Introduction
Following the activation of CD3 and CD28, the
primary and secondary signals for T-cell differentiation,
respectively, naïve T (Th0) cells have the ability to develop into
different T helper (Th) cell subsets. The recognition of other
Th-cell subsets, including type-17 Th cells (Th17) (1–3),
T-regulatory (Treg), Th9 and Th22 cells, particularly with regard
to the plasticity of these cells, gradually shifted the research
priority from that of the ratio of Th1 and Th2 cells to that of the
mutual association between various CD4+ T-cell
subsets.
Th9 cells, a more recently described subset of
effector Th cells, have promoted the general understanding of
T-cell functioning. Interleukin (IL)-9, the primary cytokine
produced by Th9 cells, is a type-2 pleiotropic cytokine that not
only regulates autoimmune and allergic reactions, but is involved
in anti-parasitic and anti-tumor responses, and the formation of
immune tolerance (4–6). However, the differentiation,
development and immunological characteristics of Th9 cells have
remained largely elusive. Previous studies have indicated that Th2
cells were able to differentiate into Th9 cells following the
addition of transforming growth factor β (TGF-β) in the presence of
IL-4 (7), and that Th9 cell
polarization was further enhanced by IL-1, IL-2 and IL-25 (8–10).
TGF-β signaling is mediated through its binding to
type I and type II receptors, and the activated ligand-receptor
complex typically activates Smad-dependent signal transduction
(11). The canonical Smad signaling
cascade is initiated by the phosphorylation of Smad2 and/or Smad3.
This allows Smad2 and/or Smad3 to bind to Smad4 with subsequent
nuclear translocation of the complex and the recruitment of
transcriptional co-activators or co-repressors to Smad-binding
elements in the promoters of TGF-β target genes (12).
The present study revealed that Th2 cells may be an
indispensable intermediate during the differentiation of Th0 cells
into Th9 cells, which depends on the activation of the Smad3/Smad4
and interferon-regulatory factor 4 (IRF-4) pathways.
Materials and methods
Animals
A total of 20 Female Balb/c mice (weight, 18–22g;
age, 6–8-weeks) were purchased from the Comparative Medicine Centre
of Yangzhou University (Yangzhou, China) and housed in a
pathogen-free facility at Jiangsu University (Zhenjiang, China).
All procedures were approved and supervised by the Animal Ethical
Committee of Jiangsu University (Zhenjiang, China).
In vitro T-cell differentiation and
flow cytometric analysis
Th0 cells were prepared from the spleens of 6–8
week-old female Balb/c mice under sterile conditions. The isolation
process was performed according to the manufacturer's protocol
(Miltenyi Biotec, Inc.). CD4+ Th0 cells were activated
with plate-bound anti-CD3 (eBiosciences; cat. no. 16-0031-85;
Thermo Fisher Scientific, Inc.) and anti-CD28 (eBiosciences; cat.
no. 16-0281-82; Thermo Fisher Scientific, Inc.) antibodies, and
supplemented with recombinant mouse IL-4 and TGF-β (Peprotech,
Inc.). After 3 or 5 days, the cultured cells were stimulated using
phorbol 12-myristate 13-acetate (50 ng/ml; Sigma-Aldrich; Merck
KGaA) and ionomycin (1 µg/ml; Sigma-Aldrich; Merck KGaA) in the
presence of monensin (2 g/ml; Sigma-Aldrich; Merck KGaA) for 4 h at
37°C in an atmosphere containing 5% CO2. The cells were
then fixed and permeabilized using permeabilization buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) for intracellular
staining, the phycoerythrin-conjugated anti-mouse IL-9 and
Per-cy5.5-conjugated anti-mouse IL-4 antibodies (1:200; cat. no.
130-102-442; Peprotech, Inc.) co-cultured with the cells at 4°C for
30 min according to the manufacturer's protocol. The labeled cells
were analyzed using an Accuri C6 flow cytometer with CFlow Sampler
software (GraphPad Prism 5; BD Biosciences). The
CD4+IL4+IL9− cell population was
considered to be Th2 cells and the
CD4+IL4−IL9+ cell population was
regarded as Th9 cells.
Reverse transcription-quantitative
(RT-q)PCR analysis
Following culture for 3 and 5 days under
Th9-polarization conditions, total RNA was extracted from T cells
using the guanidinium thiocyanate phenol chloroform method, and the
total RNA was used to generate complementary DNA with the
PrimeScript RT Reagent Kit (Takara Bio, Inc.) according to the
manufacturer's protocol. The primers were designed using Premier
5.0 software on the basis of GenBank sequences and synthesized by
Sangon Biotech Co., Ltd. The sequences of all primers used are
presented in Table I. qPCR was
performed using SYBR Premix ExTaq (Takara Bio, Inc.) according to
the manufacturer's protocol. Pre-denaturation was performed at 95°C
for 5 min, denaturation was performed at 95°C for 30 sec, annealing
was performed at 72°C for 30 sec and extension was performed at
65°C for 1 min. Fold changes in the expression of each gene
relative to β-actin were calculated using the comparative threshold
cycle (Ct) method (13). All
experiments were performed in triplicate.
 | Table I.Primer sequences for PCR. |
Table I.
Primer sequences for PCR.
| Gene | Primer sequence
(5′-3′) | Product length
(bp) |
|---|
| β-actin | F:
ATGGAAATGGGGAAGATGGTC | 349 |
|
| R:
GCGGGGAGGGTGTGAACT |
|
| IL-4 | F:
GGTCTCAACCCCCAGCTAGT | 102 |
|
| R:
GCCGATGATCTCTCTCAAGTGAT |
|
| IL-9 | F:
GGGCATCAGAGACACCAATTA | 119 |
|
| R:
AACAGTCCCTCCCTGTACTCAC |
|
| PU.1 | F:
CCCTCCATCGGATGACTTGGTT | 142 |
|
| R:
GTTGTTGTGGACATGGTGTGCG |
|
| IRF-4 | F:
GGTGTGGGAGAACGAGGAGAAG | 221 |
|
| R:
TCCTCTCGACCAATTCCTCAAA |
|
| GATA-3 | F:
ACCACGGGAGCCAGGTATG | 170 |
|
| R:
CGGAGGGTAAACGGACAGAG |
|
| Smad2 | F:
GCAGAATATCGGAGGCAGACA | 142 |
|
| R:
GATGGGTTTACGACATGCTTGA |
|
| Smad3 | F:
GGAGCAGAGTACAGGAGACA | 165 |
|
| R:
AACCCGCTCCCTTTACTCCTA |
|
| Smad4 | F:
GCTCCAGCCATCAGTCTGTC | 193 |
|
| R:
TGGTGTGCAGGACTTCATCC |
|
Statistical analysis
Values are expressed as the mean ± standard
deviation. GraphPad Prism Version 5.0 (GraphPad Software, Inc.) was
used to perform statistical analysis of the data. The unpaired
Student's t-test or Mann Whitney U-test (for RT-qPCR data) was
applied according to the results of homogeneity of variance
testing. In addition, analysis of variance and Tukey's
multiple-comparisons test were used for statistical analysis of the
flow cytometric data. P<0.05 was considered to indicate a
statistically significant difference.
Results
Variations in Th2 cell number during
the induction of Th9 cells using TGF-β and IL-4
At the optimum concentration of TGF-β (5 ng/ml), the
number of induced Th9 cells in vitro was markedly enhanced
with increasing concentrations of IL-4. As the basic prerequisite
for the generation of Th9 cells ex vivo, IL-4 and TGF-β were
used at different concentrations to induce Th9-cell differentiation
in vitro. The results suggested that the optimum cytokine
concentrations required to induce the differentiation of Th0 to Th9
cells were 30 ng/ml IL-4 and 5 ng/ml TGF-β, and that the number of
Th9 cells peaked following 5 days of induction under these
conditions (Fig. 1). It was also
revealed that Th2 cells were generated from Th0 cells at the 3-day
time-point, which then differentiated into Th9 cells at day 5 when
treated with appropriate concentrations of TGF-β and IL-4 (Fig. 2).
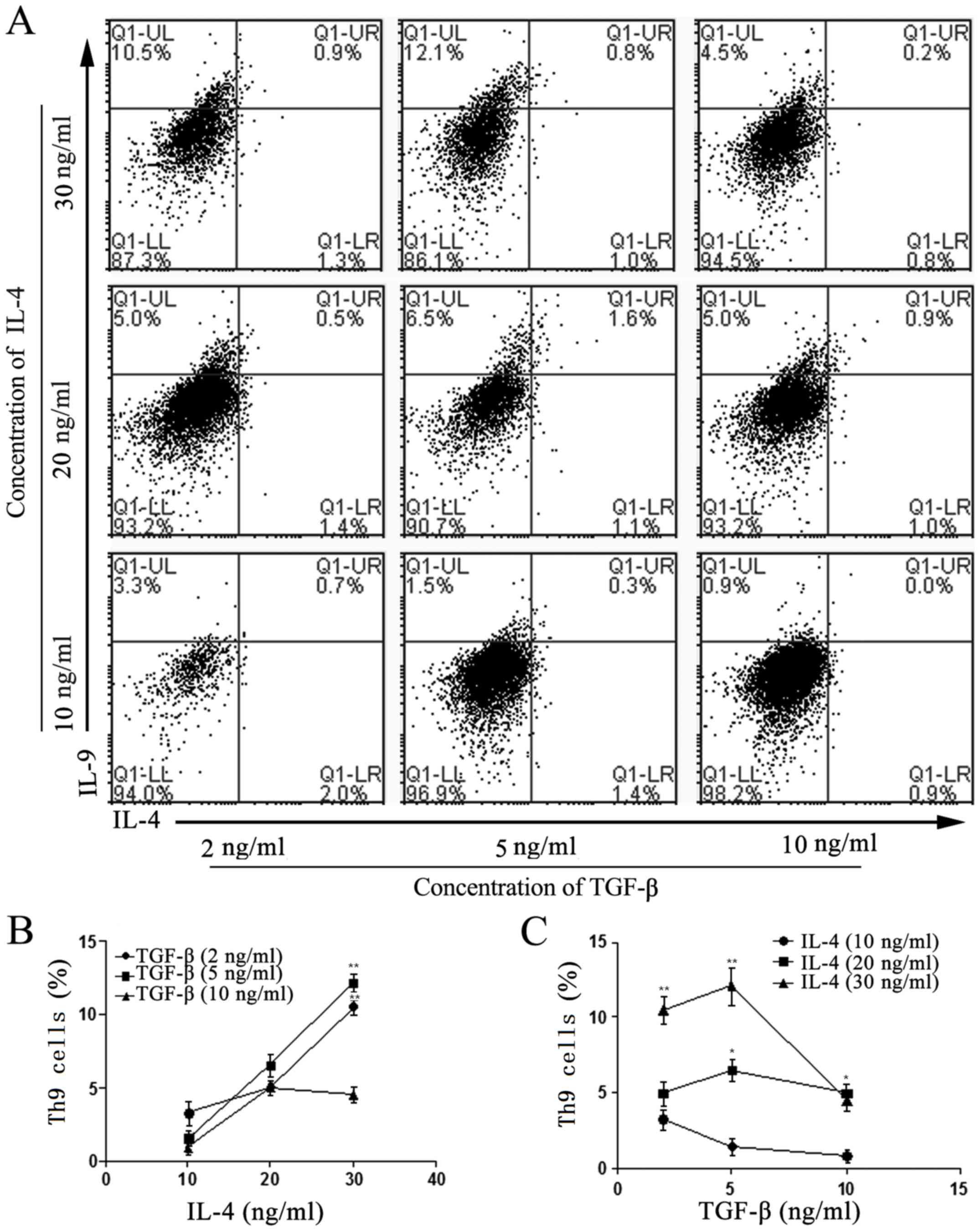 | Figure 1.Effects of IL-4 and TGF-β on the
differentiation of naïve T cells into Th9 cells. (A) Flow
cytometric analysis of the differentiation rate of Th9 cells
(IL-4− IL-9+), developed from naïve T cells
isolated from the spleens of Balb/c mice. Isolation was performed
using magnetic beads cultured with 2, 5 or 10 ng/ml TGF-β, and 10,
20 or 30 ng/ml IL-4 at day 5. (B) Effects of 10, 20 and 30 ng/ml
IL-4 on the differentiation into Th9 cells at 2, 5 and 10 ng/ml
TGF-β. **P<0.01 vs. 10 ng/ml TGF-β. (C) Effects of TGF-β (2, 5
and 10 ng/ml) on the differentiation of Th9 cells with 10, 20 and
30 ng/ml IL-4. *P<0.05 and **P<0.01 vs. 10 ng/ml
IL-4. Values are expressed as the mean ± standard deviation of
triplicate experiments. IL, interleukin; TGF-β, transforming growth
factor β; Th9 cell, type 9 T-helper cell; Q, quadrant; UL, upper
left; LR, lower right. |
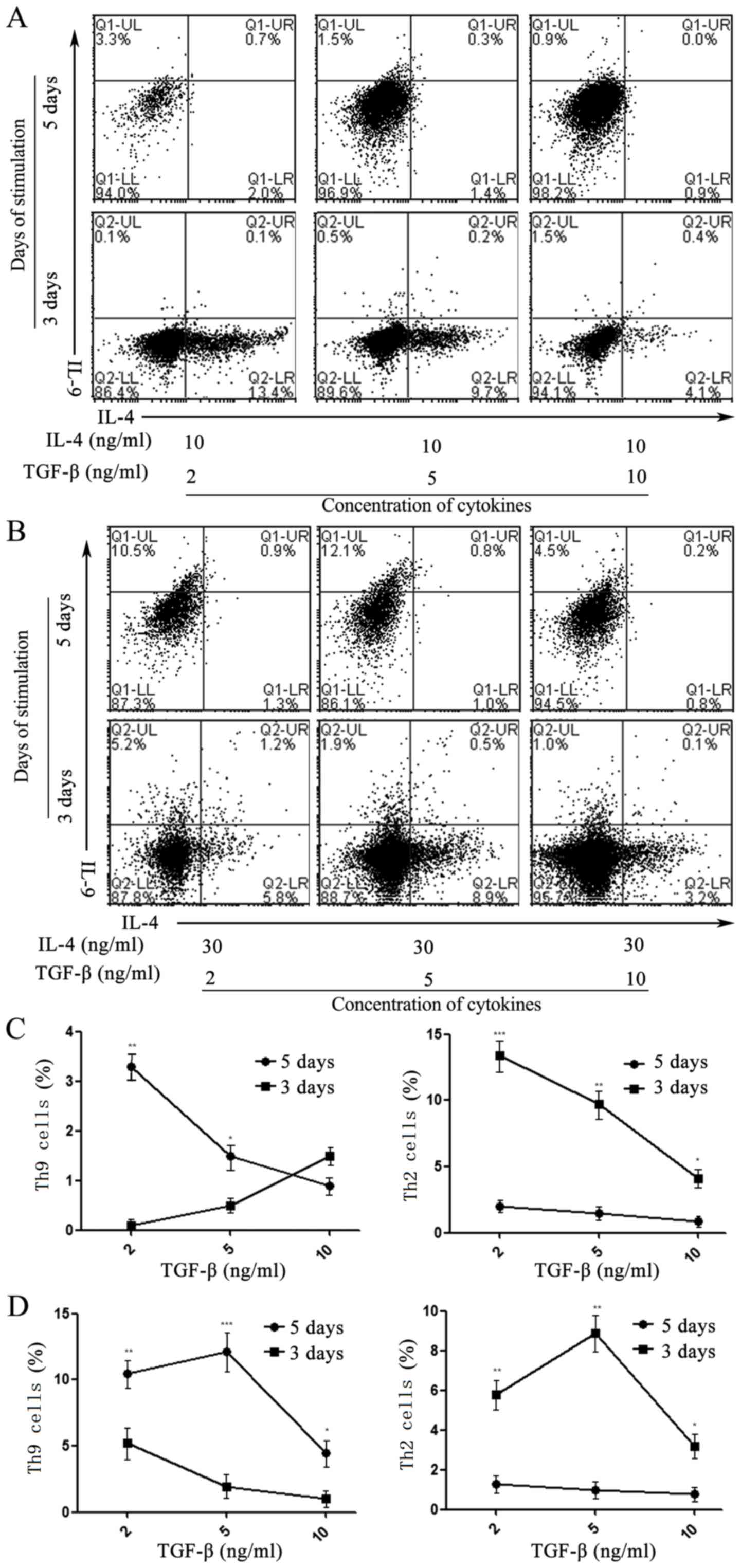 | Figure 2.Effects of induction time on the
differentiation of Th9 cells activated by IL-4 and TGF-β. Flow
cytometry indicated the differentiation rate of Th9
(IL-4− IL-9+) or Th2 (IL-4+
IL-9−) cells developed from Naïve T cells isolated using
magnetic beads, and treated with 2, 5 and 10 ng/ml TGF-β in the
presence of (A) 10 or (B) 30 ng/ml IL-4 at days 3 and 5,
respectively. The effect of induction time on the differentiation
of Th9 and Th2 cells stimulated with 2, 5 and 10 ng/ml TGF-β in the
presence of (C) 10 and (D) 30 ng/ml IL-4 at days 3 and 5. Values
are expressed as the mean ± standard deviation of triplicate
experiments. *P<0.05, **P<0.01, ***P<0.001. IL,
interleukin; TGF-β, transforming growth factor β; Th9 cell, type 9
T-helper cell; Q, quadrant; UL, upper left; LR, lower right. |
Variations in the expression levels of
Th9- and Th2-associated cytokines and transcription factors during
the generation of Th9 cells
IL-4 and IL-9 may be considered as the signature
cytokines produced by Th2 and Th9 cells, respectively. Cell
differentiation may cause alterations in the expression levels of
associated cytokines and transcription factors. In the present
study, the expression levels of IL-4 or IL-9 mRNA were evaluated;
the results indicated that expression levels were consistent with
the cell subsets cultured for 3 or 5 days, respectively, and that
the expression levels of IL-4 and IL-9 were significantly different
between these two time-points (day 3 and 5; Fig. 3A and B). Simultaneously, the mRNA
expression levels of Th9-associated transcription factors PU.1
(Sfpi1), GATA protein 3 (GATA-3) and IRF-4 were analyzed and the
data indicated that the expression of PU.1 was significantly
decreased, while that of IRF-4 was markedly elevated after 5 days
of incubation; no obvious change was observed in the expression
level of GATA-3 mRNA (Fig.
3C-E).
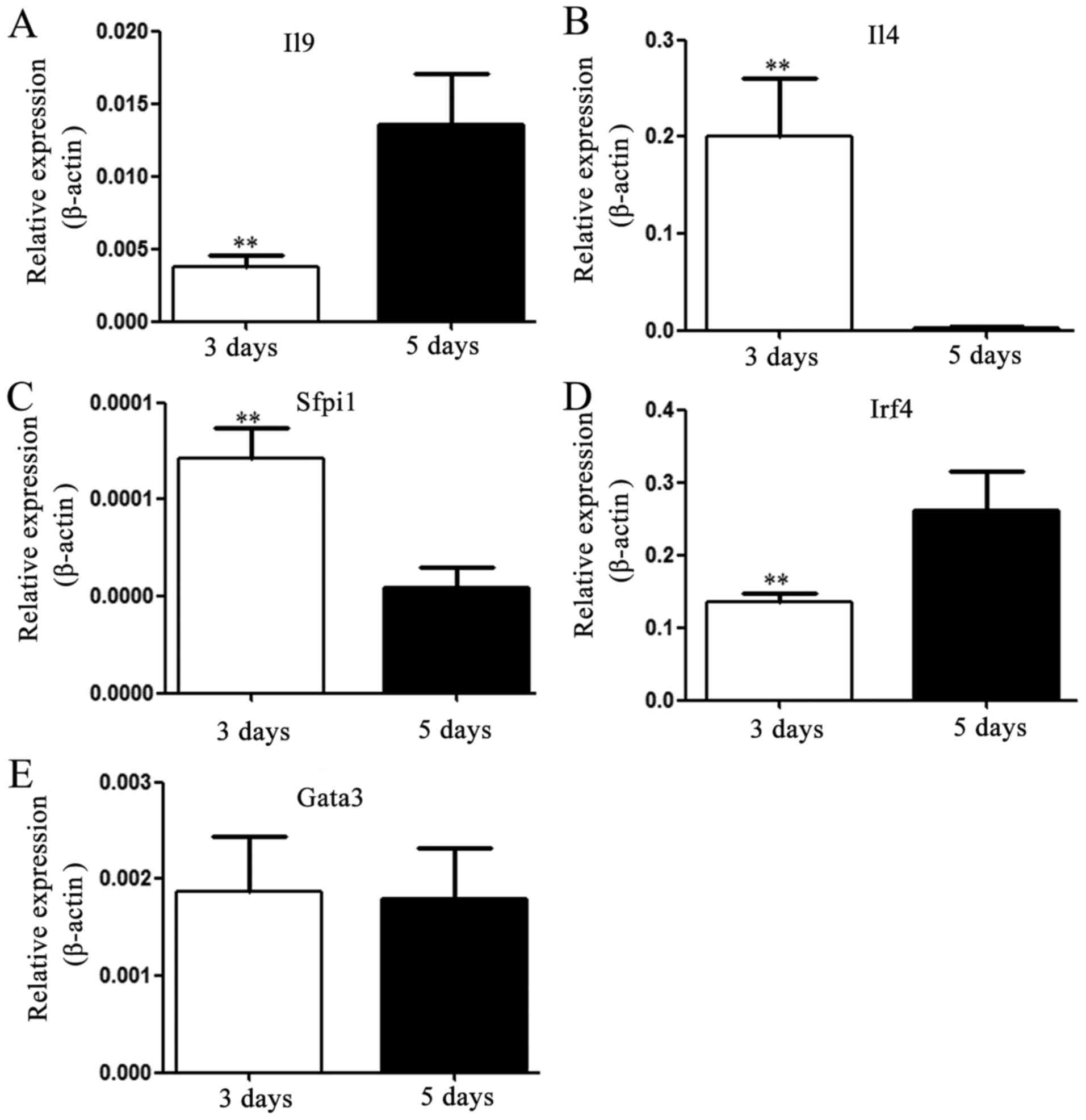 | Figure 3.Expression levels of Th9-associated
cytokines and transcription factors. After culture for 3 or 5 days,
the expression levels of IL-4, IL-9, PU.1, IRF-4 and GATA-3 mRNA
extracted from inducible naïve T cells, activated with 30 ng/ml
IL-4 and 5 ng/ml TGF-β were detected using reverse
transcription-quantitative PCR. Th9-associated cytokines: (A) IL-9
and (B) IL-4; transcription factors: (C) PU.1, (D) Irf4 and (E)
GATA-3. Values are expressed as the mean ± standard deviation of
triplicate experiments. **P<0.01 vs. day 5. IL, interleukin;
TGF-β, transforming growth factor β; Th9 cell, type 9 T-helper
cell; IRF-4, interferon-regulatory factor 4; GATA-3, GATA binding
protein 3. |
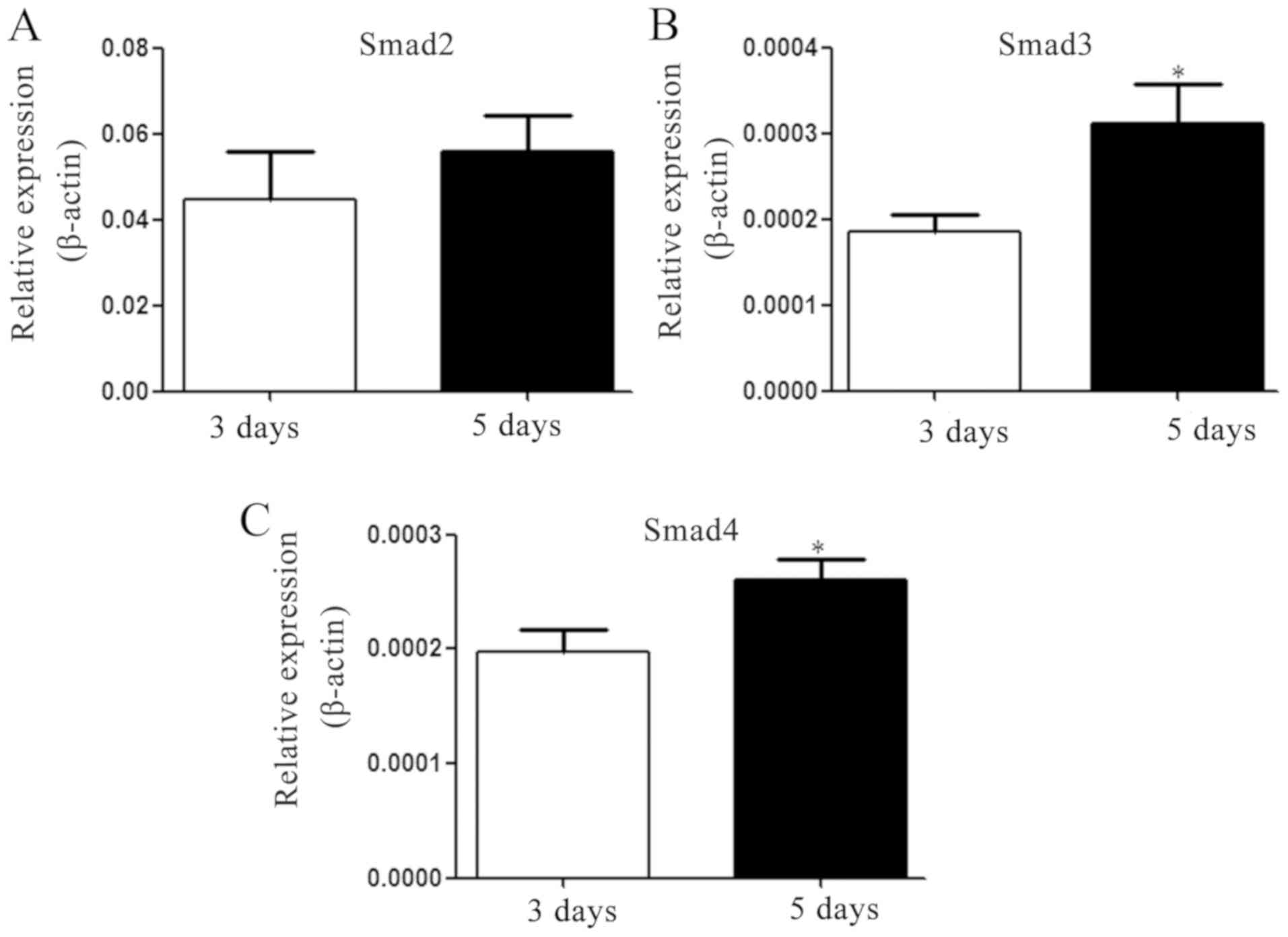
Increased expression of Smad3/Smad4 is
associated with the differentiation of Th2 to Th9 cells
TGF-β is pivotal in the induction of Th9 cells in
vivo and in vitro (9–11). As
significant components of the TGF-β signaling pathway, the mRNA
expression levels of Smad2, −3 and −4 were determined. The results
revealed that the expression levels of Smad3 and Smad4 on the 5th
day were significantly enhanced following IL-4 and TGF-β
supplementation, but no significant difference was identified in
the expression level of Smad2 (Fig.
4).
Discussion
Originally, Th9 cells were characterized by the
secretion of IL-9, and as such, were identified as an independent
Th-cell subset (7,14). As the production of IL-9 was detected
in Th9, not Th2 cells, the initial emphasis of research on
IL-9-producing Th2 cells was redirected to the occurrence and
development of cells (15). Previous
observations have revealed that the addition of TGF-β, a cytokine
with wide-ranging actions in the immune system, may alter the
characteristics of Th2 cells; this may include the loss of GATA-3
expression and the Th2-associated cytokines IL-4, IL-5 and IL-13,
resulting in the production IL-9. However, the identification of
IL-9-producing T cells as novel members of the ever-expanding
CD4+ T-cell family, has resulted in a nomenclature issue
due to the lack of unique expression profiles for T-bet, GATA-3,
RAR-related orphan receptor γt or forkhead box P3, which are known
subset-determining transcription factors associated with Th1, Th2,
Th17 and Treg cells, respectively. Among these transcription
factors, PU.1, IRF-4 and GATA-3 are notably associated with the
differentiation of Th2 cells (16–19).
Therefore, it is conceivable that the change in identification from
IL-9-producing Th2 to Th9 cells is not as simple as a change in
cytokine profiles, and that the defining mechanistic differences
between these cells require further elucidation.
Early studies of Th9 cells focused primarily on the
regulatory factors associated with IL-9 transcription, and their
influences on immune-associated diseases. A great deal of attention
has been paid to the involvement of IL-4 and TGF-β in the
transcription of the IL-9 gene in Th2 type-associated immune
disease models, including allergic airway disease (AAD) and
experimental autoimmune encephalomyelitis. The role of Th9 cells in
inflammation was documented in a Rag−/− mouse AAD model
via the adoptive transfer of these cells (17). Furthermore, PU.1 was revealed to
attenuate the expression of IL-9 in mice with a PU.1 defect
(16). This suggests that PU.1 is a
primary transcription factor associated with Th9-induced
inflammation. Concurrently, PU.1 is also associated with the
expression of IL-4 in various other cell types, including in the
survival of B cells. Simultaneously, Staudt et al (18) indicated that IRF-4 (a principal
participant in Th2-cell development) is also crucial to the
differentiation and function of Th9 cells. Previous studies have
also determined that a number of other cytokines influence the
generation of Th2 cells, including IL-2, IL-25, IFN-γ IL-21 and
IL-27, and that they may serve similar roles in the generation of
Th9 cells (20–23).
It is commonly understood that the development of
different Th subtypes relies on the appropriate external signals.
Similar to the conditions required to promote Th1-, Th2-, Th17- and
Treg-cell differentiation, Th9 cells are generated from Th0 cells
in response to TGF-β and IL-4, in addition to other cytokines in
the extracellular milieu (24). The
current consensus is that the differentiation period for Th subsets
activated using anti-CD3/CD28 differs from that of physiological
activation using specific antigen (25). It is noted that TGF-β, as an
immune-regulatory cytokine, not only regulates the differentiation
of Th-cell subsets, but is also involved in apoptosis and cell
survival (26–28). Takami et al (29) demonstrated that in the presence of
IL-4, TGF-β was able to convert p53-induced CD28-dependent
apoptosis-associated stimuli into the signal for Th9
differentiation. Therefore, TGF-β has been studied as a key
molecule involved in the generation of Th9 cells in vitro
(30).
It has been demonstrated that TGF-β redirects the
differentiation of Th0 cells from Th2 to Th9 cells (7). In light of this, the induction rates of
Th2 and Th9 cells in response to optimum Th9-cell polarization
conditions were analyzed at different time-points ex vivo.
Furthermore, changes in the expression levels of IL-4, IL-9,
GATA-3, Pu.1, IRF-4, Smad2, Smad3 and Smad4 were measured. The
results of the present study illustrated that differentiation into
Th2 cells was a necessary process for the induction of Th9 cells in
Th9-polarizing culture conditions, and that Th2 cells may be an
intermediate in the conversion of Th0 into Th9 cells in the
presence of TGF-β and IL-4 (Fig. 5).
In other words, Th9 cells were not directly generated from Th0
cells, and may represent a stable state following the intermediate
generation of Th2 cells. The present study also suggested that
although PU.1, IRF-4 and GATA-3 were expressed by Th2 and Th9
cells, the expression of IRF-4 was significantly upregulated, while
PU.1 mRNA expression was downregulated in Th9 cells. In addition,
the decrease in PU.1 expression levels was associated with the
downregulation of IL-4 expression. It was speculated that Th2 cells
may be generated in the early stage of Th9-cell differentiation,
which are then converted to Th9 cells via the Smad3/Smad4 and IRF-4
activation pathways. However, the complete mechanism of Th9-cell
generation remains elusive, and further investigation is required
to identify novel prophylactics and treatments for Th9-associated
pathologies.
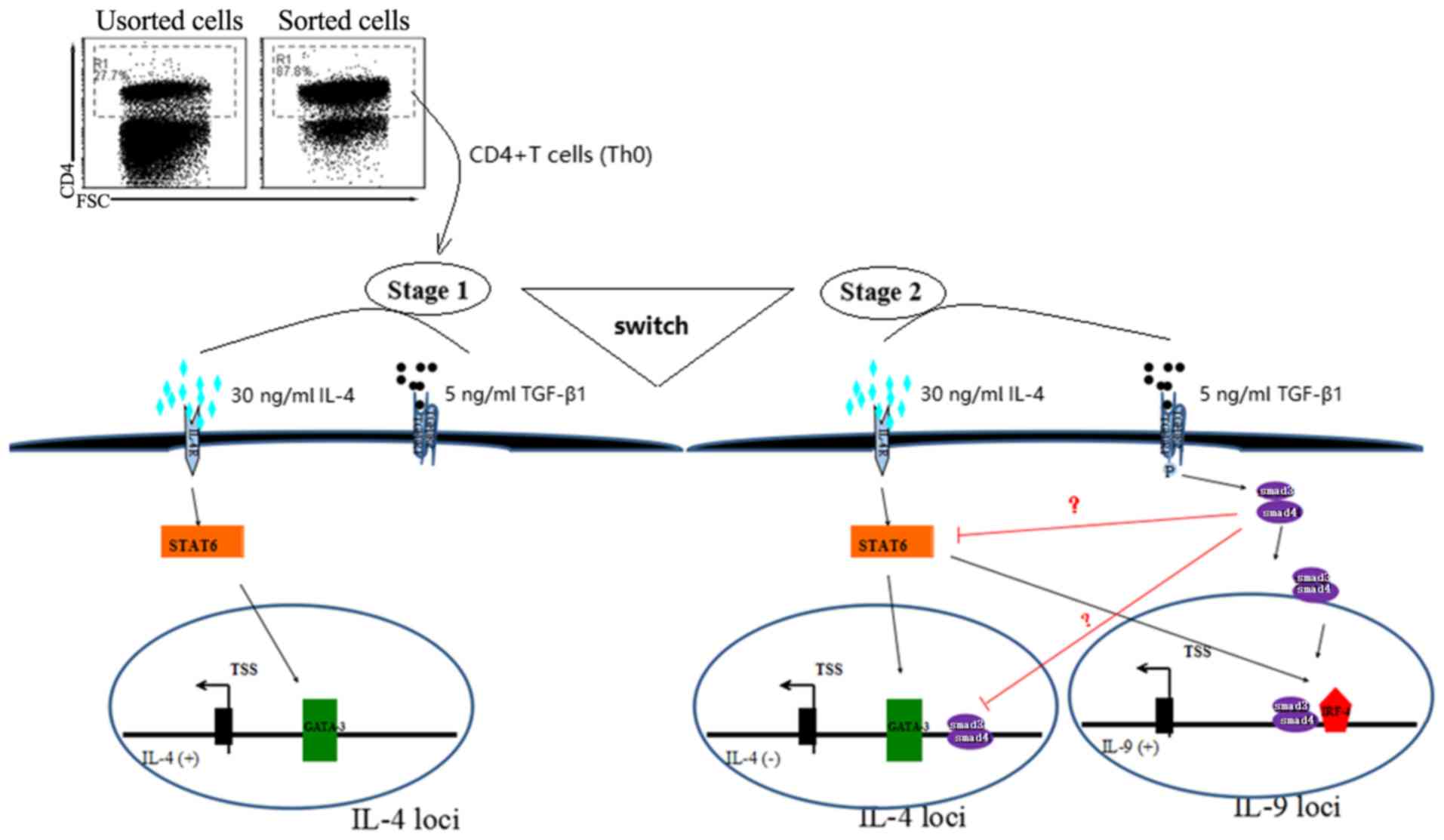 | Figure 5.Differentiation of Th2 to Th9 cells
using TGF-β and IL-4. The transformation process of Th2 to Th9
cells from Th0, in the presence of 5 ng/ml TGF-β and 30 ng/ml IL-4,
in which the Th0 cells underwent two developmental stages. In the
first stage, the Th2-specific transcription factor GATA3 was
activated by IL-4 via the STAT6 pathway, resulted in the autocrine
activation of IL-4, leading to the appearance of Th2
characteristics. In the second stage, with the prolongation of
TGF-β stimulation time, Smad3 and/or Smad4 were activated, which
further activated the transcription factor IRF-4 and may have
inhibited GATA3 activity, leading to the conversion of cytokines
secreted by cells from IL-4 to IL-9, and the emergence of Th9-cell
characteristics. Th9 cell, type 9 T-helper cell; TGF-β,
transforming growth factor β; IL, interleukin; IRF-4,
interferon-regulatory factor 4; GATA-3, GATA binding protein 3;
FSC, forward scatter, is positively correlated with the square of
cell diameter (cell size). |
Acknowledgements
The authors would like to thank Mr Qiaolin Chen
(Institute of Laboratory Medicine, Jiangsu University) and
Professor Zhijun Jiao (Affiliated Hospital of Jiangsu University)
for technical assistance.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81771756),
the Key University Science Research Project of Jiangsu Province
(grant no. 16KJA320005), the Social Development Project of Jiangsu
Province (grant no. BE2016716) and the Postdoctoral Foundation of
Jiangsu Province (grant no. 1601002C).
Availability of data and materials
The materials used during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
MHA and HX conceived and designed the present study.
MHA, HW and JC performed the experiments and analyzed the data. All
the authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were approved and supervised
by the Animal Ethical Committee of Jiangsu University (Zhenjiang,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou L, Chong MM and Littman DR:
Plasticity of CD4(+) T cell lineage differentiation. Immunity.
30:646–655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan C, Aziz MK, Lovaas JD, Vistica BP, Shi
G, Wawrousek EF and Gery I: Antigen-specific Th9 cells exhibit
uniqueness in their kinetics of cytokine production and short
retention at the inflammatory site. J Immunol. 185:6795–6801. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi G, Cox CA, Vistica BP, Tan C,
Wawrousek EF and Gery I: Phenotype switching by
inflammation-inducing polarized Th17 cells, but Not by Th1 cells. J
Immunol. 181:7205–7213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kabata H, Moro K and Koyasu S: The group 2
innate lymphoid cell (ILC2) regulatory network and its underlying
mechanisms. Immunol Rev. 286:37–52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Noelle RJ and Nowak EC: Cellular sources
and immune functions of interleukin-9. Nat Rev Immunol. 10:683–687.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Purwar R, Schlapbach C, Xiao S, Kang HS,
Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo
VK, et al: Robust tumor immunity to melanoma mediated by
interleukin-9-producing T cells. Nat Med. 18:1248–1253. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Veldhoen M, Uyttenhove C, van Snick J,
Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C and Stockinger
B: Transforming growth factor-beta ‘reprograms’ the differentiation
of T helper 2 cells and promotes an interleukin 9-producing subset.
Nat Immunol. 9:1341–1346. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmitt E, Germann T, Goedert S, Hoehn P,
Huels C, Koelsch S, Kühn R, Müller W, Palm N and Rüde E: IL-9
production of naive CD4+ T cells depends on IL-2, is
synergistically enhanced by a combination of TGF-beta and IL-4, and
is inhibited by IFN-gamma. J Immunol. 153:39891994.PubMed/NCBI
|
|
9
|
Schmitt E, Beuscher HU, Huels C, Monteyne
P, van Brandwijk R, van Snick J and Ruede E: IL-1 serves as a
secondary signal for IL-9 expression. J Immunol. 147:3848–3854.
1991.PubMed/NCBI
|
|
10
|
Angkasekwinai P, Chang SH, Thapa M,
Watarai H and Dong C: Regulation of IL-9 expression by IL-25
signaling. Nat Immunol. 11:250–256. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tamiya T, Ichiyama K, Kotani H, Fukaya T,
Sekiya T, Shichita T, Honma K, Yui K, Matsuyama T, Nakao T, et al:
Smad2/3 and IRF4 play a cooperative role in IL-9-producing t cell
induction. J Immunol. 191:2360–2371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji L, Xu J, Liu J, Amjad A, Zhang K, Liu
Q, Zhou L, Xiao J and Li X: Mutant p53 promotes tumor cell
malignancy by both positive and negative regulation of the
transforming growth factor β (TGF-β) pathway. J Biol Chem.
290:11729–11740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu P, Ji X, Wan J and Xu H: Activity of
group 2 innate lymphoid cells is associated with chronic
inflammation and dysregulated metabolic homoeostasis in type 2
diabetic nephropathy. Scand J Immunol. 87:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dardalhon V, Awasthi A, Kwon H, Galileos
G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, et
al: IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together
with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells.
Nat Immunol. 9:1347–1355. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stassen M, Schmitt E and Bopp T: From
interleukin-9 to T helper 9 cells. Ann N Y Acad Sci. 1247:56–68.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang HC, Sehra S, Goswami R, Yao W, Yu Q,
Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, et al: The
transcription factor PU.1 is required for the development of
IL-9-producing T cells and allergic inflammation. Nat Immunol.
11:527–534. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goswami R and Kaplan MH: Gcn5 Is required
for PU.1-dependent IL-9 induction in th9 cells. J Immunol.
189:3026–3033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Staudt V, Bothur E, Klein M, Lingnau K,
Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, et
al: Interferon-regulatory factor 4 is essential for the
developmental program of T helper 9 cells. Immunity. 33:192–202.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goswami R, Jabeen R, Yagi R, Pham D, Zhu
J, Goenka S and Kaplan MH: STAT6-Dependent Regulation of Th9
Development. J Immunol. 188:968–975. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao W, Lin JX and Leonard WJ: IL-2 family
cytokines: New insights into the complex roles of IL-2 as a broad
regulator of T helper cell differentiation. Curr Opin Immunol.
23:598–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin PY, Jen HY, Chiang BL, Sheu F and
Chuang YH: Interleukin-21 suppresses the differentiation and
functions of T helper 2 cells. Immunology. 144:668–676. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wong MT, Ye JJ, Alonso MN, Landrigan A,
Cheung RK, Engleman E and Utz PJ: Regulation of human Th9
differentiation by type I interferons and IL-21. Immunol Cell Biol.
88:624–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murugaiyan G, Beynon V, Pires Da Cunha A,
Joller N and Weiner HL: IFN-gamma limits Th9 mediated autoimmune
inflammation through dendritic cell modulation of IL-27. J Immunol.
253:69. 2012.
|
|
24
|
Ye J, Wang Y, Wang Z, Ji Q, Huang Y, Zeng
T, Hu H, Ye D, Wan J and Lin Y: Circulating Th1, Th2, Th9, Th17,
Th22, and treg levels in aortic dissection patients. Mediators
Inflamm. 2018:56971492018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan C, Wei L, Vistica BP, Shi G, Wawrousek
EF and Gery I: Phenotypes of Th lineages generated by the commonly
used activation with anti-CD3/CD28 antibodies differ from those
generated by the physiological activation with the specific
antigen. Cell Mol Immunol. 11:305–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jabeen R and Kaplan MH: The symphony of
the ninth: the development and function of Th9 cells. Curr Opin
Immunol. 24:303–307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heldin CH, Landström M and Moustakas A:
Mechanism of TGF-beta signaling to growth arrest, apoptosis, and
epithelial-mesenchymal transition. Curr Opin Cell Biol. 21:166–176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murillo MM, del Castillo G, Sánchez A,
Fernández M and Fabregat I: Involvement of EGF receptor and c-Src
in the survival signals induced by TGF-beta1 in hepatocytes.
Oncogene. 24:4580–4587. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takami M, Love RB and Iwashima M: TGF-beta
converts apoptotic stimuli into the signal for Th9 differentiation.
J Immunol. 188:4369–4375. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang A, Pan D, Lee YH, Martinez GJ, Feng
XH and Dong C: Cutting Edge: Smad2 and Smad4 regulate
TGF-β-Mediated Il9 gene expression via EZH2 displacement. J
Immunol. 191:4908–4912. 2013. View Article : Google Scholar : PubMed/NCBI
|