Introduction
Down's syndrome (DS), or trisomy 21, is caused by
the presence of three copies of chromosome 21 and is the most
common genetic cause of intellectual disability (1) DS has an incidence of up to 1 in 700
live births (2), and this incidence
increases with increasing maternal age (3). DS is characterized by mild-to-moderate
learning disabilities, craniofacial abnormalities and hypotonia
(4), and is typically accompanied by
more than 80 diseases such as congenital heart disease, low
immunity, leukaemia (5). DS is
associated with financial burden on the family and society, and
there is no effective therapy for the chromosomal abnormalities.
Traditional prenatal screening and diagnostic methods are
associated with several shortcomings, and the clinical application
of novel non-invasive prenatal diagnostic methods are restricted by
various factors (6). Therefore,
there is a requirement for the development of an efficient,
accurate and cost-effective screening method.
Circular RNAs (circRNAs) are a type of RNA, which
unlike linear RNA, have joined 5′ and 3′ends that form a continuous
loop (7), a structure that provides
resistance against exonucleases and further differentiates them
from other types of RNA (8).
circRNAs are a class of endogenous noncoding RNAs produced by the
non-canonical form of alternative splicing (7,9).
Developments in bioinformatics analysis and RNA deep sequencing
technology revealed that circRNAs are abundant, conserved and
stable in mammalian cells (10–16).
Previous studies demonstrated that certain circRNAs not only act as
competing endogenous RNA for gene expression regulation, but also
repress the function of microRNAs (miRNAs/miRs) by binding to miRNA
response elements (MREs), thereby regulating the expression level
of other related RNAs (13,17,18).
Studies have also shown that imbalances in the expression of
certain miRNAs may lead to extensive changes in mRNA and protein
expression, and play an important role in the development and
progression of disease (19,20). In addition, certain circRNAs serve as
a template during translation and guide the synthesis of proteins
(21,22). Moreover, circRNAs bind with specific
proteins to inhibit or regulate protein activity (23,24).
Furthermore, certain circRNAs may participate in development of
disease, particularly in cancer. circRNAs exhibit diverse functions
and may serve as diagnostic or predictive biomarkers of disease
(25,26) and novel therapeutic targets. Sun
et al (27) revealed that
hsa_circ_0000520 may be involved in the development of gastric
cancer and may therefore serve as a biomarker of the disease. Yang
et al (28) demonstrated that
differentially expressed circ-F-box, WD repeat domain containing 7
(FBXW7) and FBXW7-185aa have potential prognostic significance in
brain cancer. However, to the best of our knowledge, the
association between circRNAs and DS has not been previously
reported. Therefore, it is of significance to carry out an in-depth
and systematic research on circRNA of DS.
Materials and methods
Samples
Umbilical cord blood samples were obtained from 12
pregnant women, six (age range, 35–43 years; mean age, 39.7 years)
carrying fetuses with DS (two male and four female) and six (27–36
years, 31.3 years of mean age) carrying fetuses without DS (two
male and four female) at a gestational age of 18–22 weeks. The
participants were recruited at Shenzhen People's Hospital
(Shenzhen, China) between January 2013 and December 2014. Diagnosis
of fetal DS was performed by chromosomal examination. In addition,
peripheral blood samples were obtained from 12 children, six with
DS and six without DS (two males and four females; age range, 5–12
years, mean age, 8 years) at Guilin No. 924 Hospital (Guilin,
China) between January 2015 and December 2016. The present study
was approved by the Ethics Committees of Shenzhen People's Hospital
(Shenzhen, China) and Guilin No. 924 Hospital (Guilin, China).
Written informed consent was obtained from all pregnant women and
the parents/guardians of the children.
Sample processing
Ultrasound-guided umbilical cord blood extraction
was performed and a total of 3 ml extracted blood was placed in
EDTA-containing anticoagulant tubes. Peripheral blood (2 ml) of 12
children was extracted with a needle and placed in EDTA-containing
anticoagulant tubes. According to the single nuclear cell
extraction protocol, 1 ml umbilical cord blood mononuclear cells
(PBMCs) was extracted using lymphocyte separation solution (MD
Pacific Biotechnology Co., Ltd.), mixed with 1 ml
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), placed in cryopreservation tubes and stored at −80°C until
further use.
Extraction of total RNA
The frozen PBMCs were thawed at room temperature and
total RNA was extracted using TRIzol® reagent according
to the manufacturer's instructions. Total RNA was subsequently
treated with DNase I to remove genomic DNA contamination. The
purity and concentration of the RNA were determined using a
spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific,
Inc.). RNA integrity was assessed using standard denaturing agarose
gel electrophoresis (1%).
circRNA labelling and array
hybridization
Total RNA from each sample was amplified and
transcribed into fluorescent cRNA utilizing random primers
according to the Arraystar Super RNA Labeling protocol (Arraystar,
Inc.). The labelled cRNAs were hybridized onto Arraystar Human
circRNA arrays (cat. no. 6×7K; Arraystar, Inc.) and incubated for
17 h at 65°C in an Agilent hybridization oven (Agilent
Technologies, Inc.). After washing, slides were scanned with the
Axon GenePix 4000B Scanner (Molecular Devices, LLC).
circRNA data collection and
analysis
Scanned images were imported into GenePix Pro
software (version 6.0; Molecular Devices, LLC) for grid alignment
and raw data extraction. Quantile normalization of raw data and
subsequent data processing were performed using the R software
package heatmap.2 (version 3.2.0; http://mirrors.tuna.tsinghua.edu.cn/CRAN/). The
circRNAs that at least 1 out of 2 samples had flagged as
‘expressed’ (greater than 2 times background standard deviation)
were retained for further differential analyses. Differentially
expressed circRNAs between umbilical cord blood samples obtained
from pregnant woman carrying fetuses with DS and without DS were
identified through fold-change (FC≥2.0 or ≤-2.0) or volcano plot
filtering.
mRNA labelling and array
hybridization
In the present study, sample labelling and array
hybridization were performed according to the Agilent One-Color
Microarray-Based Gene Expression Analysis protocol (Agilent
Technologies, Inc.). Briefly, total RNA from each sample was
linearly amplified and labelled with Cy3-UTP (Enzo Life Sciences,
Inc.). The labelled cRNAs were purified using an RNeasy Mini kit
(Qiagen, Inc.). The concentration and specific activity of the
labelled cRNAs (pmol Cy3/µg cRNA) were measured using a
spectrophotometer. A total of 1 µg of each labelled cRNA was
fragmented by the addition of 11 µl 10X blocking agent (LMAI Bio)
and 2.2 µl 25X fragmentation buffer (Agilent Technologies, Inc.),
then heated at 60°C for 30 min. Subsequently, 55 µl 2X GEx
Hybridization Buffer HI-RPM (Agilent Technologies, Inc.) were added
to dilute the labelled cRNA and 100 µl hybridization solution
(Agilent Technologies, Inc.) were dispensed into the gasket slide
and assembled on the gene expression microarray slide. The slides
were incubated for 17 h at 65°C in the aforementioned Agilent
hybridization oven. The hybridized arrays were washed, fixed and
scanned using the Agilent DNA Microarray Scanner (cat. no. G2505C;
Agilent Technologies, Inc.).
Analysis and functional analysis of
differentially expressed genes
The differentially expressed genes (FC≥2.0 or ≤-2.0)
were selected using Agilent GeneSpring GX software (version 12.1;
Agilent Technologies, Inc.). Then, hierarchical clustering was
performed using scripts prepared by Aksomics Inc. Gene Ontology
(GO; http://www.geneontology.org) analysis
and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) pathway analysis were
performed using standard enrichment calculation methods.
mRNA data analysis
Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies, Inc.) was used to analyse the
acquired array images. Quantile normalization and subsequent data
processing were performed using the GeneSpring GX software package
(version 12.1; Agilent Technologies, Inc.). After quantile
normalization of the raw data, genes in which at least 1 out of 2
samples had flags in Detected (‘All Targets Value’) were chosen for
further data analysis. Differentially expressed genes between the
umbilical cord blood samples obtained from pregnant woman carrying
fetuses with and without DS were identified through FC and volcano
plot filtering. Hierarchical clustering was performed using the
aforementioned R scripts. GO and KEGG analyses were performed using
standard enrichment calculation methods.
Validation of differentially expressed
circRNA and mRNA by reverse-transcription quantitative PCR
(RT-qPCR)
Six circRNAs (Table
I) and three mRNAs (Table II)
were randomly selected to verify the accuracy of the results in
peripheral blood samples of children with and without DS by
RT-qPCR. Total RNA was extracted from the samples by TRI REAGENT BD
(Molecular Research Center, Inc.) and reverse transcribed into cDNA
using SuperScriptTM III Reverse Transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc.), 5X RT buffer, 10 mM dNTP mixture (dATP,
dGTP, dCTP and dTTP; 2.5 mM each; HyTest Ltd.) and random primers
supplied by Yingjun Biotechnology Co., Ltd., at 65°C for 5 min and
on ice for 2 min. qPCR was subsequently performed with 2X SYBRGreen
PCR master mix (Arraystar, Inc.) and primers (Table III). The following thermocycling
conditions were used: 95°C for 5 min followed by 40 cycles of a
denaturing step at 95°C for 10 sec and an annealing/extension step
at 60°C for 60 sec. All reactions were run in triplicate. mRNA
levels were quantified using the 2−∆∆Cq method (29) and normalized to the internal
reference gene β-actin.
 | Table I.Verification of the circRNA
microarray in peripheral blood samples obtained from children with
and without Down's syndrome. |
Table I.
Verification of the circRNA
microarray in peripheral blood samples obtained from children with
and without Down's syndrome.
| circRNA | Fold-change | 2−∆∆Cq
value | P-value |
|---|
|
hsa_circRNA_103135 | 4.49 | 1.23 | 0.4250 |
|
hsa_circRNA_103127 | 2.64 | 0.46 | 0.0009 |
|
hsa_circRNA_103112 | 2.04 | 0.42 | 0.0002 |
|
hsa_circRNA_103137 | −2.16 | 1.34 | 0.1530 |
|
hsa_circRNA_104907 | −4.51 | 3.29 |
1×10−6 |
|
hsa_circRNA_101116 | −5.07 | 1.17 | 0.3180 |
 | Table II.Verification of differentially
expressed genes in peripheral blood samples obtained from children
with and without Down's syndrome. |
Table II.
Verification of differentially
expressed genes in peripheral blood samples obtained from children
with and without Down's syndrome.
| Gene | Fold-change | 2−∆∆Cq
value | P-value |
|---|
| Dual specificity
tyrosine phosphorylation regulated kinase 1A | 2.64 | 1.35 | 0.190 |
| Ubiquitin specific
peptidase 25 | 2.04 | 0.84 | 0.009 |
| Spermatid
perinuclear RNA binding protein | −4.51 | 1.35 | 0.150 |
 | Table III.Primers used for PCR. |
Table III.
Primers used for PCR.
|
| Primer sequence
(5′→3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| β-actin
(Human) |
GTGGCCGAGGACTTTGATTG |
CCTGTAACAACGCATCTCATATT |
|
hsa_circRNA_103135 |
GGAGGGCTTCTACGTCATCTTC |
GTCTATGTAGGAGTGCGGGGTT |
|
hsa_circRNA_103127 |
GACCGTCGCCAGCCAAAC |
GAGTCCAGCGGCAAAACTATAA |
|
hsa_circRNA_103112 |
GCACTTCCTGGCAATGATAGAT |
GGCTTGCTGTAGTATCTGGGTG |
|
hsa_circRNA_103137 |
ATCCTGTCCTCCTAAACCTCCA |
TCTCGCTGACCAAGAACTGAATA |
|
hsa_circRNA_104907 |
TACAAAAGAGCCCACGCTAACT |
TGTCTGAAGGCTTGTTCTCTGG |
|
hsa_circRNA_101116 |
ACTGCCTACTGCTATAATTCTGAA |
GTTGTTTCTGGGCTTCTGTGAG |
| USP25 |
CCTGTTGACGATATTGACGCTAG |
CTCCCTGTTGTTCTGTTGTGCT |
| DYRK1A |
CAGGTCCAGAGTATGAGTGC |
GGCAGCGTAATCTCAACAC |
| STRBP |
GACACTCCACTCTGACACCCTC |
CTCCCTGACAAGAAACTATGCTAA |
Prediction of circRNA/microRNA
interaction
To identify circRNAs acting as miRNA sponges, the
circRNA/miRNA interactions were predicted using a miRNA target
prediction software (cat. no. AS-S-CR-H-V2.0; Arraystar, Inc.)
based on TargetScan (30) and
miRanda (31), and the
differentially expressed circRNAs were annotated with the
circRNA/miRNA interaction information.
Statistical analysis
All statistical data were analysed using SPSS
software (version 19.0). The statistical difference between the two
groups was analysed using a Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Differential expression analysis of
circRNA in umbilical cord blood
circRNAs exhibiting a FC≥2.0 or ≤-2.0 were
identified as being differentially expressed between umbilical cord
blood samples of pregnant woman carrying fetuses with and without
DS. A total of 735 differentially expressed circRNAs were detected
between the two groups, of which 414 were upregulated and 321 were
downregulated (Table SI). The
circRNA expression profile in the two groups is presented in
Fig. 1A.
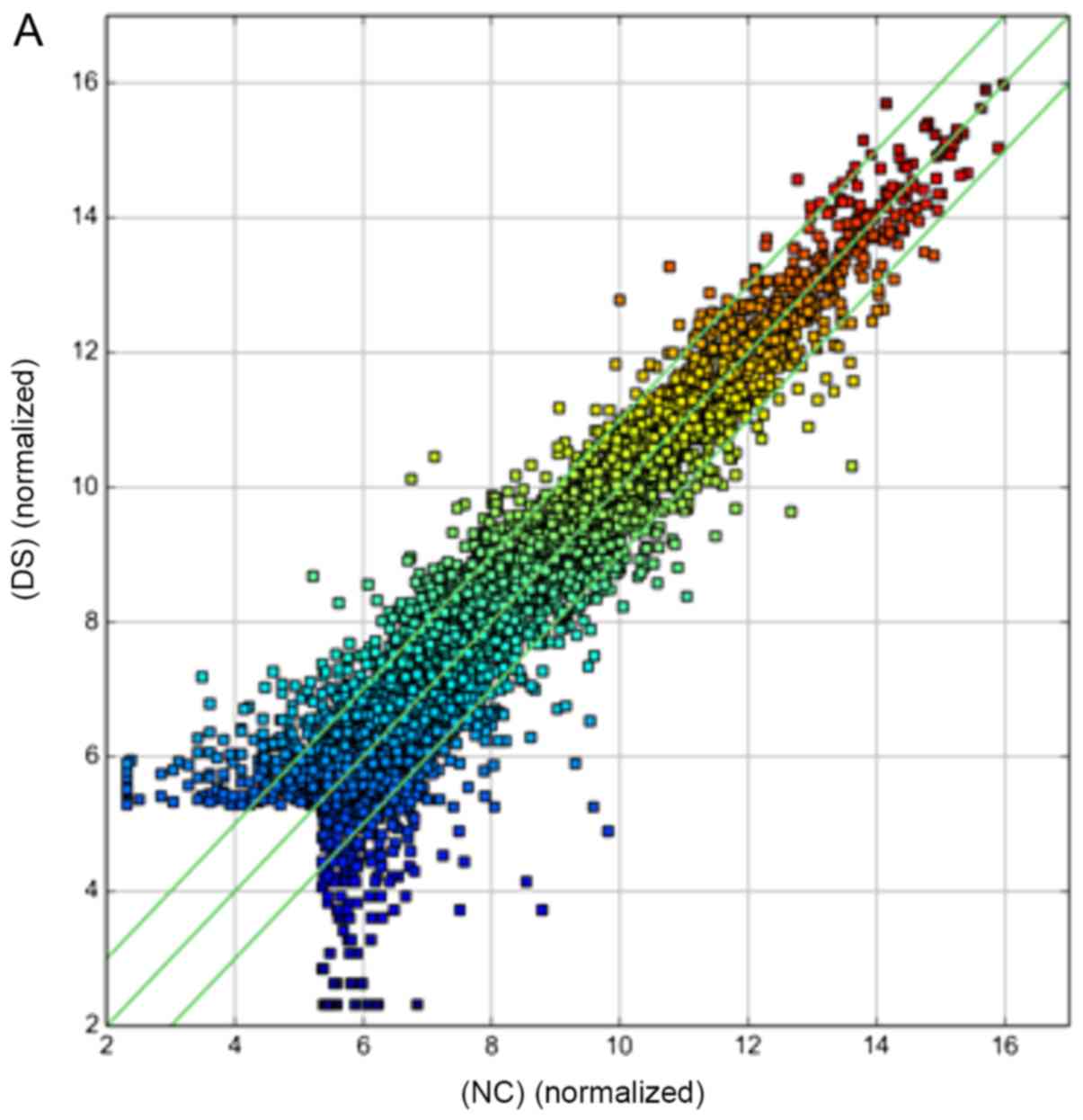
A scatter-plot was used to visualize the variation
(or reproducibility) of circRNA expression between two groups of
samples. The values of the x- and y-axes in the scatter-plot were
the normalized signal values of the samples (log2 scaled) or the
averaged normalized signal values of groups of samples (log2
scaled). The green lines represented FCs. The circRNAs above the
top green line and below the bottom green line indicated a ≥2.0-FC
in circRNA expression between the two groups.
Differential expression analysis of
mRNA in umbilical cord blood samples
mRNAs with a FC≥2.0 or ≤-2.0 were selected as the
differentially expressed ones. In the umbilical cord blood samples
obtained from pregnant woman carrying fetuses with and without DS,
6,619 differentially expressed genes were detected, of which 3,411
and 3,208 genes were upregulated and downregulated, respectively
(Table SII). The mRNA expression
profile of the two groups is shown in Fig. 1B.
RT-qPCR to validate circRNA
expression
A total of six differentially expressed circRNAs in
the microarray experiments were randomly selected and the relative
expression was calculated using the 2−∆∆Cq method
(29). The circRNAs included three
upregulated circRNAs (hsa_circRNA_103135, hsa_circRNA_103127 and
hsa_circRNA_103112) and three downregulated circRNAs
(hsa_circRNA_103137, hsa_circRNA_104907 and hsa_circRNA_101116).
The results were validated using peripheral blood samples obtained
from children with and without DS (Table
I).
As seen in Table I,
there were no significant differences in the expression levels of
hsa_circRNA_103135, hsa_circRNA_103137 and hsa_circRNA_101116
(P>0.05), but there were extremely significant differences in
the expression levels of hsa_circRNA_103127, hsa_circRNA_103112 and
hsa_circRNA_104907 (P<0.01) between six children with DS and six
children without DS.
RT-qPCR to validate the differentially
expressed genes
A total of three statistically significant genes
[dual specificity tyrosine phosphorylation regulated kinase 1A
(DYRK1A), ubiquitin specific peptidase 25 (USP25) and spermatid
perinuclear RNA binding protein (STRBP)], corresponding to three
circRNAs (hsa_circRNA_103127, hsa_circRNA_103112 and
hsa_circRNA_104907) were validated in peripheral blood samples
obtained from children with and without DS. The results are shown
in Table II.
As seen in Table II,
there was no significant difference in the expression levels of
DYRK1A and STRBP (P>0.05), but there was extremely significant
difference in the expression level of USP25 (P<0.01) between six
children with DS and six children without DS.
GO analysis of the differentially
expressed genes
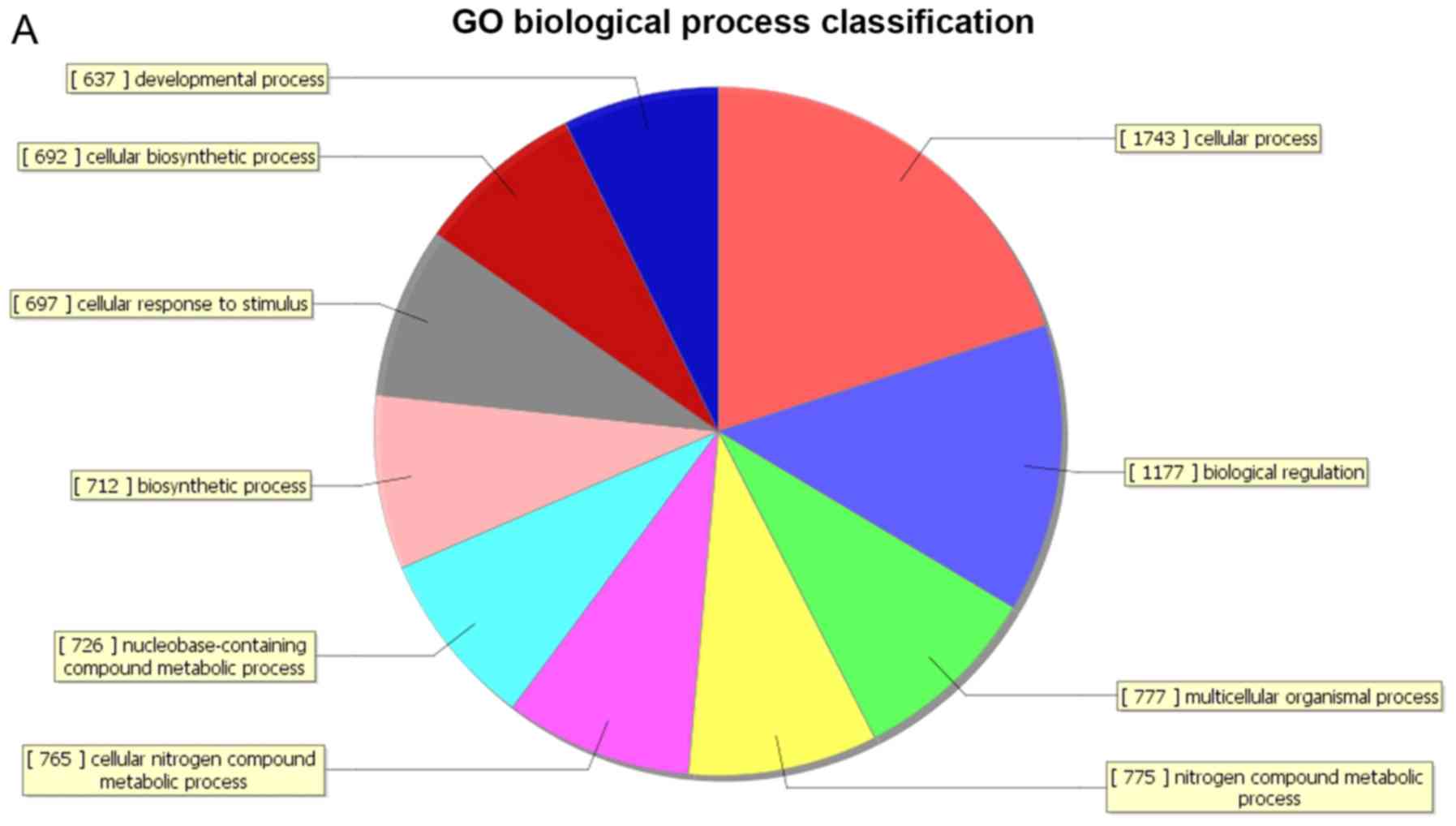
GO covers three domains: Biological process,
cellular component and molecular function. GO analysis of the
differentially expressed genes revealed that 805 genes were
associated with the biological process domain, of which 315 were
upregulated (Fig. 2A) and 490 were
down-regulated (Fig. 2B). The five
most enriched biological process terms were ‘cellular processes’,
‘primary metabolic processes’, ‘cellular metabolic processes’,
‘biological regulation’ and ‘nitrogen compound metabolic
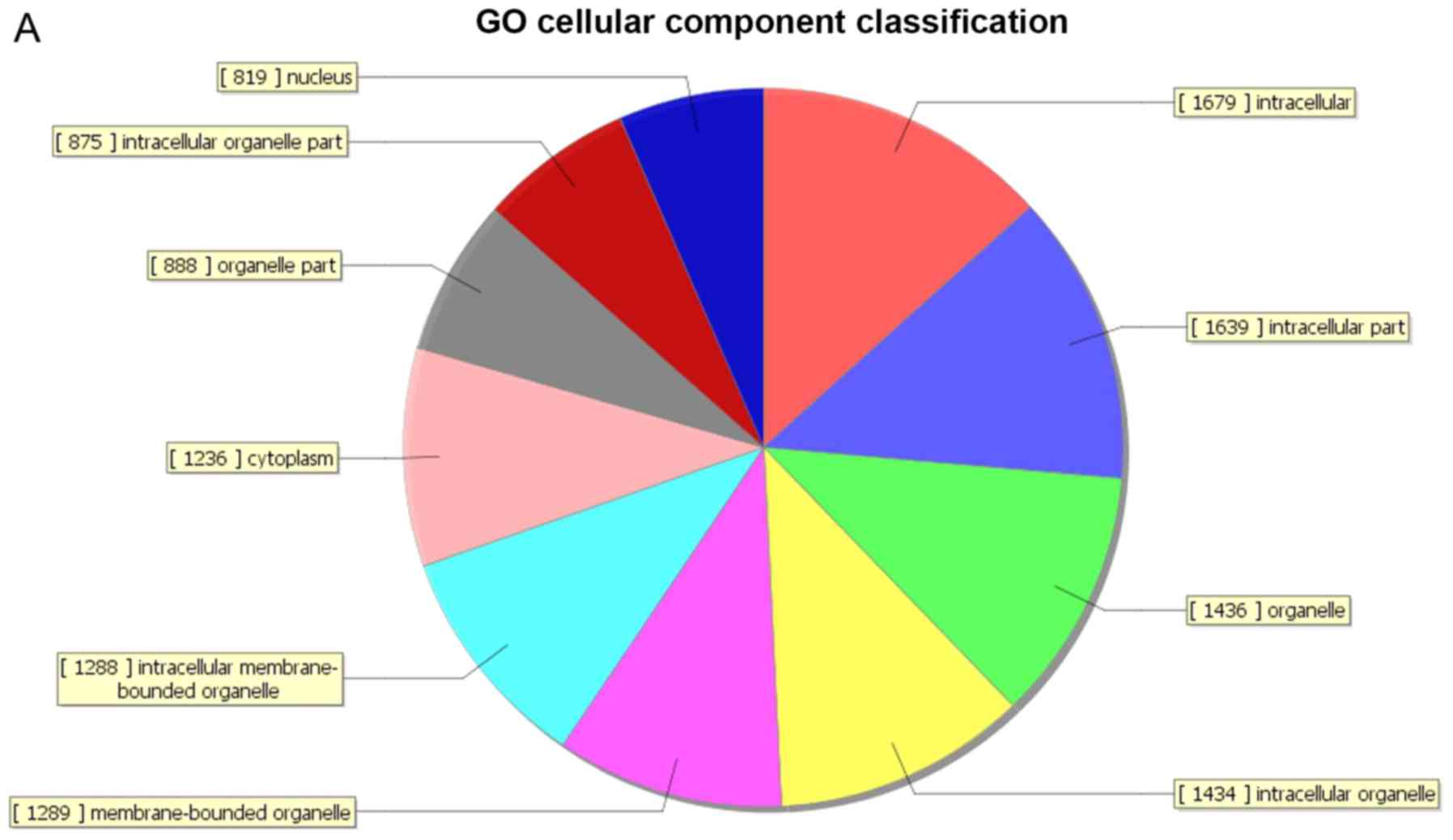
processes’. A total of 189 genes were associated with the cell
composition domain, of which 66 were upregulated (Fig. 3A) and 123 genes were downregulated
(Fig. 3B). The five most enriched
cell composition terms were ‘intracellular’, ‘intracellular part’,
‘organelle’, ‘intracellular organelle’ and ‘membrane-bound
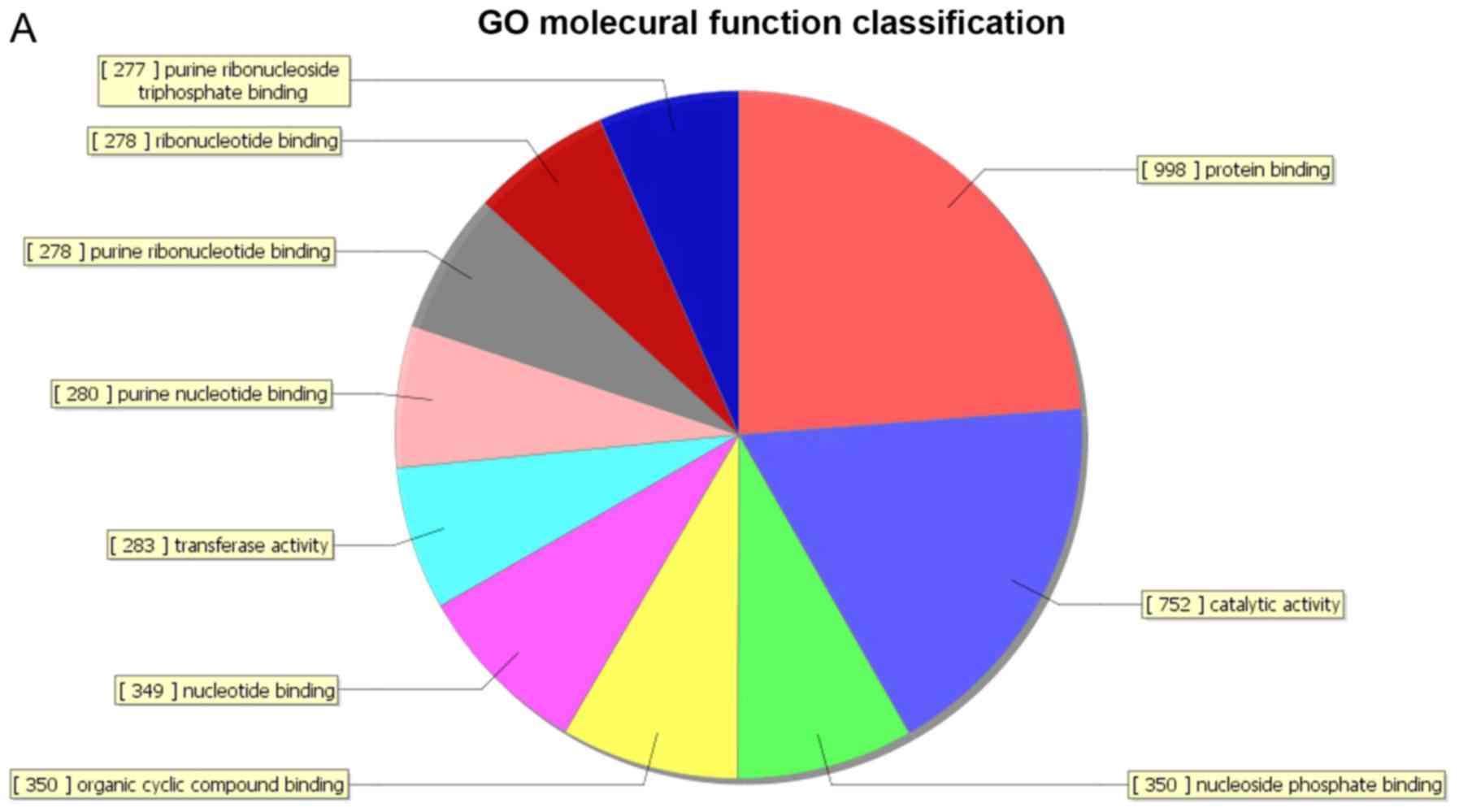
organelle’. A total of 155 genes were associated with the molecular
function domain, of which 77 were upregulated (Fig. 4A) and 78 were down-regulated
(Fig. 4B). The five most enriched
molecular function terms were ‘binding’, ‘protein binding’, ‘RNA
binding’, ‘ligase activity’ and ‘catalytic activity’.
Pathway analysis of the differentially
expressed genes
Pathway analysis of the differentially expressed
genes allows the identification of genes related to specific cell
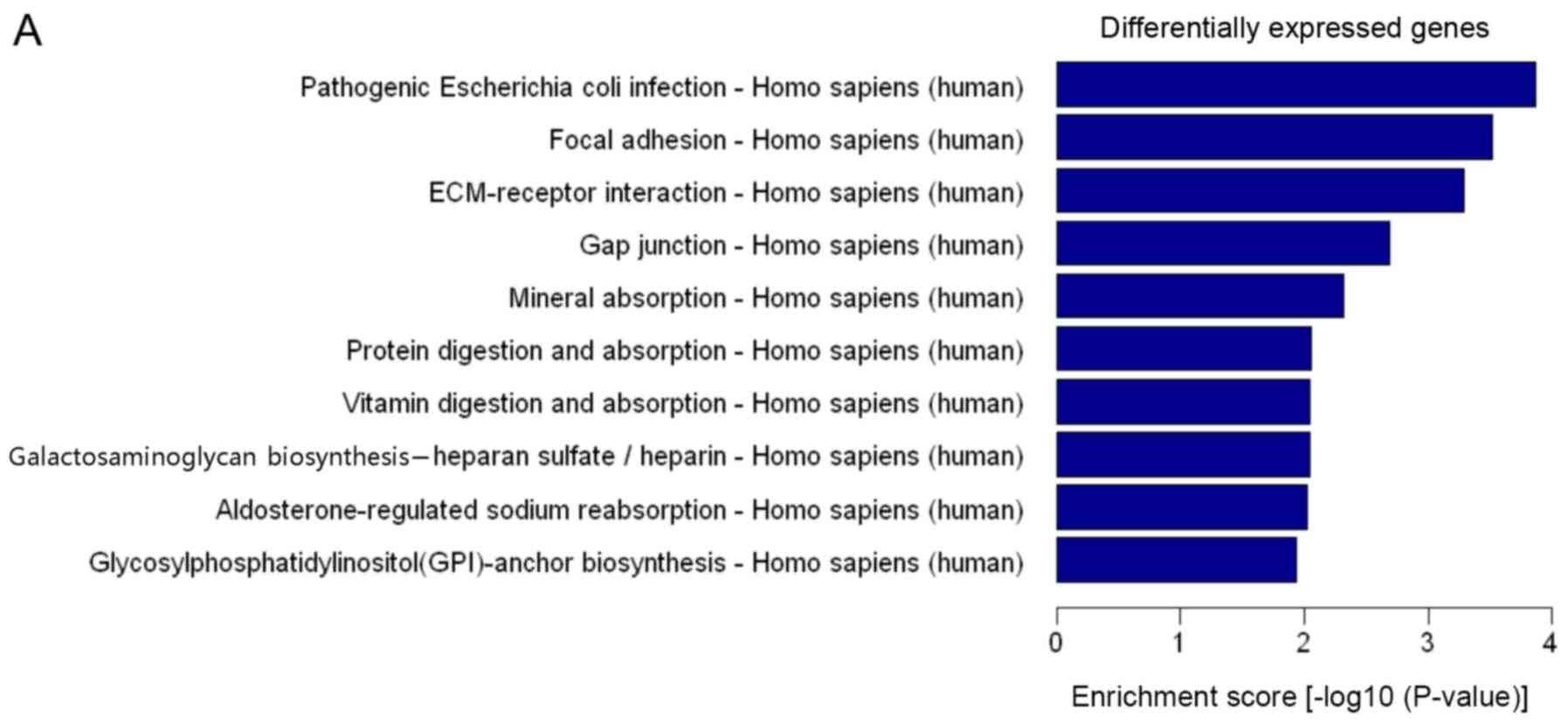
pathways. Pathway analysis revealed that the differentially
expressed genes were significantly enriched in 73 pathways
(Fig. 5A and B). The upregulated
genes were involved in 23 pathways and the downregulated genes were
involved in 50 pathways. Differentially expressed genes were mainly
involved in ‘adhesive spots’, ‘ECM-receptor interactions’, ‘gap
junctions’, ‘protein digestion and absorption’, ‘vitamin digestion
and absorption’ and ‘TNF signalling pathway’. Additional pathways
included ‘primary immunodeficiency’, ‘systemic lupus
erythematosus’, ‘rheumatoid arthritis’, ‘type I diabetes mellitus’,
‘arrhythmogenic right ventricular cardiomyopathy (ARVC)’ and
‘homologous recombination.
Interaction between differential
expression of circRNAs and miRNAs
Endogenous circRNAs function as miRNA sponges, which
means that the circRNAs bind miRNAs and repress their function
(13,18,25).
A total of 3,263 MREs of differentially expressed
circRNAs were described bioinformatically using the Arraystar miRNA
target prediction software. Analysis of the MRE sequence showed
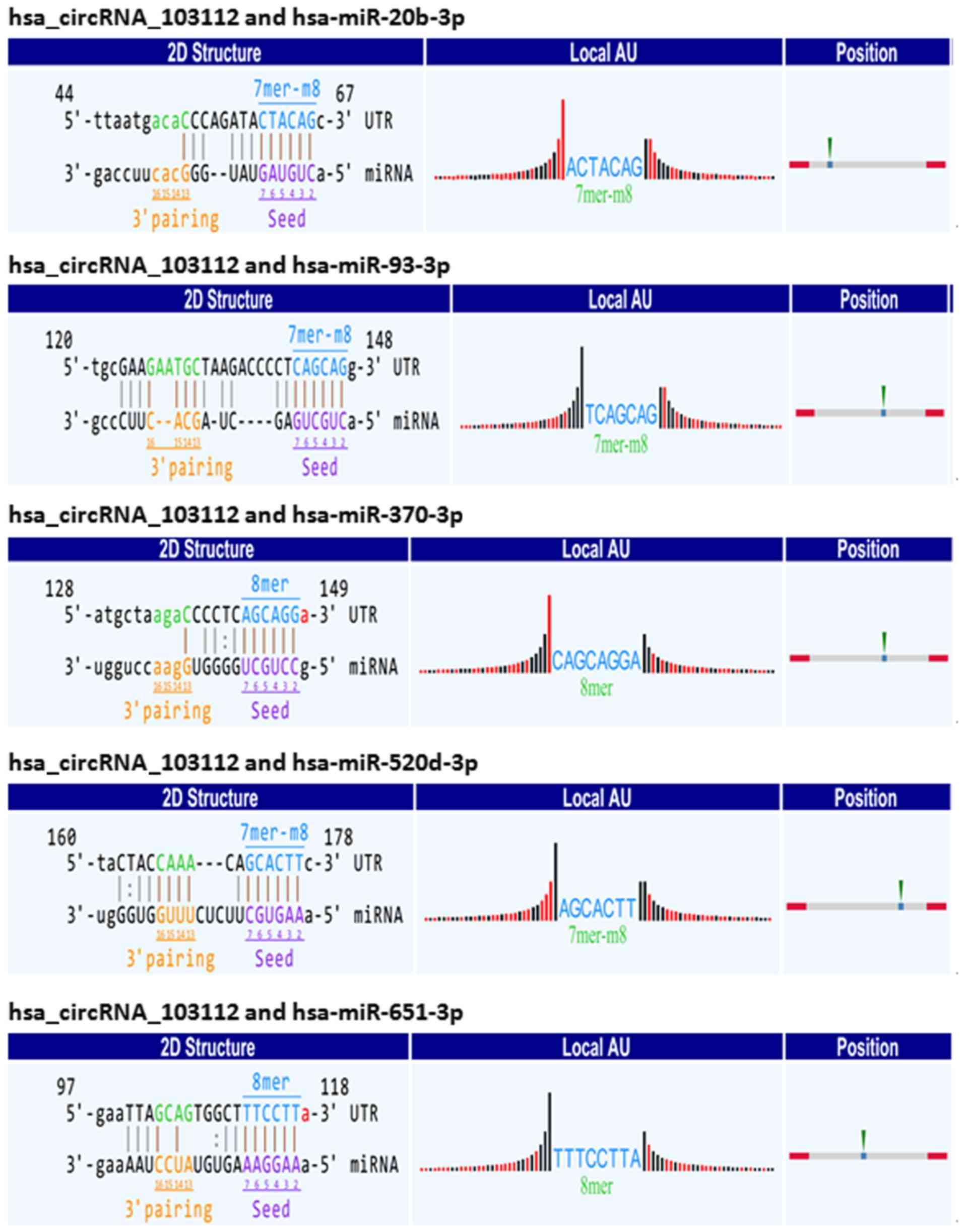
several binding sites between hsa_circRNA_103112 and various
miRNAs. There were five binding sites between hsa_circRNA_103112
and hsa-miR-20b-3p, hsa-miR-93-3p, hsa-miR-370-3p, hsa-miR-520d-3p
or hsa-miR-651-3p. The sequence of hsa_circRNA_103112 was
complementary to: i) The miR-20b-3p seed region at the 5′ end at
position 61–66 with 7mer-m8 binding; ii) the miR-93-3p seed region
at the 5′ end of the nucleotide at position 142–147 with 7mer-m8
binding; iii) the miR-370-3p seed region at the 5′ end at position
143–148 with 8mer binding; iv) the miR-520d-3p seed region at the
5′ end at position 172–177 with 7mer-m8 binding; and v) the
miR-651-3p seed region at the 5′end at position 112–117 with 8mer
binding. This indicated that hsa_circRNA_103112 could function as a
sponge for miR-20b-3p, miR-93-3p, miR-370-3p, miR-520d-3p and
miR-651-3p (Fig. 6).
The MRE sequence and the target miRNA seed type
(7mer-m8, 8mer) and 3′pairing sequence (nucleotides 13–16) are
presented in Fig. 6. The precise
base positions are shown in the alignments in the upper left and
right corners. The seed type 7mer-m8 shows an exact match to
positions 2–8 of the target miRNA (the seed + position 8), and 8mer
shows an exact match to positions 2–8 of the target miRNA (the seed
+ position 8). Adenosine/uracil (A/U) content 30 nt upstream and
downstream of the seed sequence is shown in the column Local
AU. Black bars represent guanine/cytosine and low accessibility
whereas red bars represent A/U and high accessibility of the seed.
The height of the bars shows the extent of the accessibility. The
position column presents the most likely relative MRE position on
the linear presentation of the circRNA.
Interaction between differentially
expressed circRNAs and mRNAs
circRNAs use their MRE to bind to miRNAs and
consequently repress their function. Therefore, miRNAs can identify
the mRNA of a particular coding sequence by base complementary
pairing, affecting the translation process and thus playing a role
in regulating gene expression (32).
The present study investigated the expression profiles of
differentially expressed circRNA and mRNA and conducted an
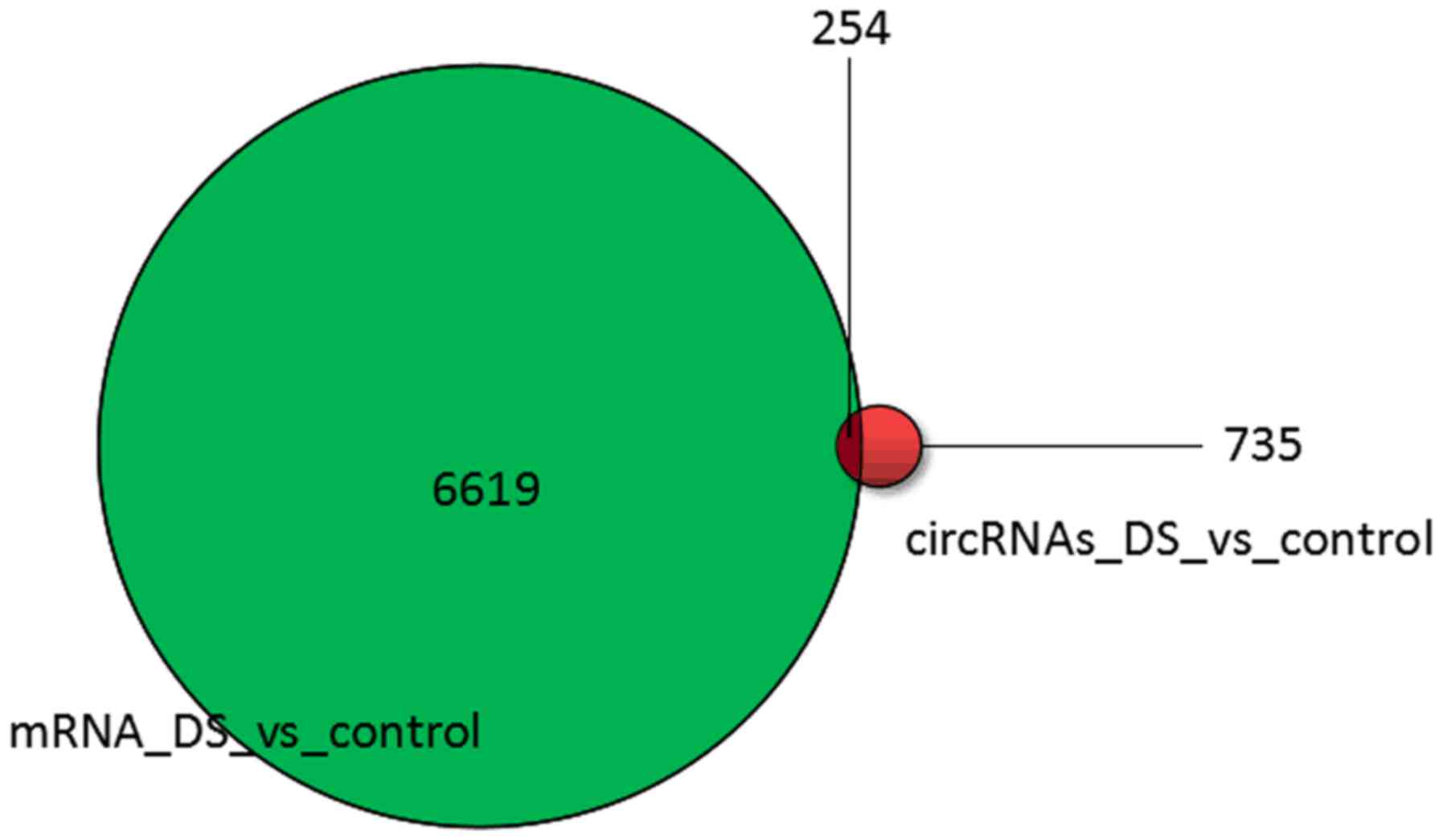
association analysis between circRNAs and mRNAs. A total of 254
circRNA transcripts were found in the corresponding differential
mRNA expression profile information. Among the 254 circRNA
associated with mRNA, there were 153 upregulated circRNAs and 101
downregulated circRNAs, indicating that these identical genes may
regulate the occurrence and development of Down's syndrome through
circRNA/miRNA/mRNA interactions. The results are presented in
Fig. 7.
Discussion
At present, prenatal screening mainly in the early
pregnancy and in the second trimester, ultrasound screening,
maternal serum screening, and joint screening programs are used to
assess the risk of fetal DS (33,34).
However, the false negative and false positive rates of traditional
prenatal screening are relatively high (35), and invasive prenatal diagnosis
methods such as amniocentesis, chorionic sampling and umbilical
cord blood collection can easily lead to bleeding, infection,
miscarriage and other complications (36). Therefore, it is necessary to identify
novel biological markers in maternal blood to further improve the
prenatal screening for DS during pregnancy.
Wu et al (37)
reported that circRNAs may be used as biomarkers for the diagnosis
of disease and evaluation of therapeutic efficacy. In addition, due
to their high stability, circRNAs are easily obtained from body
fluids (10,18). Therefore, there has been increased
interest in the in-depth and systematic research on circRNA. In
view of the aforementioned study, under physiological conditions,
the expression of circRNA in serum and plasma is relatively stable,
while under pathological conditions; specific circRNA expression is
likely to be abnormal.
The present study screened for differentially
expressed circRNAs in umbilical cord blood obtained from pregnant
women carrying fetuses with and without DS using circular gene chip
technology. As a result, 735 differentially expressed circRNAs were
detected, of which 414 were upregulated and 321 were downregulated.
RT-qPCR was subsequently used to validate the differential
expression of circRNA in peripheral blood samples obtained from
children with and without DS. A total of six differentially
expressed circRNAs, including three upregulated circRNAs
(hsa_circRNA_103135, hsa_circRNA_103127 and hsa_circRNA_103112) and
three downregulated circRNAs (hsa_circRNA_103137,
hsa_circRNA_104907 and hsa_circRNA_101116) were selected for
further analysis. Quantification using the 2−∆∆Cq method
(29) revealed that the six circRNAs
were differentially expressed between children with and without DS.
There were extremely significant differences in hsa_circRNA_103127,
hsa_circRNA_103112 and hsa_circRNA_104907 expression, but no
significant differences in the expression of hsa_circRNA_103135,
hsa_circRNA_103137 and hsa_circRNA_101116. These differences may be
related to the temporal and tissue specificity of circRNA
expression. Furthermore, analysis using Arraystar's miRNA target
prediction software revealed that hsa_circRNA_103112 was associated
with hsa-miR-20b-3p, hsa-miR-93-3p, hsa-miR-370-3p, hsa-miR-520d-3p
and hsa-miR-651-3p.
Stamova et al (38) revealed that decreased miR-93-3p
expression in the superior temporal sulcus (a region relevant to
social interaction) and primary auditory cortex, and may have a
potential impact on autism spectrum disorder (ASD). ASD is a
neurodevelopmental disorder disease, the core symptoms of which
include deficits in social communication and language, as well as
ritual stereotyped behaviour, learning disability, immune system
disturbances and sensory system alterations (39). Furthermore, Yanni et al
(40) revealed that increased
expression of endogenous miR-370-3p contributes to bradycardia
associated with heart failure, suggesting that miR-370-3p may serve
as a therapeutic target. The aforementioned phenotypic traits are
often associated with DS (41,42).
An Agilent expression profile chip was used to
obtain the differentially expressed genes between pregnant women
carrying fetuses with or without DS in the present study. A total
of 6,619 differentially expressed genes were identified, of which
3,411 were upregulated and 3,208 were downregulated. Moreover,
three genes (DYRK1A, USP25 and STRBP) corresponding to three
significantly differentially expressed circRNAs
(hsa_circRNA_103127, hsa_circRNA_103112 and hsa_circRNA_104907)
were verified in peripheral blood samples obtained from children
with and without DS. There was no significant difference in the
expression level of DYRK1A and STRBP (P>0.05). However, an
extremely significant difference in USP25 expression was observed
(P<0.01).
Valero et al (43) revealed that USP25 is a specific
ubiquitin protease gene located in the 21q11.2 region, and that an
increase in USP25 gene dosage in patients with DS disturbed the
balance between ubiquitinated and deubiquitinated substrates. USP25
is a newly described member of the ubiquitin-specific processing
proteases (UBPs) that encode the ubiquitin C-terminal hydrolases.
Ubiquitin is a highly conserved eukaryotic protein (76 aa) and
plays an important role in intracellular protein degradation. USP25
is associated with testis development and could be involved in the
defective spermiogenesis found in males with DS (43). Increasing numbers of studies have
found that disruption of ubiquitin systems can lead to a number of
human diseases, including neurodegenerative disorders and several
carcinomas (44–46). Valero et al (44) evaluated the expression level of USP25
in human fetal brains of DS and control disomic samples by RT-qPCR
analysis and found an average 1.7-fold increase in USP25 expression
in DS samples. Upregulation of USP25 in DS fetal brain indicates
that it participates in the pathogenesis of DS and has the same
gene dose effect as other UBP members associated with aneuploidy
syndromes, such as ubiquitin specific peptidase 9 X-linked in
Turner syndrome (47) and USP18 in
DiGeorge syndrome (48).
The present study revealed that circRNA had an
adsorption effect on miRNA, and the transcripts of differentially
expressed circRNA were found in the corresponding differentially
expressed gene expression profiles. A previous study demonstrated
that miRNAs identify the mRNA of a particular coding sequence by
complementary base pairing, affecting the translation process
(49). Therefore, there is an
interaction between circRNA/miRNA/mRNA, which may impact the
occurrence and development of DS.
The present study, to best of our knowledge, was the
first to reveal that circRNAs are differentially expressed between
DS and control samples. The identified differentially expressed
genes are mainly involved in cell division and development process
and are also involved in primary immunodeficiency, rheumatoid
arthritis and type I diabetes mellitus, as well as homologous
recombination processes, and may result in abnormal copies of
chromosome 21. This may disrupt normal development, causing
patients with DS to experience developmental defects of the immune
system and the emergence of various clinical symptoms (5). Most importantly, hsa_circRNA_103112 not
only showed significant expression differences in cord blood, but
also showed significant differences in the peripheral blood.
Therefore, upregulatation of hsa_circRNA_103112 and its
corresponding gene, USP25, in patients with DS patients may result
in an expression imbalance of diploid genes through
circRNA-miRNA-mRNA interactions and participates in the
pathogenesis of DS. In addition, the present study revealed that
the differential expression of hsa_circRNA_103112 in peripheral
blood may serve as a novel diagnostic marker for the non-invasive
prenatal screening for DS.
The present study had a number of limitations.
Firstly, the study was limited by the small number of samples,
which was not large enough to establish definitive conclusions.
Furthermore, the samples were collected from Shenzhen People's
Hospital and Guilin No. 924 Hospital, which may have resulted in
regional differences. Secondly, the results may not be applicable
to the general population. Thirdly, the study of circRNAs in DS is
still in its initial stages, and the functional analysis requires
further confirmation. Therefore, further circRNA expression studies
with larger sample sizes, miRNA microarray analysis, RT-qPCR
validation and circRNA knock-out or overexpression studies are
required to further evaluate the potential of circRNAs in DS.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Guangxi Science and Technology Program (grant no. 1598012-25),
Science and Technology Planning Project of Guangdong Province
(grant no. 2017B020209001), Natural Science Foundation of Guangdong
Province (grant no. 2017A030310629), Science and Technology Plan of
Shenzhen (grant no. JCYJ20170307095606266) and Natural Science
Foundation of Guangdong Province (grant no. 2016A020215027).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WS, DT and YD designed the study, analyzed the data
and drafted the manuscript. YC, MO, JC, HL, WX, YW and HH acquired
patient data and performed laboratory experiments. QG contributed
to the study design and writing of the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shenzhen People's Hospital (reference number,
LL-KT-201801184) and the Ethics Committee of Guilin No. 924
Hospital. Written informed consent was obtained from all pregnant
women and parents/guardians of the children.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kleschevnikov AM, Belichenko PV, Villar
AJ, Epstein CJ, Malenka RC and Mobley WC: Hippocampal long-term
potentiation suppressed by increased inhibition in the Ts65Dn
mouse, a genetic model of down syndrome. J Neurosci. 24:8153–8160.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roizen NJ and Patterson D: Down's
syndrome. Lancet. 361:1281–1289. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caughey AB, Washington AE and Kuppermann
M: Perceived risk of prenatal diagnostic procedure-related
miscarriage and down syndrome among pregnant women. Am J Obstet
Gynecol. 198:333 e331–338. 2008. View Article : Google Scholar
|
|
4
|
Antonarakis SE, Lyle R, Dermitzakis ET,
Reymond A and Deutsch S: Chromosome 21 and down syndrome: From
genomics to pathophysiology. Nat Rev Genet. 5:725–738. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thase ME: Longevity and mortality in downs
syndrome. J Ment Defic Res. 26:177–192. 1982.PubMed/NCBI
|
|
6
|
Cortés-López M, Gruner MR, Cooper DA,
Gruner HN, Voda AI, van der Linden AM and Miura P: Global
accumulation of circRNAs during aging in Caenorhabditis elegans.
BMC Genomics. 19:82018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salomon LJ, Alfirevic Z, Audibert F, Kagan
KO, Paladini D, Yeo G and Raine-Fenning N; ISUOG Clinical Standards
Committee, : ISUOG updated consensus statement on the impact of
cfDNA aneuploidy testing on screening policies and prenatal
ultrasound practice. Ultrasound Obstet Gynecol. 49:815–816. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Houseley JM, Garcia-Casado Z, Pascual M,
Paricio N, O'Dell KM, Monckton DG and Artero RD: Noncanonical RNAs
from transcripts of the Drosophila muscleblind gene. J Hered.
97:253–260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valdmanis PN and Kay MA: The expanding
repertoire of circular RNAs. Mol Ther. 21:1112–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaikkonen MU, Lam MT and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chu T, Mouillet JF, Hood BL, Conrads TP
and Sadovsky Y: The assembly of miRNA-mRNA-protein regulatory
networks using high-throughput expression data. Bioinformatics.
31:1780–1787. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perriman R and Ares M Jr: Circular mRNA
can direct translation of extremely long repeating-sequence
proteins in vivo. RNA. 4:1047–1054. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bohjanen PR, Colvin RA, Puttaraju M, Been
MD and Garcia-Blanco MA: A small circular TAR RNA decoy
specifically inhibits Tat-activated HIV-1 transcription. Nucleic
Acids Res. 24:3733–3738. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lawrie CH: MicroRNAs and haematology:
Small molecules, big function. Br J Haematol. 137:503–512. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan-Liang D and Xiao-Nan Y: Combined serum
and ultrasound screening for early diagnosis of Down's syndrome.
Chin J Prac Gynecol Obstetrics. 26:895–898. 2010.
|
|
34
|
Benn PA, Kaminsky LM, Ying J, Borgida AF
and Egan JF: Combined second trimester biochemical and ultrasound
screening for Down syndrome. Obstet Gynecol. 100:1168–1176. 2001.
View Article : Google Scholar
|
|
35
|
Langlois S and Brock JA; Genetics
Committee, : Current status in non-invasive prenatal detection of
down syndrome, trisomy 18, and trisomy 13 using cell-free DNA in
maternal plasma. J Obstet Gynaecol Can. 35:177–181. 2013.(In
English, French). View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kolker A and Burke BM: Grieving the wanted
child: Ramifications of abortion after prenatal diagnosis of
abnormality. Health Care for Women Int. 14:513–526. 1993.
View Article : Google Scholar
|
|
37
|
Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan
J, Li WX and Ding SW: Homology-independent discovery of replicating
pathogenic circular RNAs by deep sequencing and a new computational
algorithm. Proc Natl Acad Sci USA. 109:3938–3943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stamova B, Ander BP, Barger N, Sharp FR
and Schumann CM: Specific regional and age-related small Noncoding
RNA expression patterns within superior temporal gyrus of typical
human brains are less distinct in autism brains. J Child Neurol.
30:1930–1946. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lo YC, Chen YJ, Hsu YC, Tseng WI and Gau
SS: Reduced tract integrity of the model for social communication
is a neural substrate of social communication deficits in autism
spectrum disorder. J Child Psychol Psychiatry. 58:576–585. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yanni J, Zi M, Choudhury M, Cai X,
Logantha S, Li J, Cartwright E, Dobrzynski H, Hart G and Boyett MR:
MicroRNA 370-3p could explain the dysfunction of the cardiac
conduction system in heart failure. Proc Physiol Soc.
34:PC1582015.
|
|
41
|
Warner G, Howlin P, Salomone E, Moss J and
Charman T: Profiles of children with Down syndrome who meet
screening criteria for autism spectrum disorder (ASD): A comparison
with children diagnosed with ASD attending specialist schools. J
Intellect Disabil Res Jidr. 61:75–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Levine OR and Simpser M: Alveolar
hypoventilation and cor pulmonale associated with chronic airway
obstruction in infants with down syndrome. Clin Pediatr (Phila).
21:25–29. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Valero R, Marfany G, González-Angulo O,
González-González G, Puelles L and Gonzàlez-Duarte R: USP25, a
novel gene encoding a deubiquitinating enzyme, is located in the
gene-poor region 21q11.2. Genomics. 62:395–405. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Valero R, Bayés M, Francisca Sánchez-Font
M, González-Angulo O, Gonzàlez-Duarte R and Marfany G:
Characterization of alternatively spliced products and
tissue-specific isoforms of USP28 and USP25. Genome Biol.
2:RESEARCH00432001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ciechanover A and Brundin P: The ubiquitin
proteasome system in neurodegenerative diseases: Sometimes the
chicken, sometimes the egg. Neuron. 40:427–446. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hershko D, Bornstein G, Ben-Izhak O,
Carrano A, Pagano M, Krausz MM and Hershko A: Inverse relation
between levels of p27Kip1 and of its ubiquitin ligase subunit Skp2
in colorectal carcinomas. Cancer. 91:1745–1751. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jones MH, Furlong RA, Burkin H, Chalmers
IJ, Brown GM, Khwaja O and Affara N: The drosophila developmental
gene fat facets has a human homologue in Xp11.4 which escapes
X-inactivation and has related sequences on Yq11.2. Hum Mol Genet.
5:1695–1701. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schwer H, Liu LQ, Zhou L, Little MT, Pan
Z, Hetherington CJ and Zhang DE: Cloning and characterization of a
novel human ubiquitin-specific protease, a homologue of murine
UBP43 (USP18). Genomics. 65:44–52. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dalmay T: Mechanism of miRNA-mediated
repression of mRNA translation. Essays Biochem. 54:29–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|