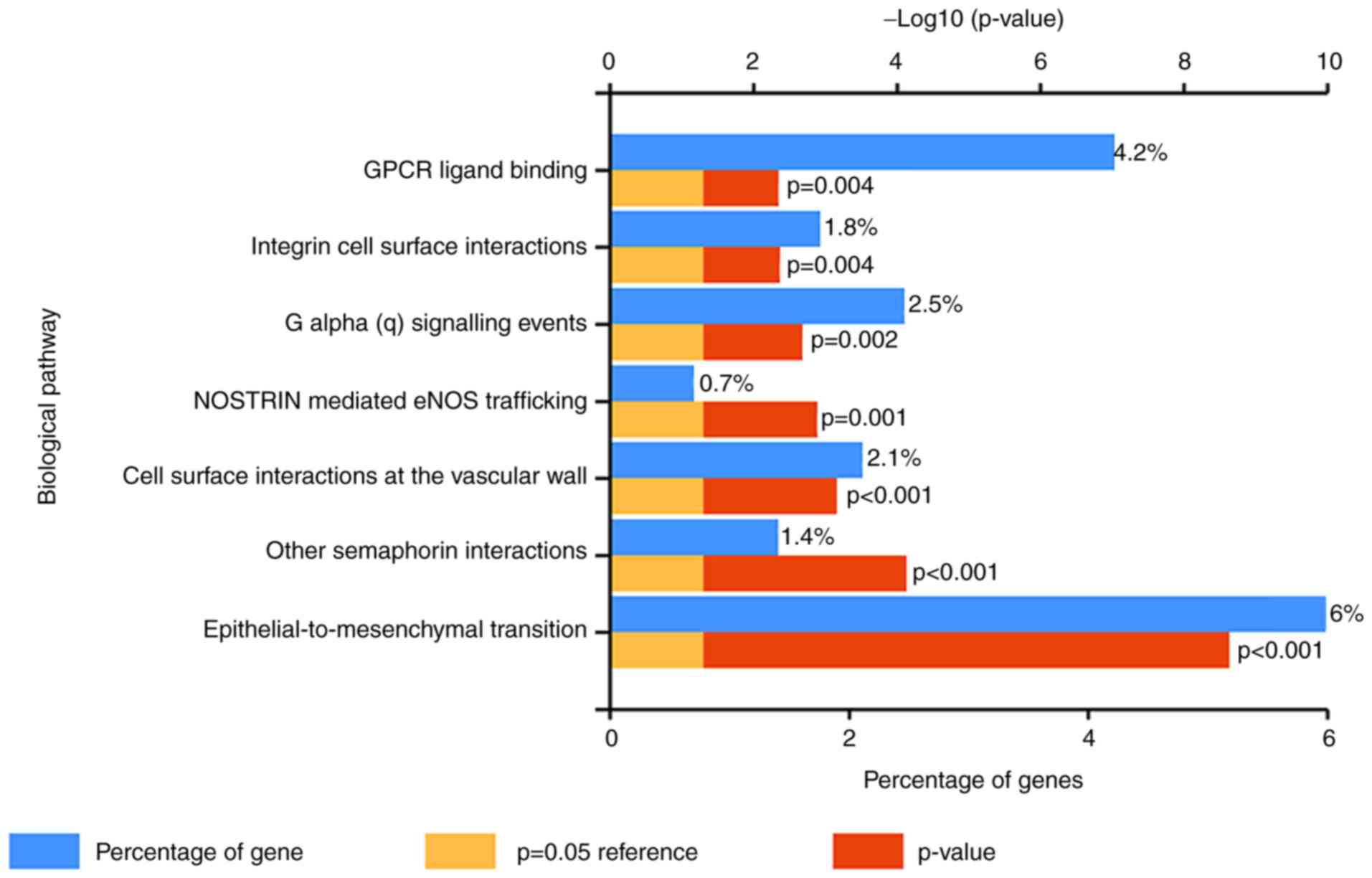
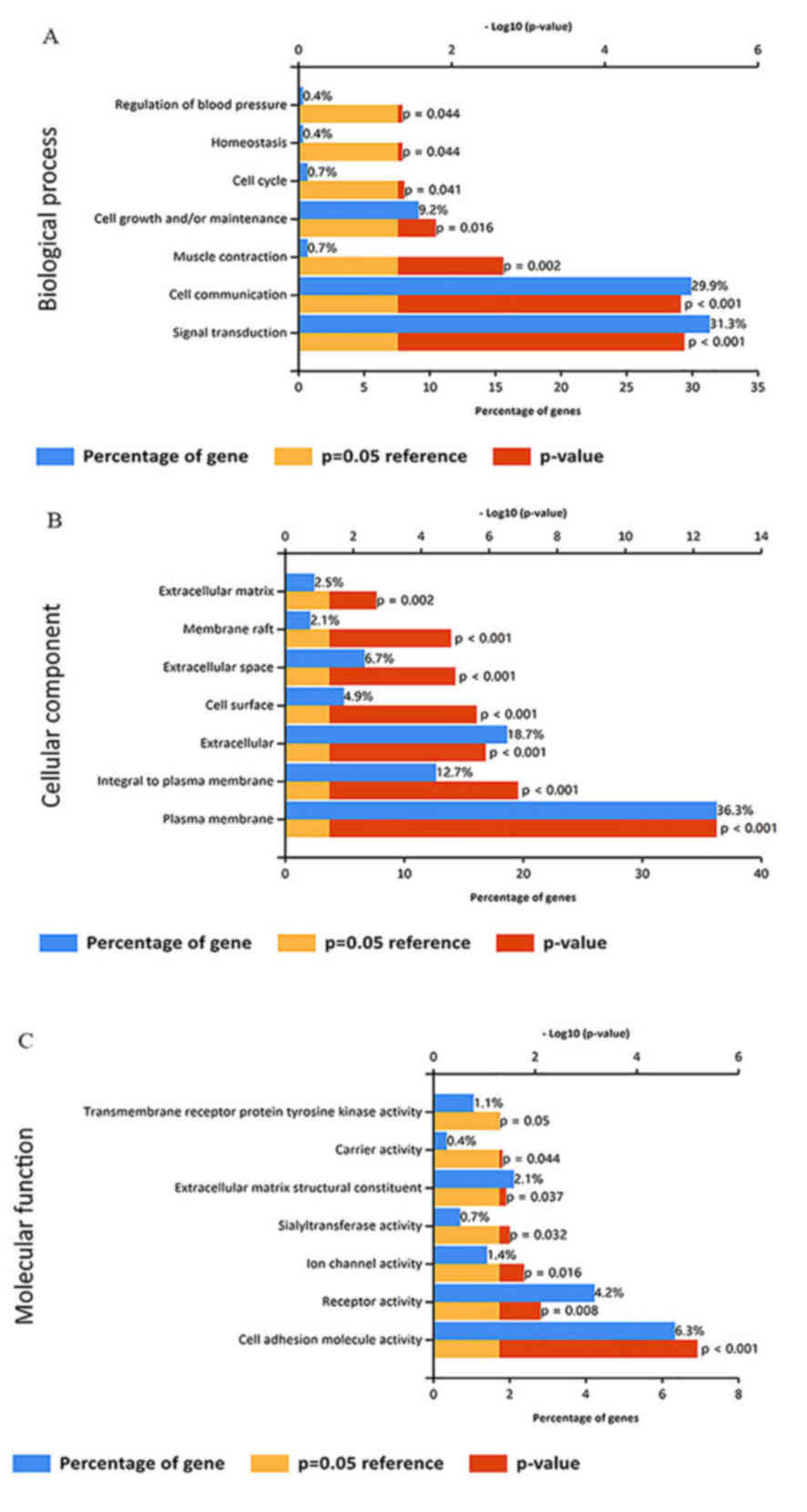
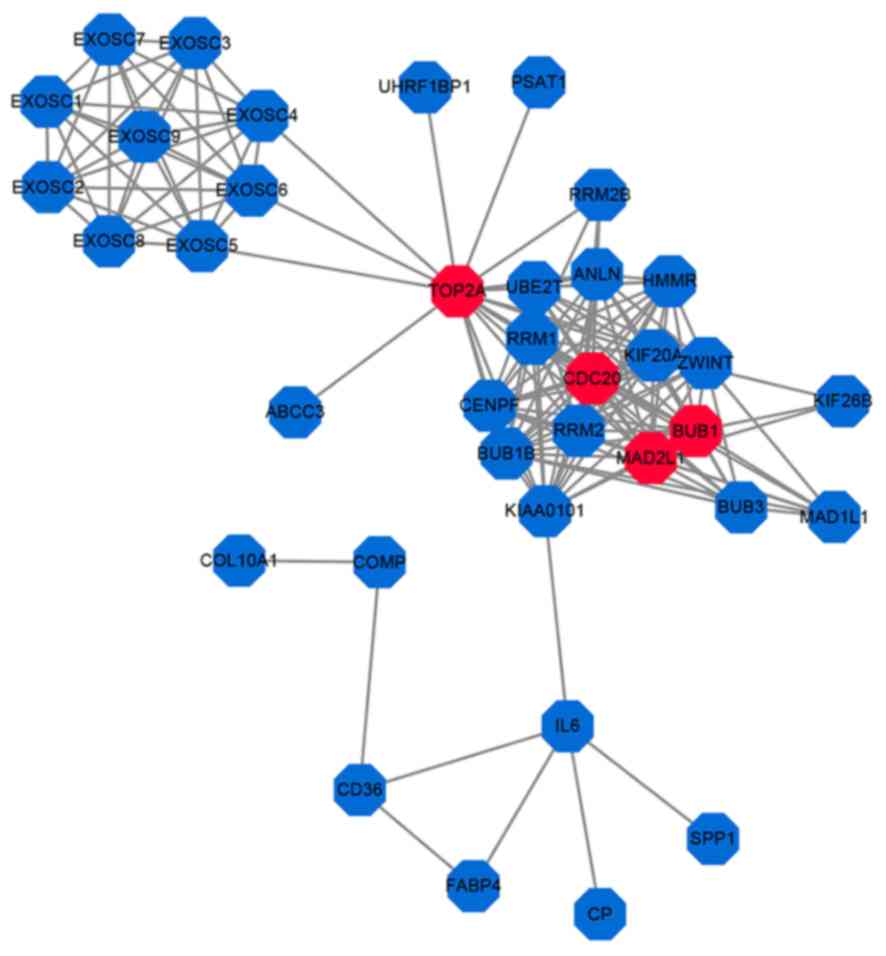
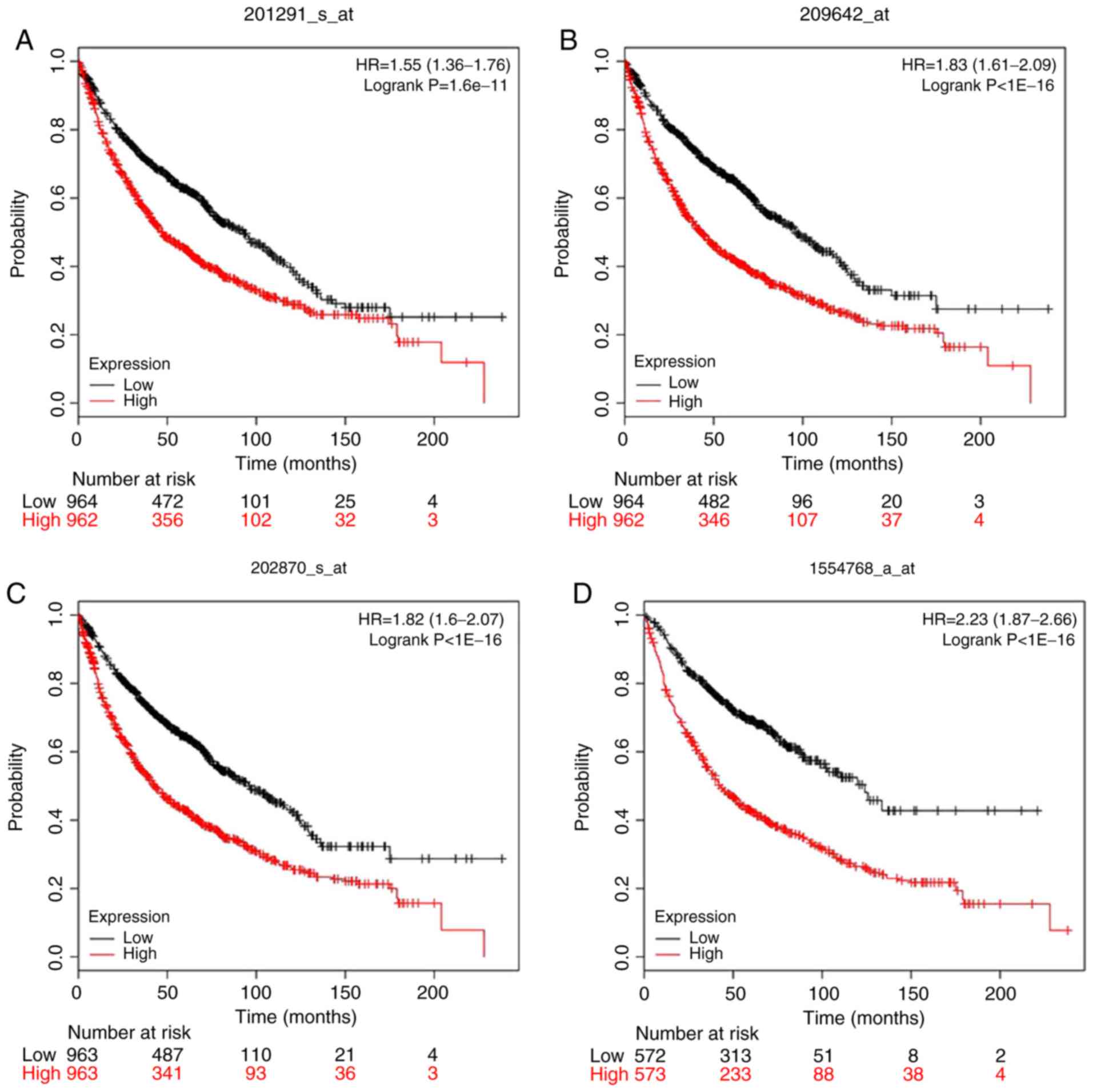
|
1
|
Centers for Disease Control and
Prevention. Lung Cancer Statistics. simplecdc.gov/cancer/lung/statistics/index.htmMarch
26–2014
|
|
2
|
Jakobsen E, Rasmussen TR and Green A:
Mortality and survival of lung cancer in Denmark: Results from the
Danish lung cancer group 2000–2012. Acta Oncol. 55 (Suppl 2):S2–S9.
2016. View Article : Google Scholar
|
|
3
|
Richards TB, Henley SJ, Puckett MC, Weir
HK, Huang B, Tucker TC and Allemani C: Lung cancer survival in the
United States by race and stage (2001–2009): Findings from the
CONCORD-2 study. Cancer. 123 (Suppl 24):S5079–S5099. 2017.
View Article : Google Scholar
|
|
4
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang Y, Ma YY, Li LL, Shen XY, Xin T,
Zhao YW and Ma R: Effect of long non-coding RNA LINC01116 on
biological behaviors of non-small cell lung cancer cells via the
hippo signaling pathway. J Cell Biochem. 1192018.DOI:
10.1002/jcb.26711.
|
|
8
|
Barrett T and Edgar R: Mining microarray
data at NCBI's Gene Expression Omnibus (GEO)*. Methods Mol Biol.
338:175–190. 2006.PubMed/NCBI
|
|
9
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC,
Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al: Protein arginine
methyltransferase 5 is a potential oncoprotein that upregulates G1
cyclins/cyclin-dependent kinases and the phosphoinositide
3-kinase/AKT signaling cascade. Cancer Sci. 103:1640–1650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei TY, Hsia JY, Chiu SC, Su LJ, Juan CC,
Lee YC, Chen JM, Chou HY, Huang JY, Huang HM and Yu CT: Methylosome
protein 50 promotes androgen- and estrogen-independent
tumorigenesis. Cell Signal. 26:2940–2950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beer DG, Kardia SL, Huang CC, Giordano TJ,
Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al:
Gene-expression profiles predict survival of patients with lung
adenocarcinoma. Nat Med. 8:816–824. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
17
|
Xing Z, Chu C, Chen L and Kong X: The use
of Gene Ontology terms and KEGG pathways for analysis and
prediction of oncogenes. Biochim Biophys Acta. 1860:2725–2734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu KZ, Wang GN, Fitzgerald J, Quachthithu
H, Rainey MD, Cattaneo A, Bachi A and Santocanale C: DDK dependent
regulation of TOP2A at centromeres revealed by a chemical genetics
approach. Nucleic Acids Res. 44:8786–8798. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klintman M, Buus R, Cheang MC, Sheri A,
Smith IE and Dowsett M: Changes in expression of genes representing
key biologic processes after neoadjuvant chemotherapy in breast
cancer, and prognostic implications in residual disease. Clin
Cancer Res. 22:2405–2416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ocaña A, Pérez-Peña J, Alcaraz-Sanabria A,
Sánchez-Corrales V, Nieto-Jiménez C, Templeton AJ, Seruga B,
Pandiella A and Amir E: In silico analyses identify gene-sets,
associated with clinical outcome in ovarian cancer: Role of mitotic
kinases. Oncotarget. 7:22865–22872. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen T, Sun Y, Ji P, Kopetz S and Zhang W:
Topoisomerase IIα in chromosome instability and personalized cancer
therapy. Oncogene. 34:4019–4031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Delgado JL, Hsieh CM, Chan NL and Hiasa H:
Topoisomerases as anticancer targets. Biochem J. 475:373–398. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An X, Xu F, Luo R, Zheng Q, Lu J, Yang Y,
Qin T, Yuan Z, Shi Y, Jiang W and Wang S: The prognostic
significance of topoisomerase II alpha protein in early stage
luminal breast cancer. BMC Cancer. 18:3312018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pei YF, Yin XM and Liu XQ: TOP2A induces
malignant character of pancreatic cancer through activating
β-catenin signaling pathway. Biochim Biophys Acta. 1864:197–207.
2018. View Article : Google Scholar
|
|
25
|
Lara-Gonzalez P, Westhorpe FG and Taylor
SS: The spindle assembly checkpoint. Curr Biol. 22:R966–R980. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raaijmakers JA, van Heesbeen RGHP, Blomen
VA, Janssen LME, van Diemen F, Brummelkamp TR and Medema RH: BUB1
is essential for the viability of human cells in which the spindle
assembly checkpoint is compromised. Cell Rep. 22:1424–1438. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ricke RM, Jeganathan KB, Malureanu L,
Harrison AM and van Deursen JM: Bub1 kinase activity drives error
correction and mitotic checkpoint control but not tumor
suppression. J Cell Biol. 199:931–949. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mukherjee A, Joseph C, Craze M,
Chrysanthou E and Ellis IO: The role of BUB and CDC proteins in
low-grade breast cancers. Lancet. 385 (Suppl 1):S722015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stahl D, Braun M, Gentles AJ, Lingohr P,
Walter A, Kristiansen G and Gütgemann I: Low BUB1 expression is an
adverse prognostic marker in gastric adenocarcinoma. Oncotarget.
8:76329–76339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hartwell LH, Culotti J and Reid B: Genetic
control of the cell-division cycle in yeast. I. Detection of
mutants. Proc Natl Acad Sci USA. 66:352–359. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Quek LS, Grasset N, Jasmen JB, Robinson KS
and Bellanger S: Dual role of the anaphase promoting
Complex/Cyclosome in regulating Stemness and differentiation in
human primary keratinocytes. J Invest Dermatol. 138:1851–1861.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kapanidou M, Curtis NL and Bolanos-Garcia
VM: Cdc20: At the Crossroads between chromosome segregation and
mitotic exit. Trends Biochem Sci. 42:193–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Taniguchi K, Momiyama N, Ueda M, Matsuyama
R, Mori R, Fujii Y, Ichikawa Y, Endo I, Togo S and Shimada H:
Targeting of CDC20 via small interfering RNA causes enhancement of
the cytotoxicity of chemoradiation. Anticancer Res. 28:1559–1563.
2008.PubMed/NCBI
|
|
34
|
Jiang J, Jedinak A and Sliva D:
Ganodermanontriol (GDNT) exerts its effect on growth and
invasiveness of breast cancer cells through the down-regulation of
CDC20 and uPA. Biochem Biophys Res Commun. 415:325–329. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang DZ, Ma Y, Ji B, Liu Y, Hwu P,
Abbruzzese JL, Logsdon C and Wang H: Increased CDC20 expression is
associated with pancreatic ductal adenocarcinoma differentiation
and progression. J Hematol Oncol. 5:152012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choi JW, Kim Y, Lee JH and Kim YS: High
expression of spindle assembly checkpoint proteins CDC20 and MAD2
is associated with poor prognosis in urothelial bladder cancer.
Virchows Arch. 463:681–687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kato T, Daigo Y, Aragaki M, Ishikawa K,
Sato M and Kaji M: Overexpression of CDC20 predicts poor prognosis
in primary non-small cell lung cancer patients. J Surg Oncol.
106:423–430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karra H, Repo H, Ahonen I, Löyttyniemi E,
Pitkänen R, Lintunen M, Kuopio T, Söderström M and Kronqvist P:
Cdc20 and securin overexpression predict short-term breast cancer
survival. Br J Cancer. 110:2905–2913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Han Y, Jin X, Zhou H and Liu B:
Identification of key genes associated with bladder cancer using
gene expression profiles. Oncol Lett. 15:297–303. 2018.PubMed/NCBI
|
|
40
|
He X, Zhang C, Shi C and Lu Q:
Meta-analysis of mRNA expression profiles to identify
differentially expressed genes in lung adenocarcinoma tissue from
smokers and non-smokers. Oncol Rep. 39:929–938. 2018.PubMed/NCBI
|
|
41
|
Wang Z, Katsaros D, Shen Y, Fu Y, Canuto
EM, Benedetto C, Lu L, Chu WM, Risch HA and Yu H: Biological and
clinical significance of MAD2L1 and BUB1, genes frequently
appearing in expression signatures for breast cancer prognosis.
PLoS One. 10:e01362462015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kato T, Daigo Y, Aragaki M, Ishikawa K,
Sato M, Kondo S and Kaji M: Overexpression of MAD2 predicts
clinical outcome in primary lung cancer patients. Lung Cancer.
74:124–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Asmis TR, Ding K, Seymour L, Shepherd FA,
Leighl NB, Winton TL, Whitehead M, Spaans JN, Graham BC and Goss
GD; National Cancer Institute of Canada Clinical Trials Group, :
Age and comorbidity as independent prognostic factors in the
treatment of non small-cell lung cancer: A review of National
Cancer Institute of Canada Clinical Trials Group trials. J Clin
Oncol. 26:54–59. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zou ZQ, Du YY, Sui G and Xu SN: Expression
of TS, RRM1, ERCC1, TUBB3 and STMN1 genes in tissues of non-small
cell lung cancer and its significance in guiding postoperative
adjuvant chemotherapy. Asian Pac J Cancer Prev. 16:3189–3194. 2015.
View Article : Google Scholar : PubMed/NCBI
|