Introduction
Esophageal carcinoma is one of the most common tumor
types with a high grade of malignancy. Esophageal adenocarcinoma is
common in European and American countries, while esophageal
squamous cell carcinoma (ESCC) frequently occurs in Asian
countries, including China (1).
Esophageal carcinoma is mostly diagnosed at the advanced stage. In
spite of the continuous improvement of treatments, including
surgery and radiochemotherapy, the 5-year survival rate of
esophageal carcinoma patients remains dismal due to delayed
diagnosis and high recurrence rate (2,3).
Furthermore, the etiology and pathogenesis of esophageal carcinoma
have remained to be fully elucidated, and effective molecular
markers and targets for targeted therapy are currently lacking.
Therefore, illustration of the molecular mechanisms of the genesis
and metastasis of esophageal carcinoma, discovery of genes for
specific and effective molecular targeted therapy and development
of effective methods for early diagnosis, precise evaluation and
prognostication are of great significance.
microRNAs (miRNAs/miRs) are a class of endogenous
and highly conserved single-stranded non-coding small RNAs of 19–22
nucleotides in length. They are able to inhibit target gene
expression at the post-transcriptional level through specifically
binding with the 3′-untranslated region (3′-UTR) of their target
mRNAs, thus exerting biological functions (4,5). Studies
have verified that miRNAs have important regulatory roles in cell
proliferation, apoptosis, development and apoptosis, processes that
are also closely associated with tumor pathogenesis, development
and prognosis (6,7). miR-940 is an important member of the
miRNA family, which has been reported to be abnormally expressed in
tumorous tissues and to have an important regulatory role (8,9).
However, to the best of our knowledge, miR-940 expression in ESCC
has not been previously reported, and its association with
prognosis remains elusive.
In the present study, miR-940 expression was
assessed in ESCC cell lines with various degrees of
differentiation, as well as in a normal esophageal cell line, and
in ESCC tissues and para-carcinoma tissues by using reverse
transcription-quantitative (RT-q)PCR. The association of miR-940
expression with the clinicopathological features and prognosis in
ESCC patients was analyzed. The possible mechanistic roles of
miR-940 in the occurrence and development of ESCC were also
investigated.
Materials and methods
Tissue specimens and clinical
data
A total of 210 fresh surgically resected ESCC
tissues and matched para-carcinoma tissues (at least 5 cm away from
the tumor margin; 150 males and 60 females; aged 52–79 years old)
were collected at Weihai Central Hospital (Weihai, China) between
January 2011 and January 2012. Following excision, the tissue
specimens were washed in saline within 30 min, placed in liquid
nitrogen and preserved in a refrigerator at −80°C. None of the
patients had received any anti-tumor therapies, including
radiotherapy, chemotherapy or biological immunotherapy;
furthermore, they were not complicated by any primary tumors in
other organs. All specimens were confirmed as squamous cell
carcinoma by pathologists after surgery. A total of 52 cases
received adjuvant radiotherapy and chemotherapy after the
operation. The clinical features of the patients are presented in
Table I. Their clinicopathological
stage and criteria of tumor differentiation were evaluated
according to the TNM classification system of the American Joint
Committee on Cancer-International Union Against Cancer staging
manual (10). The criteria for
determining the differentiation of tumors are as follows: i)
Well-differentiated-prominent keratinization with pearl formation
and a minor component of non-keratinizing basal-like cells, tumor
cells arranged in sheets and low mitotic counts; ii) moderately
differentiated-variable histologic features ranging from
parakeratotic to poorly keratinizing lesions and pearl formation
generally absent; iii) poorly differentiated-predominantly
consisting of basal-like cells forming large and small nests with
frequent central necrosis and with the nests consisting of sheets
or pavement-like arrangements of tumor cells that are occasionally
punctuated by small numbers of parakeratotic or keratinizing cells.
Overall survival (OS) time was defined as the date of surgery to
the date of death or completion of follow-up. Completion of
follow-up, loss to follow-up and death of other causes were treated
as the endpoint data. All patients had provided written informed
consent to participate in the study. The present study was approved
by the Medical Ethics Committee of Weihai Central Hospital (Weihai,
China).
 | Table I.Associations of miR-940 expression
with clinicopathological characteristics of patients with
esophageal squamous cell carcinoma. |
Table I.
Associations of miR-940 expression
with clinicopathological characteristics of patients with
esophageal squamous cell carcinoma.
| Clinicopathological
feature | Cases (n) | miR-940 low
(n=100) | miR-940 high
(n=110) | P-value |
|---|
| Sex |
|
|
| 0.275 |
| Male | 150 | 75 (50.00) | 75 (50.00) |
|
|
Female | 60 | 25 (41.67) | 35 (58.33) |
|
| Age (years) |
|
|
| 0.778 |
|
>60 | 162 | 78 (48.15) | 84 (51.85) |
|
| ≤60 | 48 | 22 (45.83) | 26 (54.17) |
|
| Tumor size (cm) |
|
|
| 0.179 |
|
>5 | 111 | 48 (43.24) | 63 (56.76) |
|
| ≤5 | 99 | 52 (52.53) | 47 (47.47) |
|
| Tumor
differentiation |
|
|
| 0.034 |
|
Well/moderate | 125 | 52 (41.60) | 73 (58.40) |
|
| Poor | 85 | 48 (56.47) | 37 (43.53) |
|
| TNM stage |
|
|
| 0.009 |
| I/II | 110 | 43 (39.09) | 67 (60.91) |
|
| III | 100 | 57 (57.00) | 43 (43.00) |
|
| Tumor location |
|
|
| 0.588 |
|
Upper | 28 | 12 (42.86) | 16 (57.14) |
|
|
Middle+lower | 182 | 88 (48.35) | 94 (51.65) |
|
| Lymphatic
metastasis |
|
|
| 0.001 |
| Yes | 135 | 76 (56.30) | 59 (43.70) |
|
| No | 75 | 24 (32.00) | 51 (68.00) |
|
Cell culture and cell
transfection
The human ESCC cell lines KYSE410 (poorly
differentiated invasive ESCC, TE-13 (poorly differentiated),
ECA-109 (moderately differentiated) and KYSE-510 (highly
differentiated), as well as the normal human esophageal cell line
Het-1A, were purchased from the Shanghai Cell Institute of the
Chinese Academy of Sciences. In addition, the TE-13 cell line used
in the present study was authenticated by STR profiling. All cells
were placed in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) and cultured in an incubator containing 5%
CO2 at 37°C. KYSE410 and TE-13 cells in the logarithmic
growth phase were harvested and transfected with either miR-940
mimics (cat. no. B02002; Shanghai GenePharma Co., Ltd.) and
miR-negative control (NC) (cat. no. B1012; Shanghai GenePharma Co.,
Ltd.), in strict accordance with the manufacturer's protocol for
the Lipofectamine™ 2000 kit (Invitrogen; Thermo Fisher Scientific,
Inc.). At 24 h following transfection, miR-940 expression in cells
was detected through RT-qPCR, so as to determine the efficiency of
overexpression.
Total RNA extraction and RT-qPCR
Total RNA was extracted using a TRIzol kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and the purity of the extracted RNA was
detected with an ultraviolet spectrophotometer. RNA with a 260/280
nm absorbance value ratio of 1.7–2.0 served as the qualification
standard for subsequent experiments. Reverse transcription was
performed immediately; alternatively, the RNA was stored in a
refrigerator at −80°C. Reverse transcription of total RNA to
complementary DNA was performed using a Moloney murine leukemia
virus RT kit (Promega Corp.) according to the manufacturer's
protocol. Specific primers for miR-940 and internal reference U6
were designed and synthesized by RiboBio. The primer sequences used
were as follows: miR-940 forward, 5′-GTATAAAGGGCCCCCGCT-3′ and
reverse, 5′-AGGGTCCGAGGTATTCGCACT-3′; U6 forward,
5′-CATCACCATCAGGAGAGTCG-3′ and reverse, 5′-TGACGCTTGCCCACAGCCTT-3′.
qPCR was performed using the SYBR Green Realtime PCR Master Mix
(Fermentas) and specific primers, with the complementary DNA used
as the template. The LightCycle 96 fluorescent quantitative PCR
machine (Roche) was used for the reaction and detection. The
reaction conditions were as follows: 2 min at 94°C, 20 sec at 94°C
and 30 sec at 60°C for 40 cycles. The relative expression of
miR-940 was calculated according to the 2−∆∆Cq method
(11), with U6 used as the internal
reference. All assays were performed in triplicate.
Cell viability detected by MTT
assay
Cells transfected for 24 h were added to the 96-well
cell culture plate (5,000 cells/well), and 20 µl MTT
(Sigma-Aldrich; Merck KGaA) was added to each well at 24, 48, 72
and 96 h, followed by incubation for another 4 h at 37°C. The
supernatant was carefully aspirated, and subsequently, 150 µl DMSO
was added to each well, followed by agitation for 15 min to fully
dissolve the crystals that had formed. The OD value was determined
using a microplate reader (Bio-Tech Instruments) at a wavelength of
450 nm. Three duplicate wells were set for each group.
Cell cycle analysis by flow
cytometry
Cells transfected for 48 h were collected through
digestion with 0.25% trypsin solution (EDTA-free) and washed with
PBS for three times. The supernatant was discarded after
centrifugation, and cells were re-suspended with 70% ethanol, in
which they were fixed at 4°C for 18 h. The cells were then
collected through centrifugation (1,000 × g; 5 min; 4°C) and washed
with PBS twice, and the supernatant was discarded following
centrifugation. Cells were then re-suspended with 400 µl RNAse A
(BD Biosciences)-containing propidium iodide (PI) staining
solution, incubated in the dark at 4°C for 1 h and the cell cycle
was detected using a flow cytometer.
Cell apoptosis detected by Annexin
V-FITC/PI double-staining
After digestion with 0.25% trypsin solution
(EDTA-free), cells transfected for 48 h were collected using
centrifugation (111.8 × g; 5 min; 4°C). Binding buffer (500 µl) was
added to re-suspend the cells in accordance with the protocol of
the Annexin V-FITC/PI double-staining cell apoptosis detection kit
(Beyotime Institute of Biotechnology). Subsequently, 5 µl Annexin
V-FITC was added and the suspension was mixed sufficiently,
followed by addition of 5 µl PI, sufficient mixing and incubation
in the dark at room temperature for 5–15 min. Finally, cell
apoptosis was detected with a flow cytometer. The Annexin
V-FITC+/PI− cells were considered apoptotic
and counted.
Statistical analysis
Data were analyzed using SPSS 20.0 statistical
software (IBM Corp.). Measurement data were expressed as the mean ±
standard deviation and were compared using Student's t-test or
one-way analysis of variance. A Student-Newman-Keuls test was used
as a post-hoc test following ANOVA. The associations between
miR-940 expression and various clinicopathological characteristics
were assessed using a chi-square test. OS was assessed using the
Kaplan-Meier method and groups were compared using the log-rank
test. Univariate and multivariate analysis regarding OS was
performed using the Cox proportional hazards regression model.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-940 expression in ESCC tissues and
cell lines
miR-940 expression in 210 pairs of ESCC tissues and
matched para-carcinoma tissues was detected using RT-qPCR. As
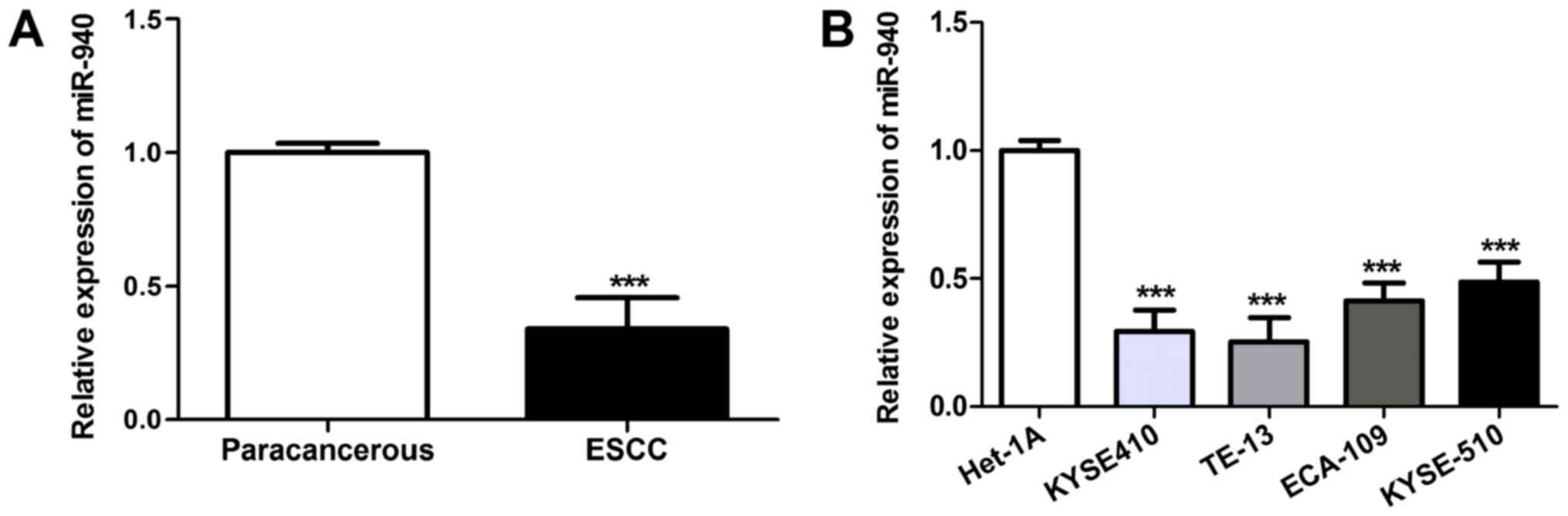
presented in Fig. 1A, compared with
that in para-carcinoma tissues, miR-940 expression in ESCC tissues
was markedly downregulated (P<0.001). Furthermore, miR-940
expression was analyzed in the human ESCC cell lines KYSE410,
TE-13, ECA-109 and KYSE-510, as well as in the normal human
esophageal cell line Het-1A. As presented in Fig. 1B, miR-940 expression in the human
ESCC cell lines KYSE410, TE-13, ECA-109 and KYSE-510 was distinctly
downregulated relative to that in the normal human esophageal cell
line Het-1A (P<0.001), which was particularly obvious in the
poorly-differentiated KYSE410 and TE-13 cell lines.
Association of miR-940 with
clinicopathological parameters and prognosis in ESCC patients
To confirm the role of miR-940 expression in the
development of ESCC, the associations of miR-940 with the
clinicopathological parameters and prognosis of ESCC patients were
then assessed. The ESCC patients were divided into 2 subgroups
according to their miR-940 expression status. The high miR-940
expression group included patients whose miR-940 level was higher
than the average for all patients, while the low miR-940 expression
group included patients whose miR-940 level was lower than the
average for all patients. As indicated in Table I, low miR-940 expression was closely
associated with poor tumor differentiation, positive lymph node
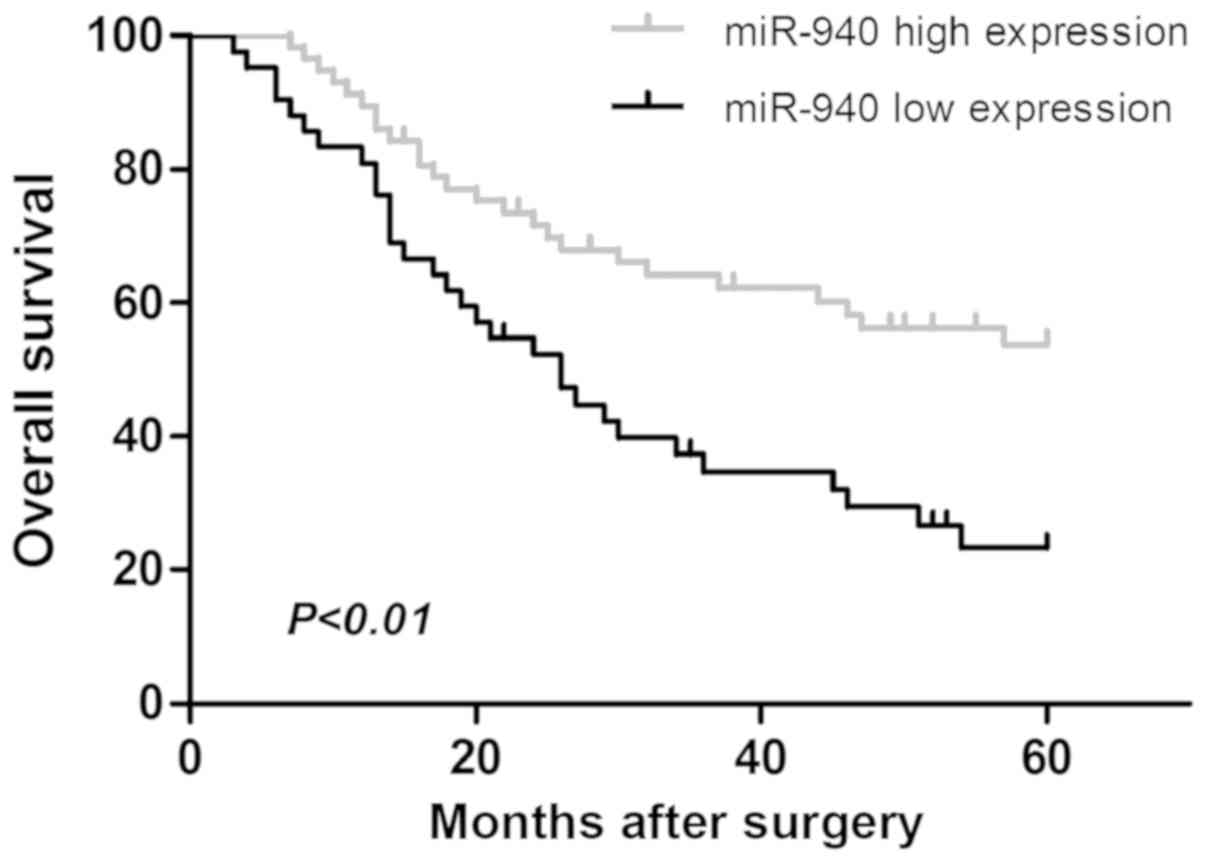
metastasis and advanced clinical stage (P<0.05). Kaplan-Meier
survival analysis suggested that the low miR-940 expression group
had poor prognosis (P<0.01; Fig.
2). Univariate Cox regression analysis was performed for all
clinicopathological parameters (Table
II). The tumor differentiation degree, lymph node metastasis,
TNM stage and miR-940 expression were determined as risk factors
affecting patient prognosis. Furthermore, risk factors with
statistical significance were incorporated into the multivariate
Cox regression model analysis. The results suggested that lymph
node metastasis, TNM stage and miR-940 expression were independent
risk factors affecting patient prognosis (P<0.05).
 | Table II.Univariate and multivariate analysis
of overall survival in 210 patients with ESCC. |
Table II.
Univariate and multivariate analysis
of overall survival in 210 patients with ESCC.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | RR (95% CI) | P-value | RR (95% CI) | P-value |
|---|
| Sex |
| (male
vs. female) | 0.98
(0.90~1.21) | 0.210 | – | – |
| Age |
| (>60
vs. ≤60) | 1.19
(0.94~1.51) | 0.148 | – | – |
| Tumor size |
| (>5
vs. ≤5) | 1.33
(0.95~1.87) | 0.099 | – | – |
| Tumor
differentiation |
|
(Well/moderate vs. poor) | 2.88
(1.23~6.75) | 0.015 | 1.65
(0.95~2.87) |
0.076 |
| TNM stage |
| (I/II
vs. III) | 3.11
(1.58~6.11) | 0.001 | 2.87
(1.30~6.33) |
0.009 |
| Tumor location |
| (upper
vs. female) | 1.25
(0.94~1.66) | 0.124 | – | – |
| Lymphatic
metastasis |
| (no vs.
yes) | 3.34
(1.82~6.13) | <0.001 | 3.58
(1.67~7.65) | 0.001 |
| miR-940
expression |
| (high
vs. low) | 3.01
(1.37~6.61) | 0.006 | 2.44
(1.14~5.20) | 0.021 |
Effect of miR-940 overexpression on
cell viability, cell cycle and apoptotic rate of ESCC cells
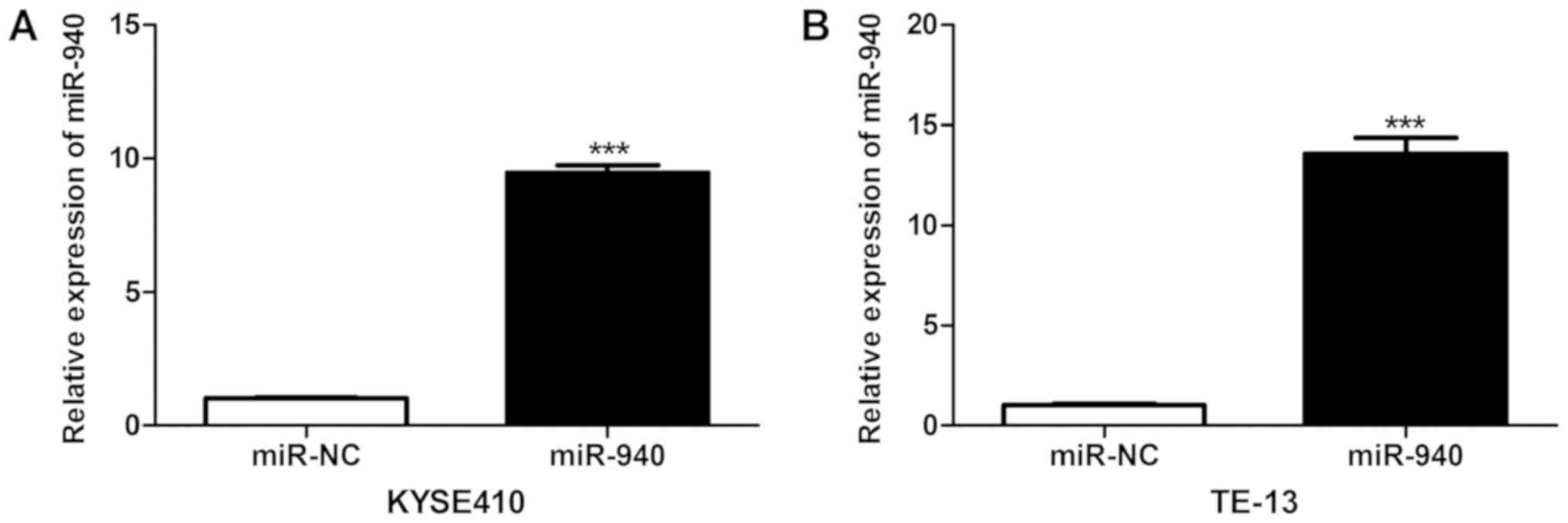
As presented in Fig.
3, RT-qPCR analysis confirmed that at 24 h after transfection,
the miR-940 expression levels in KYSE410 and TE-13 cells in the
miR-940 mimics group were markedly elevated (P<0.001),
suggesting successful miR-940 mimics transfection and high
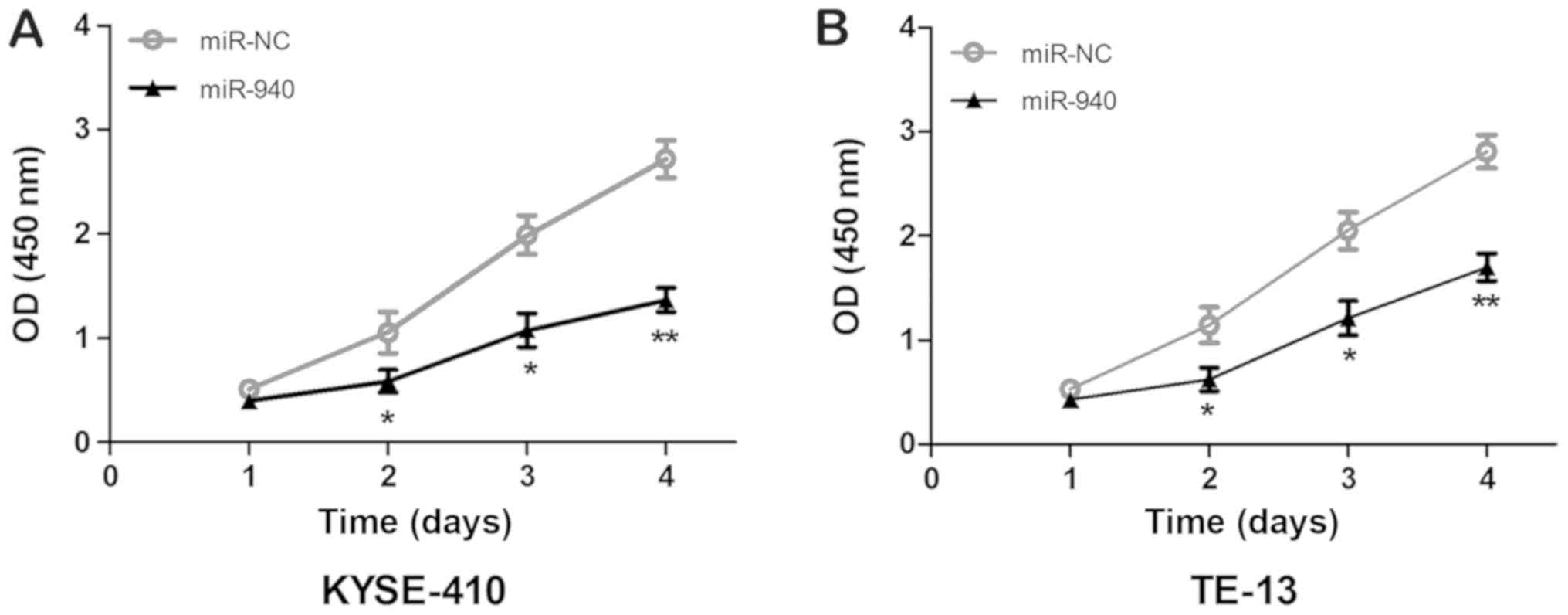
efficiency of overexpression. The results of the MTT assay are
presented in Fig. 4. It was observed
that, compared with miR-NC group, the OD 450 nm values of KYSE410
and TE-13 cells transfected with miR-940 mimics were significantly
decreased on days 2, 3 and 4 (P<0.05), indicating that
upregulation of miR-940 expression markedly suppressed the
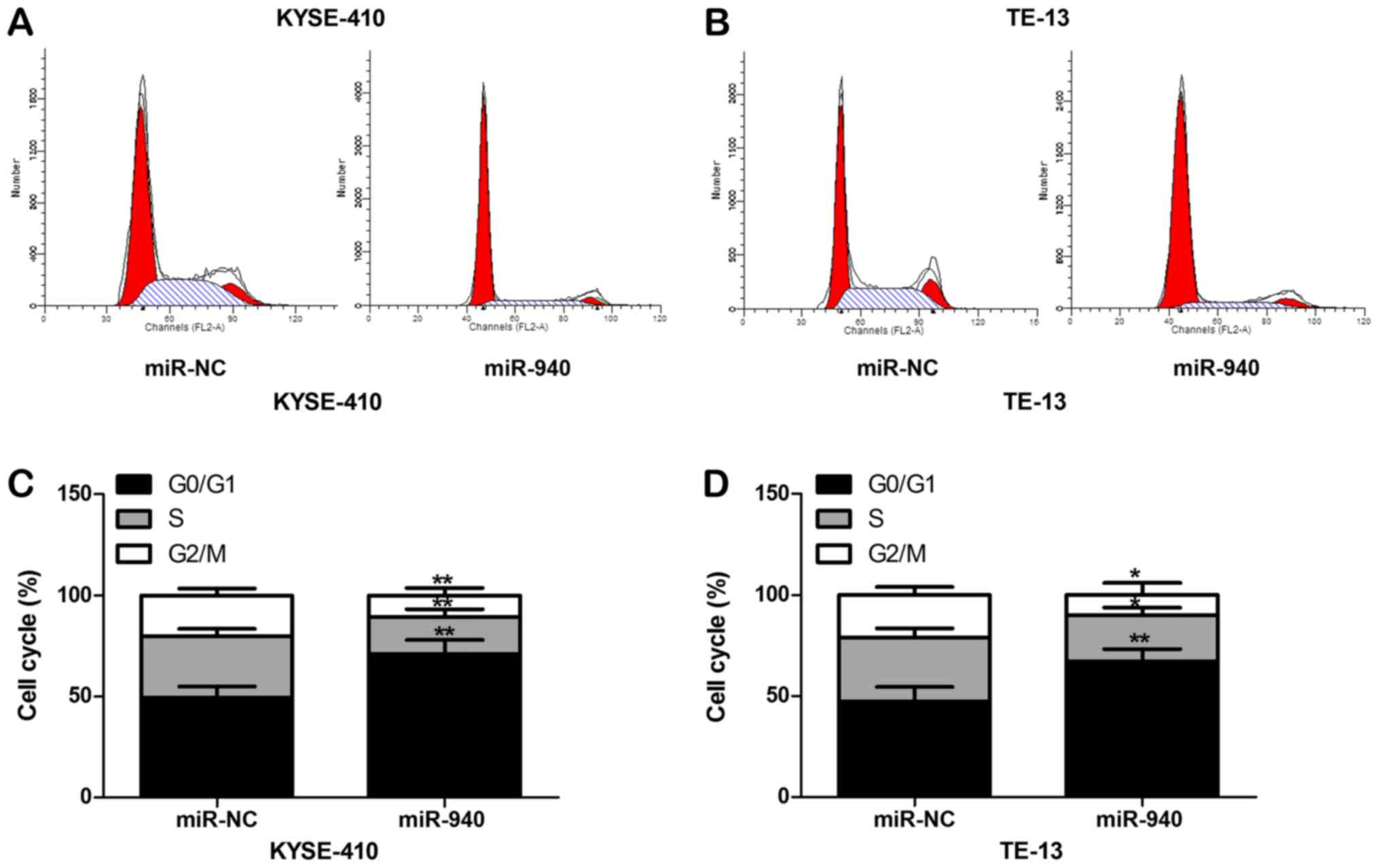
viability of ESCC cells. The cell cycle was detected by flow
cytometry and the results are presented in Fig. 5. The G0/G1-phase population of
KYSE410 and TE-13 cells with up-regulated miR-940 expression was
significantly increased (P<0.01), while the proportions of cells
in the G2/M- and S-phase were evidently decreased (P<0.05),
indicating that overexpression of miR-940 caused cell cycle arrest
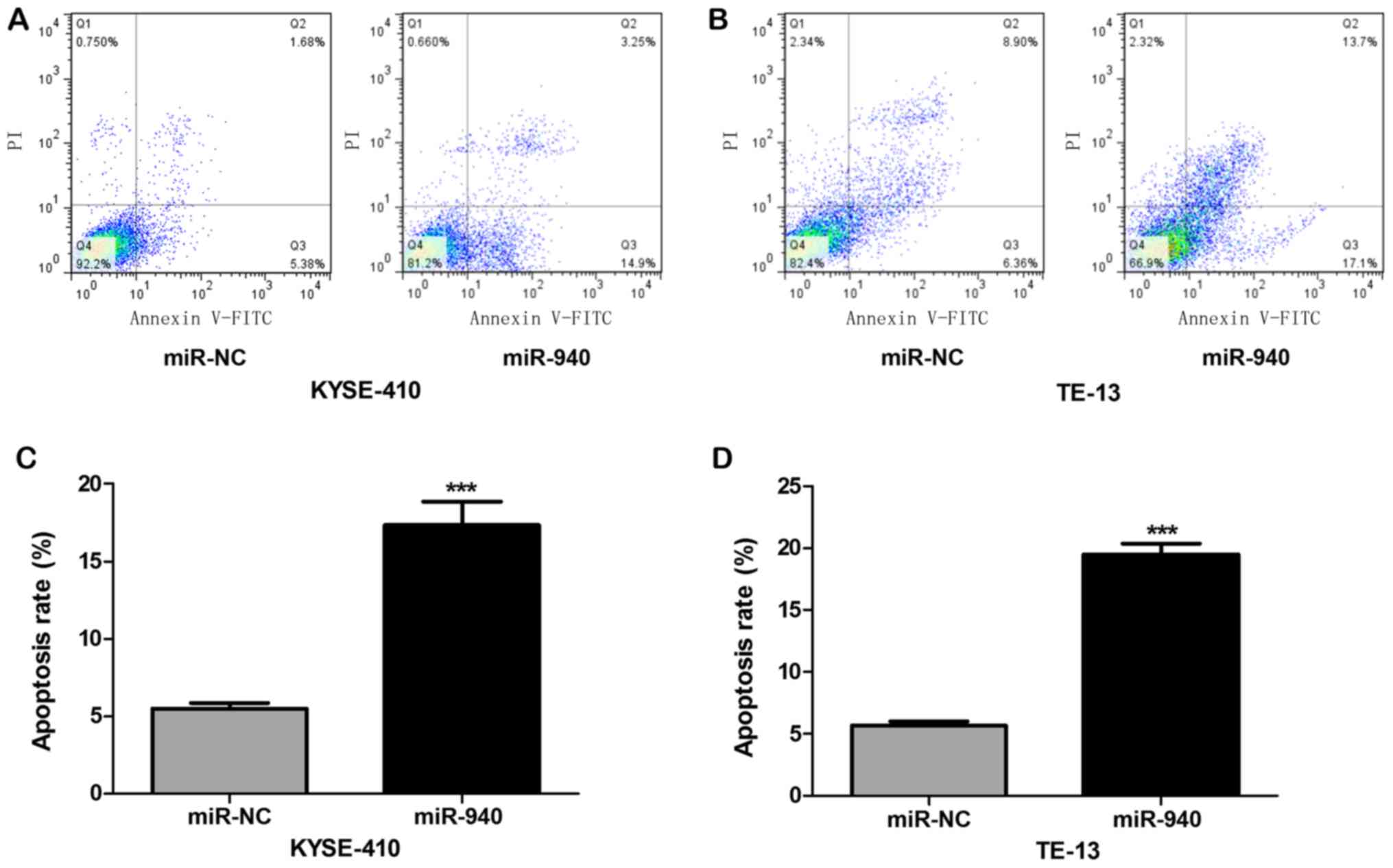
of ESCC cells at the G0/G1 phase. The results of the flow
cytometric analysis of apoptotic cells with Annexin V-FITC/PI
double-staining are displayed in Fig.
6. The apoptotic rates of KYSE410 and TE-13 cells with
overexpression of miR-940 were evidently elevated (P<0.01),
indicating that miR-940 mimics promote apoptosis of ESCC cells.
Discussion
In spite of the progress made in the surgical and
pharmacological therapy of esophageal carcinoma in recent years,
the prognosis for esophageal carcinoma remains poor (12). Local recurrence and metastasis of
esophageal carcinoma are major bottlenecks restricting the efficacy
of esophageal carcinoma therapy and affecting the improvement of
survival of esophageal carcinoma patients. To date, the mechanisms
of the genesis and development of esophageal carcinoma have
remained to be fully elucidated and no effective targeted
therapeutics are available at present. Therefore, it is urgently
required to illustrate the precise mechanisms of the genesis,
development and metastasis of esophageal carcinoma at the molecular
level and search for effective gene targets for targeted therapy of
esophageal carcinoma.
Aberrant expression of various miRNAs has been
detected in multiple cancer types, which have crucial effects on
tumor cell proliferation, apoptosis, invasion and migration.
Therefore, differentially expressed miRNAs have become a hotspot in
current research on tumors and gene therapy (13). Furthermore, numerous miRNAs have been
verified to be involved in the genesis, development, invasion and
metastasis of esophageal carcinoma (14,15).
miR-940 is one of the most important members of the miRNA family
discovered recently. It has been indicated that miR-940 has an
important role in various diseases. Liang et al (16) reported that downregulation of miR-940
had a key role in the development of tetralogy of Fallot in humans.
Xu et al (17) indicated that
serum miR-940 levels were significantly increased after exhaustive
exercise in patients with congestive heart failure, which may be
used as a marker for adaptive exercise in such patients. The
expression pattern of miR-940 in malignant tumors is different from
that in the above scenario. It has been reported that miR-940
expression is reduced in various types of malignant tumor,
including ovarian cancer (9),
prostate cancer (18),
nasopharyngeal carcinoma (19) and
hepatocellular carcinoma (20).
Furthermore, overexpression of miR-940 may inhibit the
proliferation, invasion and migration of tumor cells, suggesting
that the role of miR-940 is that of a tumor suppressor gene in some
malignant tumors (9,18–20).
Conversely, it has been indicated that the expression of miR-940 is
increased in gastric cancer (21),
bladder cancer (22) and pancreatic
cancer (23), where it has the role
of an oncogene. However, to date, miR-940 expression in ESCC has
not been previously reported, to the best of our knowledge. The
present study first reported on the abnormal expression of miR-940
in ESCC. The expression levels of miR-940 in 210 ESCC tissues and
matched para-carcinoma tissues, as well as in the human ESCC cell
lines KYSE410, TE-13, ECA-109 and KYSE-510, and in the normal human
esophageal cell line Het-1A, were detected using RT-qPCR. The
present results suggested that miR-940 expression in ESCC tissues
and cells was decreased. Further statistical analysis indicated
that low miR-940 expression was closely associated with poor tumor
differentiation, positive lymph node metastasis and advanced
clinical stage, suggesting that reduced miR-940 expression was
closely linked to the development and metastasis of ESCC.
Studies on the association between miR-940 and
prognosis for patients with malignant tumors are currently sparse.
It has been reported that low miR-940 expression in hepatocellular
carcinoma is associated with poor prognosis for patients (24). However, it has also been indicated
that high expression of miR-940 is associated with poor prognosis
in patients with gastric cancer (21). In the present study, post-operative
follow-up was performed, the results of which indicated that
patients in the low miR-940 expression group had a shorter OS and
that low miR-940 levels are therefore associated with poor
prognosis. Further multivariate analysis using the Cox regression
model suggested that lymph node metastasis, clinical stage and
miR-940 expression were independent risk factors affecting patient
prognosis.
Excessive cell proliferation and blocked cell
apoptosis are the basic biological characteristics of malignant
tumor cells. To study the underlying mechanisms of action of
miR-940 in ESCC, miR-940 mimics were transfected into ESCC cells to
perform a gain-of-function experiment. The MTT assay suggested that
miR-940 mimics suppressed the viability of ESCC cells. Cell cycle
progression is a key factor regulating cell proliferation. The flow
cytometric cell cycle analysis performed in the present study
indicated that miR-940 mimics made the cells stay at G0 phase or
blocked the cell cycle progression from G1- to S-phase. In
addition, the proportion of cells in G0/G1 phase was markedly
increased, suggesting that miR-940 suppresses the viability of ESCC
cells partially through blocking the cell cycle. Finally, cell
apoptosis was also detected through Annexin V-FITC/PI
double-staining, and the results suggested that miR-940 mimics
increased apoptosis of ESCC cells. Each miRNA has several
regulatory target genes, and the final effects of a miRNA are the
consequences of the comprehensive action of the complex downstream
network of multiple genes. The precise targeted regulatory
mechanisms by which miR-940 affects the biological characteristics
of ESCC cells remain to be fully elucidated, which should be
predicted by bioinformatics methods and assessed/verified through
further cytological experiments.
In conclusion, the present study indicated that
miR-940 expression is decreased in ESCC, and low miR-940 expression
is significantly associated with poor tumor differentiation,
advanced clinical stage, lymph node metastasis and poor prognosis.
miR-940 is also an independent risk factor affecting patient
prognosis. Overexpression of miR-940 in ESCC cells markedly reduced
the cell viability, blocked the cell cycle at G0/G1-phase and
promoted cell apoptosis. Therefore, miR-940 is a promising novel
anti-cancer target and prognostic biomarker in ESCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS designed the experiments. HW, TS and YQ performed
the experiments. JS and HW analyzed the data. HW wrote the
manuscript. All authors have read and approved the manuscript.
Ethical approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Weihai Central Hospital (Weihai, China). Informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zeng H, Fan Y,
Qiao Y and Zhou Q: Esophageal cancer incidence and mortality in
China, 2010. Thorac Cancer. 5:343–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shen J, Stass SA and Jiang F: MicroRNAs as
potential biomarkers in human solid tumors. Cancer Lett.
329:125–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sorel O and Dewals BG: MicroRNAs in large
herpesvirus DNA genomes: Recent advances. Biomol Concepts.
7:229–239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paulmurugan R: MicroRNAs-a new generation
molecular targets for treating cellular diseases. Theranostics.
3:927–929. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rashed MH, Kanlikilicer P,
Rodriguez-Aguayo C, Pichler M, Bayraktar R, Bayraktar E, Ivan C,
Filant J, Silva A, Aslan B, et al: Exosomal miR-940 maintains
SRC-mediated oncogenic activity in cancer cells: A possible role
for exosomal disposal of tumor suppressor miRNAs. Oncotarget.
8:20145–20164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Wang Z, Gu X and Cui J: miR-940
upregulation suppresses cell proliferation and induces apoptosis by
targeting PKC-δ in ovarian cancer OVCAR3 cells. Oncol Res.
25:107–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rice TW, Ishwaran H, Ferguson MK,
Blackstone EH and Goldstraw P: Cancer of the esophagus and
esophagogastric junction: An eighth edition staging primer. J
Thorac Oncol. 12:36–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schimittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pereira DM, Rodrigues PM, Borralho PM and
Rodrigues CM: Delivering the promise of miRNA cancer therapeutics.
Drug Discov Today. 18:282–289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang D, Xu X, Deng F, Feng J, Zhang H,
Liu Y, Zhang Y, Pan L, Liu Y, Zhang D, et al: miRNA-940 reduction
contributes to human Tetralogy of Fallot development. J Cell Mol
Med. 18:1830–1839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu T, Zhou Q, Che L, Das S, Wang L, Jiang
J, Li G, Xu J, Yao J, Wang H, et al: Circulating miR-21, miR-378,
and miR-940 increase in response to an acute exhaustive exercise in
chronic heart failure patients. Oncotarget. 7:12414–12425.
2016.PubMed/NCBI
|
|
18
|
Rajendiran S, Parwani AV, Hare RJ,
Dasgupta S, Roby RK and Vishwanatha JK: MicroRNA-940 suppresses
prostate cancer migration and invasion by regulating MIEN1. Mol
Cancer. 13:2502014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma J, Sun F, Li C, Zhang Y, Xiao W, Li Z,
Pan Q, Zeng H, Xiao G, Yao K, et al: Depletion of intermediate
filament protein nestin, a target of microRNA-940, suppresses
tumorigenesis by inducing spontaneous DNA damage accumulation in
human nasopharyngeal carcinoma. Cell Death Dis. 5:e13772014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo
L, Shao Z and Li X: miR-940 Suppresses tumor cell invasion and
migration via regulation of CXCR2 in hepatocellular carcinoma.
Biomed Res Int. 2016:76183422016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu
Z, Tang W, Gan L, Sun M and Li J: MicroRNA-940 promotes tumor cell
invasion and metastasis by downregulating ZNF24 in gastric cancer.
Oncotarget. 6:25418–25428. 2015.PubMed/NCBI
|
|
22
|
Wang R, Wu Y, Huang W and Chen W:
MicroRNA-940 targets INPP4A or GSK3β and activates the
Wnt/β-catenin pathway to regulate the malignant behavior of bladder
cancer cells. Oncol Res. 26:145–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang HW, Liu GH, Liu YQ, Zhao HC, Yang Z,
Zhao CL, Zhang XF and Ye H: Over-expression of microRNA-940
promotes cell proliferation by targeting GSK3β and sFRP1 in human
pancreatic carcinoma. Biomed Pharmacother. 83:593–601. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan B, Liang Y, Wang D and Luo F: MiR-940
inhibits hepatocellular carcinoma growth and correlates with
prognosis of hepatocellular carcinoma patients. Cancer Sci.
106:819–824. 2015. View Article : Google Scholar : PubMed/NCBI
|