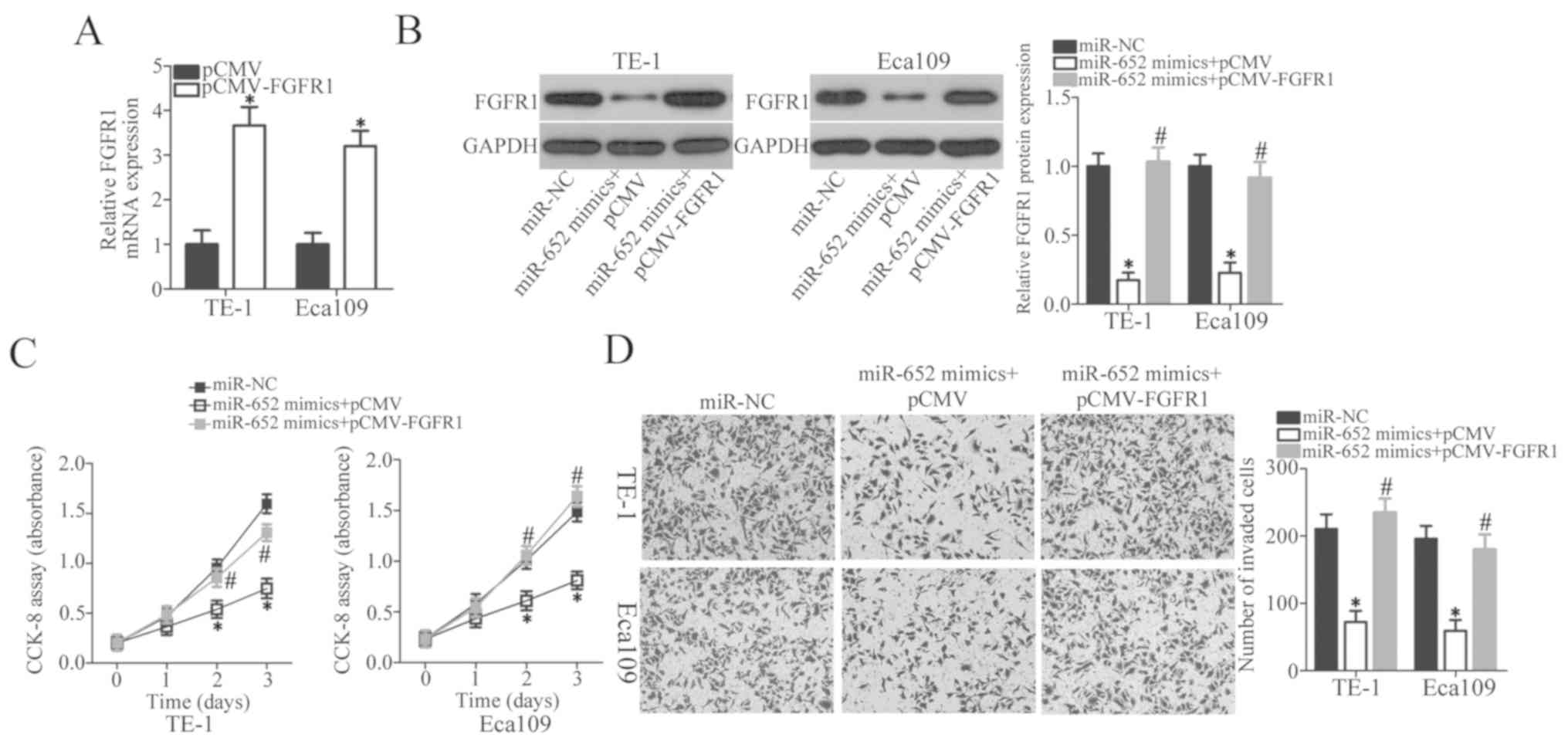
FGFR1 restoration counteracts the
suppressive effects of miR-652 in ESCC cells
Rescue experiments were performed by transfecting
FGFR1 overexpression vector lacking 3′-UTR pCMV-FGFR1 or empty pCMV
plasmid into TE-1 and Eca109 cells treated with miR-652 mimics.
After transfection, RT-qPCR was firstly performed to determine
FGFR1 expression in TE-1 and Eca109 cells after pCMV or pCMV-FGFR1
transfection. FGFR1 mRNA expression significantly increased in the
pCMV-FGFR1-transfected TE-1 and Eca109 cells compared with the
empty vector group (Fig. 5A,
P<0.05). Western blot analysis demonstrated that the FGFR1
protein expression levels were significantly decreased by miR-652
mimics transfection compared with the miR-NC group and recovered in
cells co-transfected with miR-652 mimics and pCMV-FGFR1 (Fig. 5B; P<0.05). Restoring FGFR1
expression attenuated the effect of miR-652 mimics on inhibition of
proliferation (Fig. 5C; P<0.05)
and invasion (Fig. 5D; P<0.05),
respectively. These results suggest that miR-652 inhibits ESCC cell
proliferation and invasion, at least partly, by inhibiting FGFR1
expression.
Discussion
Numerous studies have demonstrated that that a
number of miRNAs may be aberrantly expressed in ESCC (32–34).
Changes in miRNA expression may be associated with ESCC formation
and progression, and involved in the regulation of various
pathophysiological processes (35–37).
Therefore, identification of ESCC-related miRNAs may help elucidate
the mechanisms underlying the pathogenesis of ESCC, which may be
useful for the development of improved therapeutic approaches for
treating patients with ESCC. In the present study, miR-652 was
examined in ESCC tissues and cell lines for the first time, to the
best of our knowledge. Furthermore, the detailed roles of miR-652
in ESCC progression and the associated underlying mechanisms were
examined. These results demonstrated that miR-652 may suppress
proliferation and invasion of ESCC cells by targeting FGFR1
directly.
miR-652 is dysregulated in several different types
of cancer. For example, miR-652 is upregulated in endometrial
cancer tissues and cell lines (22–25).
Patients with endometrial cancer with increased expression levels
of miR-652 often have reduced overall survival rates and experience
earlier recurrence compared with patients with lower expression
levels of miR-652 (22). miR-652 is
also overexpressed in non-small cell lung cancer, and associated
with lymph node metastasis, Tumor-Node-Metastasis stage and
prognosis in non-small cell lung cancer (23). miR-652 expression is often decreased
in pediatric acute lymphoblastic leukemia (24) and pancreatic cancer (25). Decreased expression levels of miR-652
are associated with pancreatic cancer stage, lymphatic invasion and
metastasis (25). However, to the
best of our knowledge, the expression pattern of miR-652 in ESCC
has not been reported. In the present study, a total of 37 pairs of
ESCC tissues and adjacent non-tumor tissues were collected, and
RT-qPCR analysis was performed to detect miR-652 expression levels.
The data demonstrated that miR-652 was downregulated in ESCC
tissues and cell lines.
miR-652 is implicated as an oncogene or a tumor
suppressor gene in the initial stages and progression of human
malignancies depending on the characteristic of its target genes.
For instance, upregulation of miR-652 promotes endometrial cancer
cell growth and metastasis in vitro and in vivo
(22). Similarly, in non-small cell
lung cancer, miR-652 overexpression increases cell proliferation
and motility, and suppresses cell apoptosis in vitro
(23). However, miR-652 transfection
inhibits the acidity-induced epithelial-mesenchymal transition of
pancreatic cancer cells in vitro (25). In pediatric acute lymphoblastic
leukemia, upregulation of miR-652 improves the sensitivity to
vincristine and cytarabine and induces apoptosis in vitro
and in vivo (24). To the
best of our knowledge, the biological functions of miR-652 in ESCC
cells were not previously studied. In the present study, functional
assays of cell behaviors associated with cancer showed that
upregulation of miR-652 decreased ESCC cell proliferation and
invasion in vitro.
Several genes, including nuclear receptor ROR-α in
endometrial cancer (22), lethal
(2) giant larvae in non-small cell
lung cancer (23) and zinc finger
E-box-binding homeobox 1 in pancreatic cancer (25), have been identified as direct targets
of miR-652. In the present study, the molecular mechanisms
underlying the tumor-suppressing effects of miR-652 in ESCC cells
were explored. FGFR1, a member of the fibroblast growth factor
family (38), was demonstrated to be
a direct target gene of miR-652 in ESCC cells. Increased expression
of FGFR1 is observed in multiple types of human cancer, such as
gastric (39), bladder (40), lung (41) and breast (42) cancer. In ESCC, FGFR1 is upregulated
in tumor tissues (27). ESCC
patients with increased FGFR1 expression exhibit reduced
disease-free survival and overall survival rates compared with
patients with lower levels of FGFR1 (28–30).
FGFR1 plays oncogenic roles in the progression and development of
ESCC (31).
In conclusion, the present study demonstrated that
the expression of miR-652 was downregulated in ESCC tissues and
cell lines. Upregulation of miR-652 attenuated cell proliferation
and invasion in ESCC. In addition, FGFR1 was validated as a direct
target of miR-652 and the roles of miR-652 in ESCC cells were
primarily mediated by suppressing FGFR1 expression. miR-652 may
have multiple target genes; however, only FGFR1 was identified as a
direct target of miR-652 in ESCC in the current study. The
association between miR-652 expression and the survival of ESCC
patients was not investigated. These limitations should be
addressed in future studies. Additionally, the ability of miR-652
to alter cell biological behaviors in vivo should be
investigated.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Fund for
Scientific Research Activities Shengjing Hospital of China Medical
University (Liaoning, China).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and CZ designed the study, and performed the
experiments. JH performed the experiments and the statistical
analysis. All authors have read approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Shengjing Hospital of China Medical University
(Liaoning, China), and was performed in accordance with the
Declaration of Helsinki and the guidelines of the Ethics Committee
of Shengjing Hospital of China Medical University. Written informed
consent was obtained from all patients recruited in the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cools-Lartigue J, Spicer J and Ferri LE:
Current status of management of malignant disease: Current
management of esophageal cancer. J Gastrointest Surg. 19:964–972.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X,
Ma J, Yin X, Mao A and Shang M: Overexpression of phosphoserine
aminotransferase 1 (PSAT1) predicts poor prognosis and associates
with tumor progression in human esophageal squamous cell carcinoma.
Cell Physiol Biochem. 39:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Xie X, Zhou C, Peng S, Rao D and Fu
J: Which factors are associated with actual 5 year survival of
oesophageal squamous cell carcinoma? Eur J Cardiothorac Surg.
41:e7–e11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toh Y, Egashira A and Yamamoto M:
Epigenetic alterations and their clinical implications in
esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg.
61:262–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nature Rev Cancer. 6:857–866. 2006. View Article : Google Scholar
|
|
9
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mei LL, Qiu YT, Zhang B and Shi ZZ:
MicroRNAs in esophageal squamous cell carcinoma: Potential
biomarkers and therapeutic targets. Cancer Biomark. 19:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Askandar Iqbal M, Arora S, Prakasam G,
Calin GA and Syed MA: MicroRNA in lung cancer: Role, mechanisms,
pathways and therapeutic relevance. Mol Aspects Med.
17:300065–300067. 2018.
|
|
14
|
Yang S, Sun Z, Zhou Q, Wang W, Wang G,
Song J, Li Z, Zhang Z, Chang Y, Xia K, et al: MicroRNAs, long
noncoding RNAs, and circular RNAs: Potential tumor biomarkers and
targets for colorectal cancer. Cancer Manag Res. 10:2249–2257.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan HL, Wang T and Zhang KH: MicroRNAs as
potential biomarkers for diagnosis, therapy and prognosis of
gastric cancer. OncoTargets Ther. 11:3891–3900. 2018. View Article : Google Scholar
|
|
16
|
Sharma N and Baruah MM: The microRNA
signatures: Aberrantly expressed miRNAs in prostate cancer. Clin
Transl Oncol. 21:126–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee
NPY, Law S, Xu LY, Li EM, Chan KW, et al: MicroRNA-377 suppresses
initiation and progression of esophageal cancer by inhibiting CD133
and VEGF. Oncogene. 36:3986–4000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lindner K, Eichelmann AK, Matuszcak C,
Hussey DJ, Haier J and Hummel R: Complex epigenetic regulation of
chemotherapy resistance and biohlogy in esophageal squamous cell
carcinoma via microRNAs. Int J Mol Sci. 19:E4992018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mayne GC, Hussey DJ and Watson DI:
MicroRNAs and esophageal cancer-implications for pathogenesis and
therapy. Curr Pharm Des. 19:1211–1226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xue L, Nan J, Dong L, Zhang C, Li H, Na R,
He H and Wang Y: Upregulated miR-483-5p expression as a prognostic
biomarker for esophageal squamous cell carcinoma. Cancer Biomark.
19:193–197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Islam F, Gopalan V, Law S, Tang JC, Chan
KW and Lam AK: MiR-498 in esophageal squamous cell carcinoma:
Clinicopathological impacts and functional interactions. Human
Pathol. 62:141–151. 2017. View Article : Google Scholar
|
|
22
|
Sun X, Dongol S, Qiu C, Xu Y, Sun C, Zhang
Z, Yang X, Zhang Q and Kong B: MiR-652 promotes tumor proliferation
and metastasis by targeting RORA in endometrial cancer. Mol Cancer
Res. 16:1927–1939. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang W, Zhou C, Luo M, Shi X, Li Y, Sun Z,
Zhou F, Chen Z and He J: MiR-652-3p is upregulated in non-small
cell lung cancer and promotes proliferation and metastasis by
directly targeting Lgl1. Oncotarget. 7:16703–16715. 2016.PubMed/NCBI
|
|
24
|
Jiang Q, Lu X, Huang P, Gao C, Zhao X,
Xing T, Li G, Bao S and Zheng H: Expression of miR-652-3p and
effect on apoptosis and drug sensitivity in pediatric acute
lymphoblastic leukemia. BioMed Res Int. 5:57246862018.
|
|
25
|
Deng S, Li X, Niu Y, Zhu S, Jin Y, Deng S,
Chen J, Liu Y, He C, Yin T, et al: MiR-652 inhibits acidic
microenvironment-induced epithelial-mesenchymal transition of
pancreatic cancer cells by targeting ZEB1. Oncotarget.
6:39661–39675. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sugiura K, Ozawa S, Kitagawa Y, Ueda M and
Kitajima M: Co-expression of aFGF and FGFR-1 is predictive of a
poor prognosis in patients with esophageal squamous cell carcinoma.
Oncol Rep. 17:557–564. 2007.PubMed/NCBI
|
|
28
|
Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG,
Hur J, Chung H, Park JC, Jung DH, Shin SK, et al: Fibroblast growth
factor receptor 1 gene amplification is associated with poor
survival in patients with resected esophageal squamous cell
carcinoma. Oncotarget. 6:2562–2572. 2015.PubMed/NCBI
|
|
29
|
Song Q, Liu Y, Jiang D, Wang H, Huang J,
Xu Y, Sujie A, Zeng H, Xu C and Hou Y: High amplification of FGFR1
gene is a delayed poor prognostic factor in early stage ESCC
patients. Oncotarget. 8:74539–74553. 2017.PubMed/NCBI
|
|
30
|
Wang D, Du L, Wang Z, Liu X, Qin Y, Wang
Q, Yang Z, Yao Z, Shi M, Shang B, et al: Association of fibroblast
growth factor receptor 1 gene amplification with poor survival in
patients with esophageal squamous cell carcinoma. Oncotarget.
8:88857–88869. 2017.PubMed/NCBI
|
|
31
|
Chen B, Liu S, Gan L, Wang J, Hu B, Xu H,
Tong R, Yang H, Cristina I, Xue J, et al: FGFR1 signaling
potentiates tumor growth and predicts poor prognosis in esophageal
squamous cell carcinoma patients. Cancer Biol Ther. 19:76–86. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma T, Zhao Y, Lu Q, Lu Y, Liu Z, Xue T and
Shao Y: MicroRNA-30c functions as a tumor suppressor via targeting
SNAI1 in esophageal squamous cell carcinoma. Biomed Pharmacothe.
98:680–686. 2018. View Article : Google Scholar
|
|
33
|
Qi B, Wang Y, Chen ZJ, Li XN, Qi Y, Yang
Y, Cui GH, Guo HZ, Li WH and Zhao S: Down-regulation of
miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell
proliferation by activating the Wnt signaling pathway. World J
Gastroenterol. 23:7965–7977. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui XB, Peng H, Li RR, Mu JQ, Yang L, Li
N, Liu CX, Hu JM, Li SG, Wei Y, et al: MicroRNA-34a functions as a
tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh
esophageal squamous cell carcinoma. Oncotarget. 8:92454–92469.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang B, Xie R, Wu SN, Gao CC, Yang XZ and
Zhou JF: MicroRNA-615-5p targets insulin-like growth factor 2 and
exerts tumor-suppressing functions in human esophageal squamous
cell carcinoma. Oncol Rep. 39:255–263. 2018.PubMed/NCBI
|
|
36
|
Zuo J, Zhu K, Wang Y and Yu Z:
MicroRNA-34a suppresses invasion and metastatic in esophageal
squamous cell carcinoma by regulating CD44. Mol Cell Biochem.
443:139–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao X, Xie Z, Wang Z, Cheng K, Liang K and
Song Z: Overexpression of miR-191 predicts poor prognosis and
promotes proliferation and invasion in esophageal squamous cell
carcinoma. Yonsei Med J. 58:1101–1110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Turner CA, Calvo N, Frost DO, Akil H and
Watson SJ: The fibroblast growth factor system is downregulated
following social defeat. Neurosci Lett. 430:147–150. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schäfer MH, Lingohr P, Sträßer A, Lehnen
NC, Braun M, Perner S, Höller T, Kristiansen G, Kalff JC and
Gütgemann I: Fibroblast growth factor receptor 1 gene amplification
in gastric adenocarcinoma. Human Pathol. 46:1488–1495. 2015.
View Article : Google Scholar
|
|
40
|
Abdul-Maksoud RS, Shalaby SM, Elsayed WS
and Elkady S: Fibroblast growth factor receptor 1 and cytokeratin
20 expressions and their relation to prognostic variables in
bladder cancer. Gene. 591:320–326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang K, Ji W, Yu Y, Li Z, Niu X, Xia W and
Lu S: FGFR1-ERK1/2-SOX2 axis promotes cell proliferation,
epithelial-mesenchymal transition, and metastasis in
FGFR1-amplified lung cancer. Oncogene. 27:5340–5354. 2018.
View Article : Google Scholar
|
|
42
|
Cheng CL, Thike AA, Tan SY, Chua PJ, Bay
BH and Tan PH: Expression of FGFR1 is an independent prognostic
factor in triple-negative breast cancer. Breast Cancer Res Treat.
151:99–111. 2015. View Article : Google Scholar : PubMed/NCBI
|